Abstract
The guanosine triphosphatases (GTPases) of the immunity-associated protein (GIMAP) family of putative GTPases has been implicated in the regulation of T-lymphocyte development and survival. A mouse conditional knockout allele was generated for the immune GTPase gene GIMAP1. Homozygous loss of this allele under the influence of the lymphoid-expressed hCD2-iCre recombinase transgene led to severe (> 85%) deficiency of mature T lymphocytes and, unexpectedly, of mature B lymphocytes. By contrast there was little effect of GIMAP1 deletion on immature lymphocytes in either B or T lineages, although in vitro studies showed a shortening of the survival time of both immature and mature CD4+ single-positive thymocytes. These findings show a vital requirement for GIMAP1 in mature lymphocyte development/survival and draw attention to the nonredundant roles of members of the GIMAP GTPase family in these processes.
Introduction
The guanosine triphosphatases (GTPases) of the immunity-associated proteins (GIMAPs) are a family of guanosine nucleotide–binding proteins implicated in the regulation of T-lymphocyte development and survival.1 They are encoded in humans, rats, and mice by a cluster of 7 to 8 autosomal genes. They are expressed strongly in lymphoid tissues but are also readily detectable in some other tissues such as kidney, lungs, and heart.2-4 Recent evidence indicates that the GIMAP genes are targets of Notch regulation.5 With the exception of GIMAP5 and GIMAP4, there is little information on the in vivo function(s) of these proteins. GIMAP5, which has prosurvival properties in T lymphocytes, is the best-studied family member as a result of a spontaneous recessive mutation within the rat gene orthologue. This mutation predisposes to autoimmunity and gave rise to the BB rat model of type 1 diabetes and, more recently, to a model of T-helper 2 (Th2)–type inflammatory bowel disease.2,6-8 Furthermore, autoimmunity-related traits in humans have been associated with a GIMAP5 gene variant.9,10 The mutation in rat GIMAP5 causes peripheral T lymphopenia (reviewed in Ramanathan and Poussier11 ), but thymic abnormalities in the mutant animals are limited and largely confined to the mature CD4+ and CD8+ single-positive (SP) compartments.12 No effect on B-lymphocyte populations has been reported in these rats. Recently, a similar T-cell deficiency has been reported for a GIMAP5 knockout mouse strain,13 although deficiency in a minor population of B cells, the splenic marginal zone (MZ) B cells, was also detected. Short hairpin RNA knockdown of GIMAP5 and GIMAP3 expression in mouse fetal thymic organ culture has implicated these genes in, respectively, the generation of CD4+CD8+ double-positive (DP) thymocytes and T cell–positive selection.1
In contrast with GIMAP5, GIMAP4 has “prodeath” properties. Knocking out the GIMAP4 gene had no deleterious effect on in vivo lymphocyte development; instead, ex vivo, T lymphocytes exhibited a slowing of the apoptotic process when subjected to a number of death-inducing treatments.14 In combination, these data imply diverse but related roles for the GIMAP genes in T-cell development and survival.
GIMAP1 is a close relative of GIMAP5 and lies adjacent to it within the GIMAP gene cluster. Like GIMAP5, it encodes a protein that contains a putative transmembrane domain at its carboxyl terminus, indicative of targeting to intracellular membranes. GIMAP1 mRNA has been found in all stages of T-cell development, without major variations in level,1,14,15 although differential expression of human T cell GIMAP1 under conditions of Th1 or Th2 polarization in vitro has recently been reported.16 We now report on the novel phenotype of mice in which GIMAP1 has been knocked out conditionally in lymphocytes. Our data show critical requirements for this gene in lymphocyte development, not only in the T-cell lineage but also, unexpectedly, in the B-cell lineage.
Methods
Animals
Mice were bred and maintained in specific pathogen-free conditions at The Babraham Institute. Husbandry and experimentation complied with existing United Kingdom Home Office licenses and local standards, as approved by the Babraham Institute Animal Welfare, Ethics, and Experimentation Committee.
Generation of mice bearing a conditional GIMAP1 allele
A 129 mouse P1 artificial chromosome library17 was screened for the presence of GIMAP genes. Recombineering18 was used to develop a targeting vector for GIMAP1 exon 3, using P1 artificial chromosome no. RP21-609H15 as the source of the relevant DNA sequences. Exon 3 encodes all but the first 14 residues of the predicted GIMAP1 polypeptide, so the conditional removal of this exon was expected to abolish the function of the gene. In the final targeting construct, LoxP sites flanked GIMAP1 exon 3 and, 3′ of that, a neomycin-resistance cassette was located between FRT sites (Figure 1A). The construct was electroporated into embryonic day 14 embryonic stem (ES) cells.19 After antibiotic selection with G418, ES cell colonies were picked, expanded and analyzed for the targeting event by Southern blotting of SpeI- and BamHI-digested DNA. Correctly targeted ES cells were injected into C57BL/6Tyr−/− blastocysts that were transferred to pseudopregnant recipients. Chimeras generated in this way were bred to obtain germline transmission, confirmed by analysis of tail biopsy DNA. Polymerase chain reaction A (PCR-A) distinguishes between wild-type GIMAP1 and the floxed allele. Mice positive for the GIMAP1 floxed allele were paired with mice expressing the FlpE recombinase to remove the Frt-flanked NeoR cassette and this was assessed with the use of PCR-B (Figure 1A). Mice with this neo-less conditional GIMAP1 allele were then bred with animals carrying the hCD2-iCre transgene20 to produce GIMAP1fl/fl,hCD2iCre+/− mice. The mice used for the analyses reported here were on a mixed 129-C57BL/6 background. The PCR primers for PCR-A and PCR-B were as follows: PCR-A forward, 5′-CCAGTCCTAGAGTGAGCTTCACTCATCAGG-3′; PCR-A reverse, 5′-GGATTCTCTGTGCACCTGCAGATGCTCCTG-3′; PCR-B forward, 5′-GATTCACAGTGCAGTGACTTGGCATCAACT-3′; PCR-B reverse, 5′-CTGAGTCGACGTGTGTTCTCTTTGCGCTGTGA-3′.
Flow cytometric analysis
Single-cell suspensions from thymus, spleen, lymph nodes (axillary, brachial, and inguinal), bone marrow (BM), and peritoneal flushings were stained for flow cytometry. For peritoneal cells and splenocytes, cells were first incubated with Fc block (2.4G2; eBioscience). Biotinylated or fluorochrome-conjugated antibodies against CD3, CD4, CD8, CD19, CD21, CD23, CD24, CD25, CD93, T-cell receptor β (TCRβ), B220, immunoglobulin M (IgM), c-kit, Thy1.2, and Qa2, as well as fluorochrome-conjugated streptavidin as a second-stage reagent (eBioscience; BD Biosciences) were used. After staining, cells were washed and resuspended in 25 ng/mL 4′,6-diamidino-2-phenylindole before analysis on an LSRII Flow Cytometer (BD Biosciences). Analysis was performed using FlowJo (TreeStar Inc) software.
Intracellular staining
For pSTAT5 analysis, cells were first stimulated with culture medium containing 10% interleukin-7 (IL-7) supernatant at 37°C for 30 minutes. Cells were then fixed with 2% paraformaldehyde at 37°C. Subsequently, cells were permeabilized on ice with the use of 90% methanol in phosphate-buffered saline. After washing twice in fluorescence-activated cell sorting (FACS) buffer, cells were then stained for 1 hour with anti–pSTAT5-PE before analysis on a LSRII Flow cytometer (BD Biosciences).
Fluorescence-activated cell sorting
Single-cell suspensions of thymocytes, BM cells, or splenocytes were stained as described under “Flow cytometric analysis,” and cell populations were sorted using a FACSAria (BD Biosciences). Purities were between 90% and 99.9%.
Cell survival assays
FACS-purified CD4 SP thymocytes were washed and resuspended in RPMI (10% fetal calf serum; with or without IL-7 supplementation) and plated at 5 to 7 × 105/well in 96-well plates and incubated at 37°C. At set times, cells were harvested and stained with Annexin V, anti-Qa2, and anti-CD24 antibodies before analysis by flow cytometry.
Western blotting of cell lysates
Cell lysates were prepared, and Western blots were performed essentially as described elsewhere.4 For the analyses in Figure 7A and B, 105 cell equivalents were run per track. For Figure 7C, 5 × 105 cell equivalents were used. Details of the derivation and characteristics of the anti-GIMAP monoclonal antibodies used are given in Table 1.
Derivation and characteristics of the anti-GIMAP monoclonal antibodies
| GIMAP no. . | MAb name . | Rat γ isotype . | Rat strain . | ♀/♂ . | Fusion partner . | Immunogen . | Vector/bacteria . | Immunogen preparation . |
|---|---|---|---|---|---|---|---|---|
| 1 | MAC420* | 2a | LOU/C | ♀ | Y3Ag1.2.3† | GST-mGIMAP1aa1-238 | pDEST15/RosettaDE3 | Glutathione bead-eluted/gel-purified |
| 4 | MAC417‡ | 2b | LOU/C | ♀ | Y3Ag1.2.3 | GST-mGIMAP4aa1-310 | pGEX.4T1/Rosetta2 | Glutathione bead-eluted |
| 5 | MAC421 | 2a | LEW | ♀ | Y3Ag1.2.3 | GST-mGIMAP5aa1-270 | pDEST15/RosettaDE3 | Glutathione bead-eluted |
| 6 | MAC432 | 2b | DA | ♂ | IR983F§ | GST-mGIMAP6 full length | pGEX.4T1/Rosetta | Inclusion bodies |
| 9‖ | MAC433 | 2a | DA | ♂ | Y3Ag1.2.3 | GST-mGIMAP9 full length | pGEX.4T1/Rosetta | Inclusion bodies |
| GIMAP no. . | MAb name . | Rat γ isotype . | Rat strain . | ♀/♂ . | Fusion partner . | Immunogen . | Vector/bacteria . | Immunogen preparation . |
|---|---|---|---|---|---|---|---|---|
| 1 | MAC420* | 2a | LOU/C | ♀ | Y3Ag1.2.3† | GST-mGIMAP1aa1-238 | pDEST15/RosettaDE3 | Glutathione bead-eluted/gel-purified |
| 4 | MAC417‡ | 2b | LOU/C | ♀ | Y3Ag1.2.3 | GST-mGIMAP4aa1-310 | pGEX.4T1/Rosetta2 | Glutathione bead-eluted |
| 5 | MAC421 | 2a | LEW | ♀ | Y3Ag1.2.3 | GST-mGIMAP5aa1-270 | pDEST15/RosettaDE3 | Glutathione bead-eluted |
| 6 | MAC432 | 2b | DA | ♂ | IR983F§ | GST-mGIMAP6 full length | pGEX.4T1/Rosetta | Inclusion bodies |
| 9‖ | MAC433 | 2a | DA | ♂ | Y3Ag1.2.3 | GST-mGIMAP9 full length | pGEX.4T1/Rosetta | Inclusion bodies |
Results
A conditional knockout of the mouse GIMAP1 gene
GIMAP genes are expressed most strongly in lymphoid cells and tissues, but expression of some family members is also detectable elsewhere, eg, in lung, kidney, and heart. For this reason, a conditional targeting approach to the reverse genetic analysis of GIMAP1 was selected. In addition, the availability of a conditional allele of this gene would provide flexibility in future studies of its function in different cell lineages.
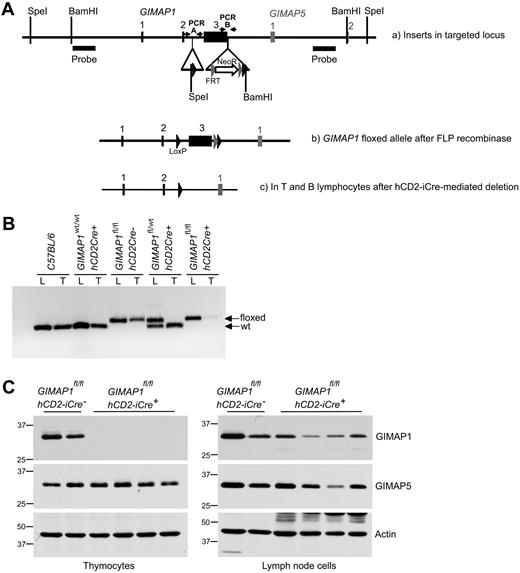
A conditional targeting vector for exon 3 of mouse GIMAP1 was developed and used to generate mouse ES cells with a modified GIMAP1 allele (see “Methods”; Figure 1A). Mice bearing the modified allele were successfully derived and then crossed with mice bearing the hCD2-iCre transgene. This transgene allows Cre recombinase expression in the T-cell lineage from the CD4−CD8− double-negative 1 (DN1) thymocyte stage onward (and possibly also in prethymic precursors); in the B lineage it is expressed from at least the late pro-B stage onward.20 Efficient hCD2-iCre–mediated excision within the targeted GIMAP1 gene was shown by PCR analysis of thymus DNA (Figure 1B). Western blots detected no GIMAP1 protein in the thymus of GIMAPfl/fl,hCD2iCre+/− mice (Figure 1C), in which T and B lineage–specific excision of GIMAP1 was expected to occur. These mice are designated “GIMAP1/CD2-cKO” in the rest of the text.
Conditional deletion of the “floxed” GIMAP1 allele with the use of the hCD2-iCre transgene. (A) Gene targeting strategy. The targeting vector introduced LoxP sites (black arrowheads) to flank GIMAP1 exon 3. A neomycin resistance cassette flanked by FRT sites (gray arrowheads) was located downstream of the LoxP sites. Southern probes used to determine correct targeting of ES cells after SpeI and BamHI DNA digests are indicated. (B) PCR analysis to detect the floxed and wild-type alleles of GIMAP1 in genomic lung (L) and thymus (T) DNA. (C) Western blot analysis of GIMAP1 and GIMAP5 expression in thymocytes and lymph node cells.
Conditional deletion of the “floxed” GIMAP1 allele with the use of the hCD2-iCre transgene. (A) Gene targeting strategy. The targeting vector introduced LoxP sites (black arrowheads) to flank GIMAP1 exon 3. A neomycin resistance cassette flanked by FRT sites (gray arrowheads) was located downstream of the LoxP sites. Southern probes used to determine correct targeting of ES cells after SpeI and BamHI DNA digests are indicated. (B) PCR analysis to detect the floxed and wild-type alleles of GIMAP1 in genomic lung (L) and thymus (T) DNA. (C) Western blot analysis of GIMAP1 and GIMAP5 expression in thymocytes and lymph node cells.
Because of the compact nature of the GIMAP gene region, the modifications to the GIMAP1 gene were in close proximity to the neighboring GIMAP5 gene (Figure 1A). For this reason, and because some (rare) transcripts of GIMAP5 including exon 1 of GIMAP1 have been reported,24 Western blots were also performed to assess the expression of the GIMAP5 protein. GIMAP5 expression was intact in the thymus of GIMAP1/CD2-cKO mice (Figure 1C), indicating that the gene targeting had not affected the expression of the GIMAP5 gene.
Some GIMAP1 protein was detected in peripheral lymphoid tissues of GIMAP1/CD2-cKO mice (eg, lymph nodes; Figure 1C) but at lower levels than in controls. Some of this may have been expressed by myeloid cells (in which the Cre recombinase would not be expressed), but part was due to lymphocytes, and this is discussed later.
Profound lymphopenia in GIMAP1/CD2-cKO mice
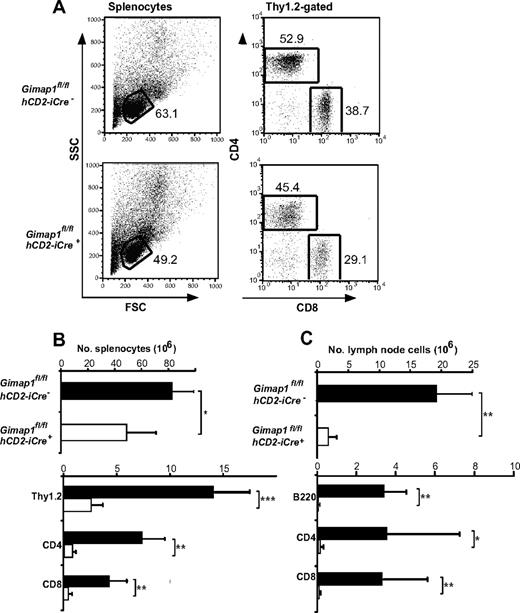
When the lymphoid tissues of GIMAP1/CD2-cKO mice were analyzed, the outstanding feature was a massive deficiency of lymphoid cells in the peripheral compartments (Figure 2). Given the T cell–selective effects of GIMAP5 mutations described previously,11,13 we were surprised that the lesion in GIMAP1/CD2-cKO mice affected not only T cells but also B cells (Figure 2C). In the case of the spleen, despite the lymphopenia, the overall cellularity was reduced by only approximately 40% in the conditional knockout (Figure 2B) because of increases in both neutrophils (Gr1+CD11b+) and monocyte/macrophages (Gr1−CD11b+; data not shown; evident in the forward scatter vs side scatter plots in Figure 2A). The cellularity of lymph nodes, in which a large-scale expansion of myeloid cell numbers did not occur, was more severely reduced (Figure 2C).
Defective peripheral lymphoid compartment in GIMAP1/CD2-cKO mice. Lymphoid subsets from spleen (A-B) and lymph nodes (C) of GIMAP1fl/flhCD2-iCre− and GIMAP1fl/flhCD2-iCre+ mice were enumerated with the use of flow cytometry. (A) Representative FACS plots of splenocytes for each genotype. (B-C) Mean numbers (± SD) of cell subsets positive for the indicated surface markers (GIMAP1fl/flhCD2-iCre−, n = 9; GIMAP1fl/flhCD2-iCre+, n = 5). *P < .05; **P < .01; ***P < .001 (unpaired 2-tailed Student t test).
Defective peripheral lymphoid compartment in GIMAP1/CD2-cKO mice. Lymphoid subsets from spleen (A-B) and lymph nodes (C) of GIMAP1fl/flhCD2-iCre− and GIMAP1fl/flhCD2-iCre+ mice were enumerated with the use of flow cytometry. (A) Representative FACS plots of splenocytes for each genotype. (B-C) Mean numbers (± SD) of cell subsets positive for the indicated surface markers (GIMAP1fl/flhCD2-iCre−, n = 9; GIMAP1fl/flhCD2-iCre+, n = 5). *P < .05; **P < .01; ***P < .001 (unpaired 2-tailed Student t test).
The T-cell lineage in GIMAP1/CD2-cKO mice.
Both spleen and lymph nodes exhibited reductions of 90% or greater in the numbers of CD4+ and CD8+ T cells enumerated with the use of flow cytometry (Figure 2B-C). Severe T lymphopenia was also evident in the peritoneum (Figure 5C) and in the blood (data not shown), the latter finding ruling out a defect affecting lymphocyte emigration from the blood to lymph nodes or spleen as the cause of the phenotype.
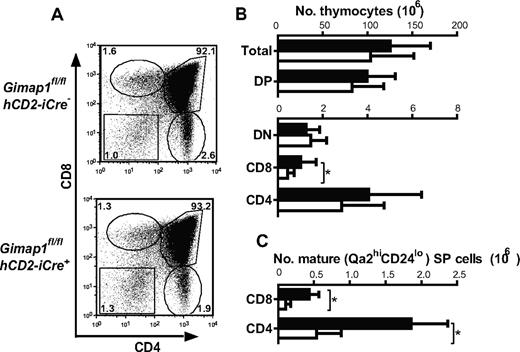
By contrast, thymi from GIMAP1/CD2-cKO mice were of broadly normal size and appearance. Detailed analysis, however, showed important effects of the conditional GIMAP1 deletion (Figure 3). The total number of cells in GIMAP1/CD2-cKO thymi tended to be lower than controls, although this trend did not reach statistical significance. When thymocyte subsets were enumerated, however, a significant reduction was seen in the CD8 SP subset (Figure 3B). Investigation of maturation markers within the SP subsets also indicated a numerical reduction in the most mature (Qa2hiCD24lo) subsets of CD4 SP and CD8 SP cells (Figure 3C), although the proportions of these within the overall CD4 SP and CD8 SP compartments, respectively, were not significantly affected. Thus, the main effect of GIMAP1 loss on the T lineage takes effect from the late thymic stage onward, as has been found for the GIMAP5-deficient models.11,13 Additional evidence for the functional maturity of GIMAP1-deficient SP thymocytes came from their high level of expression of CD5 (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article), a property that is regulated developmentally by engagement of the αβTCR at positive selection,25,26 and from their up-regulation of CD40L in vitro in response to stimulation through the TCR with the anti-CD3 antibody27 (with or without anti-CD28 costimulation; supplemental Figure 1B) both of which were equivalent to wild-type controls.
Thymocyte development in GIMAP1/CD2-cKO mice. (A) Representative FACS plots of thymocytes from GIMAP1fl/flhCD2-iCre− and GIMAP1fl/flhCD2-iCre+ mice stained for CD4, CD8, and TCR (the latter to exclude CD8+ intermediate SP cells). (B) Mean numbers (± SD) of thymocyte subsets (■ indicates GIMAP1fl/flhCD2-iCre−, n = 22; □, GIMAP1fl/flhCD2-iCre+, n = 18). (C) Mean numbers (± SD) of mature (Qa2hiCD24−) CD4 and CD8 SP thymocytes (■ indicates GIMAP1fl/flhCD2-iCre−, n = 6; □, GIMAP1fl/flhCD2-iCre+, n = 4).
Thymocyte development in GIMAP1/CD2-cKO mice. (A) Representative FACS plots of thymocytes from GIMAP1fl/flhCD2-iCre− and GIMAP1fl/flhCD2-iCre+ mice stained for CD4, CD8, and TCR (the latter to exclude CD8+ intermediate SP cells). (B) Mean numbers (± SD) of thymocyte subsets (■ indicates GIMAP1fl/flhCD2-iCre−, n = 22; □, GIMAP1fl/flhCD2-iCre+, n = 18). (C) Mean numbers (± SD) of mature (Qa2hiCD24−) CD4 and CD8 SP thymocytes (■ indicates GIMAP1fl/flhCD2-iCre−, n = 6; □, GIMAP1fl/flhCD2-iCre+, n = 4).
The B-cell lineage in GIMAP1/CD2-cKO mice.
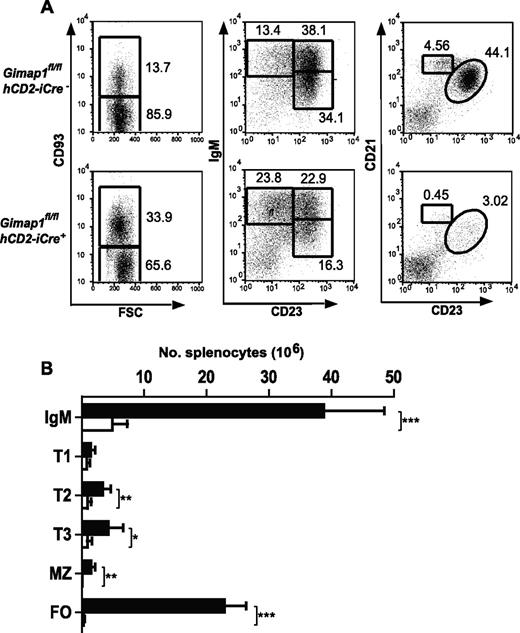
Severe reductions in B-cell numbers were seen in both lymph nodes (> 90%) and spleen (∼ 85%; Figure 4A-B). Flow cytometric analysis for the markers CD93, IgM, CD21, and CD23 was used to enumerate B-cell subsets in GIMAP1/CD2-cKO and control spleens. In the CD93− fraction, the normally predominant follicular B-cell population (CD21intCD23hi) was massively reduced (by > 95%) in the conditional knockout as were the MZ B cells (CD21hiCD23int). Immature splenic B cells of the transitional T1, T2, and T3 stages were identified in the CD93+ population by the markers surface IgM and CD23. T2 and T3 populations were significantly reduced in GIMAP1/CD2-cKO mice relative to controls, whereas any reduction in the T1 population did not reach statistical significance.
Peripheral B-cell analysis. Splenocytes from GIMAP1fl/flhCD2-iCre− and GIMAP1fl/flhCD2-iCre+ mice were stained for flow cytometric analysis to identify B-cell subsets. (A) Representative FACS plots for each genotype. The numbers given inside the component panels in A are the percentages of lymphocyte events contained within the indicated gates. (B) Mean numbers (± SD) of B-cell subsets (■ indicates GIMAP1fl/flhCD2-iCre−, n = 6; □, GIMAP1fl/flhCD2-iCre+, n = 6). *P < .05; **P < .01; ***P < .001 (unpaired 2-tailed Student t test). FSC indicates forward scatter; and FO, follicular.
Peripheral B-cell analysis. Splenocytes from GIMAP1fl/flhCD2-iCre− and GIMAP1fl/flhCD2-iCre+ mice were stained for flow cytometric analysis to identify B-cell subsets. (A) Representative FACS plots for each genotype. The numbers given inside the component panels in A are the percentages of lymphocyte events contained within the indicated gates. (B) Mean numbers (± SD) of B-cell subsets (■ indicates GIMAP1fl/flhCD2-iCre−, n = 6; □, GIMAP1fl/flhCD2-iCre+, n = 6). *P < .05; **P < .01; ***P < .001 (unpaired 2-tailed Student t test). FSC indicates forward scatter; and FO, follicular.
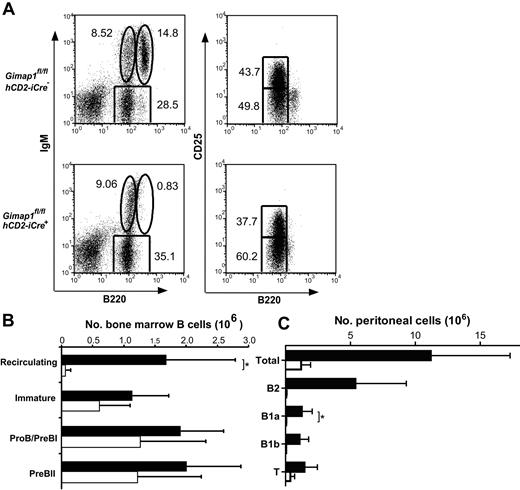
B-lineage cells were also analyzed in the BM (Figure 5A-B). IgM versus B220 FACS plots showed a severe deficit of mature, recirculating B cells (IgM+B220hi). No statistically significant differences were seen between GIMAP1/CD2-cKO and control mice in the number of IgM−B220intCD25− (pro-B/pre-BI) cells, IgM−B220intCD25+ (pre-BII) cells, or immature (IgM+B220int) B cells. The number of B cells in the peritoneum was also strongly reduced, and this was true of both conventional B2 B cells (CD19+B220hiCD5−) and the B1a (CD19+B220intCD5+) and B1b (CD19+B220intCD5−) subsets (Figure 5C).
BM and peritoneal cell analysis. BM (A-B) and peritoneal (C) cells from GIMAP1fl/flhCD2-iCre− and GIMAP1fl/flhCD2-iCre+ mice were stained for flow cytometric analysis to identify B-lineage subsets. (A) Representative FACS plots for each genotype. The numbers given inside the component panels in A are the percentages of lymphoctye events contained within the indicated gates. (B) Mean numbers (± SD) of B-lineage subsets in BM (■ indicates GIMAP1fl/flhCD2-iCre−,n = 6; □, GIMAP1fl/flhCD2-iCre+, n = 6). (C) Mean numbers (± SD) of lymphocyte populations in peritoneum (■ indicates GIMAP1fl/flhCD2-iCre−, n = 6; □, GIMAP1fl/flhCD2-iCre+, n = 4). *P < .05 (unpaired 2-tailed Student t test).
BM and peritoneal cell analysis. BM (A-B) and peritoneal (C) cells from GIMAP1fl/flhCD2-iCre− and GIMAP1fl/flhCD2-iCre+ mice were stained for flow cytometric analysis to identify B-lineage subsets. (A) Representative FACS plots for each genotype. The numbers given inside the component panels in A are the percentages of lymphoctye events contained within the indicated gates. (B) Mean numbers (± SD) of B-lineage subsets in BM (■ indicates GIMAP1fl/flhCD2-iCre−,n = 6; □, GIMAP1fl/flhCD2-iCre+, n = 6). (C) Mean numbers (± SD) of lymphocyte populations in peritoneum (■ indicates GIMAP1fl/flhCD2-iCre−, n = 6; □, GIMAP1fl/flhCD2-iCre+, n = 4). *P < .05 (unpaired 2-tailed Student t test).
Thus, the main effect on B lymphocytes of the deletion of GIMAP1 takes effect from the late BM stage onward, with all the peripheral B-cell types tested being affected, with the possible exception of the splenic T1 B cells. It should be noted that a substantial proportion of the few B (and T) cells found in the periphery of GIMAP1/CD2-cKO mice express GIMAP1 protein and so are probably from rare clones in which the Cre-mediated ablation of GIMAP1 has failed to occur28 (Figure 1C; data not shown). Hence, the severity of the lymphopenia recorded here probably underestimates the dependence of peripheral lymphocytes on GIMAP1.
GIMAP1 ablation reduces the in vitro survival capacity of CD4+ SP thymocytes
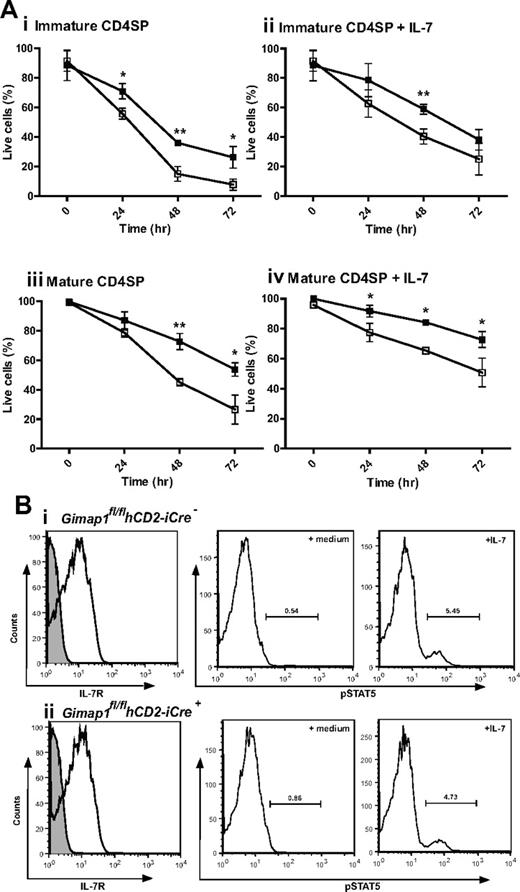
The most dramatic effect of GIMAP1 loss in the T-cell lineage is seen from the late thymic stage onward. One of the characteristics acquired by developing thymocytes during the medullary stage, as CD4+CD8− or CD4−CD8+ SP cells, is the acquisition of improved longevity/survival capacity,29 a property also characteristic of peripheral, naive, T cells. This is reflected in a relatively long average survival of SP thymocytes in tissue culture. When GIMAP1/CD2-cKO CD4+CD8− SP thymocytes were assayed for this property, they performed significantly worse than their GIMAP1-sufficient counterparts; this difference applied to both “immature” (Qa2loCD24hi) and “mature” (Qa2hiCD24lo) cells within the CD4+CD8− SP compartment (Figure 6A). Although cell survival could be improved by supplementing the tissue culture medium with IL-7, the difference in CD4+CD8− SP thymocyte survival because of the GIMAP1 deletion remained. When similar in vitro experiments were conducted to examine the survival of CD4−CD8+ SP thymocytes, the cells from GIMAP1/CD2-cKO mice again appeared to perform worse than wild type, but in this case the effect did not reach statistical significance (supplemental Figure 2A). Ex vivo GIMAP1/CD2-cKO splenic T cells showed increased Annexin V binding, indicative of impaired survival properties of the few peripheral T cells that were present (data not shown).
In vitro analysis of CD4 SP thymocyte survival. (A) CD4 SP thymocytes from GIMAP1fl/flhCD2-iCre− (■) and GIMAP1fl/flhCD2-iCre+ (□) mice were FACS-purified and cultured with (ii,iv) or without (i,iii) IL-7. Immature (i,ii) and mature (iii,iv) CD4 SP cells were defined on the basis of Qa2 and CD24 expression. Live cells were determined by annexin V staining. Plots show mean percentage of live cells (± SD) for 3 mice of each genotype. *P < .05; **P < .01; ***P < .001 (unpaired 2-tailed Student t test). (B) IL-7 receptor expression and function in thymocytes. (Left) Expression of IL-7R (CD127; white histogram) compared with control stain (gray histogram) on CD4 SP thymocytes (as determined by flow cytometry) from (i) GIMAP1fl/flhCD2-iCre− and (ii) GIMAP1fl/flhCD2-iCre+ mice. (Right) Responses of total thymocytes to IL-7 as determined by pSTAT5 induction. Total thymocytes were incubated at 37°C in medium with or without IL-7 for 30 minutes before fixing, permeabilizing, staining with anti–phospho-STAT5 antibody and analyzing by flow cytometry.
In vitro analysis of CD4 SP thymocyte survival. (A) CD4 SP thymocytes from GIMAP1fl/flhCD2-iCre− (■) and GIMAP1fl/flhCD2-iCre+ (□) mice were FACS-purified and cultured with (ii,iv) or without (i,iii) IL-7. Immature (i,ii) and mature (iii,iv) CD4 SP cells were defined on the basis of Qa2 and CD24 expression. Live cells were determined by annexin V staining. Plots show mean percentage of live cells (± SD) for 3 mice of each genotype. *P < .05; **P < .01; ***P < .001 (unpaired 2-tailed Student t test). (B) IL-7 receptor expression and function in thymocytes. (Left) Expression of IL-7R (CD127; white histogram) compared with control stain (gray histogram) on CD4 SP thymocytes (as determined by flow cytometry) from (i) GIMAP1fl/flhCD2-iCre− and (ii) GIMAP1fl/flhCD2-iCre+ mice. (Right) Responses of total thymocytes to IL-7 as determined by pSTAT5 induction. Total thymocytes were incubated at 37°C in medium with or without IL-7 for 30 minutes before fixing, permeabilizing, staining with anti–phospho-STAT5 antibody and analyzing by flow cytometry.
The improved in vitro survival of GIMAP1/CD2-cKO CD4+8− SP thymocytes brought about by IL-7 supplementation indicated that signaling through the IL-7 receptor was intact in the mutant thymocytes, and this was confirmed by flow cytometric analysis showing normal expression of IL-7Rα (Figure 6B). In addition, GIMAP1/CD2-cKO and wild-type thymocytes exhibited similar downstream phosphorylation of STAT5 after in vitro exposure to IL-7 (Figure 6B). A further indication that IL-7 signaling was intact in GIMAP1/CD2-cKO SP thymocytes was their undiminished level of Bcl-2 expression (data not shown), a property that is under the control of IL-7 signaling.30
GIMAP1/CD2-cKO B cells express BAFF-R
Given the potent effect of GIMAP1 loss on mature B-cell survival in vivo, we investigated the expression of the key cytokine receptor involved in B-cell maintenance, namely the B cell–activating factor receptor (BAFF-R).31,32 This receptor was found expressed at approximately normal levels in GIMAP1/CD2-cKO immature B and splenic B cells (supplemental Figure 2B). This result requires cautious interpretation (particularly in the case of the splenic B cells) because, as described under “The B-cell lineage in GIMAP1/CD2-cKO mice,” some of the B lymphocytes in GIMAP1/CD2-cKO spleen are rare survivors in which GIMAP1 expression has not been extinguished. Nevertheless, the flow cytometric profile did suggest that BAFF-R was being expressed by all of the splenic B cells, ie, potentially both GIMAP1-expressing and nonexpressing cells, and therefore that its expression was not disrupted by GIMAP1 gene loss. This conclusion was supported by normal BAFF-R expression in the immature BM B cells.
Expression of GIMAP proteins in the T and B lineages
The results described indicate parallel behavior in the T- and B-cell lineages in response to GIMAP1 loss. In both cases there are, at most, only minor effects apparent on the earlier developmental stages examined but major effects on the mature cell populations in the periphery. This points to a critical, nonredundant role for GIMAP1 in the transition between primary and secondary lymphoid stages. To what extent does this dependence of cells on GIMAP1 match the expression of this gene and its product? Previously, we reported real-time PCR studies of GIMAP gene expression in the B-cell lineage.4 For 7 of the 8 mouse GIMAP genes low expression in the early BM stages was followed by a sharp rise in expression at either the immature CD25−B220+IgD−IgM+ B-cell stage (for GIMAP1, GIMAP5, GIMAP9) or the mature CD25−B220+IgD+IgM+ stage (for GIMAP3, GIMAP4, GIMAP6, GIMAP8). This indicates a generally good correlation with the stage(s) at which B-lineage cells display sensitivity to GIMAP1 loss (see Figures 4,5).
We were interested to confirm this relationship at the protein level. Using monoclonal antibodies against 5 of the mouse GIMAPs, we found that in cells from normal C57BL/6 mice the levels of all these proteins were low in pre-BII and immature B cells and then rose sharply (eg, at the T1 or T2 stages) to high levels in the more mature B-cell stages (Figure 7A-B). A similar analysis was performed on subsets of thymocytes, including division of DP cells into 3 populations on the basis of TCR expression (Figure 7C). In the case of GIMAP4, it was already known that, after a sharp rise at the DN3 to DN4 stage, expression is extinguished in DP thymocytes and reappears at the mature SP stage.14,33 A similar, but less absolute, pattern was seen for most of the GIMAP family, with DP thymocytes showing low expression before a substantial rise in the mature SP subsets. GIMAP1, however, was an exception, displaying a roughly constant level of expression throughout, and this confirmed previous findings.4
Analysis of GIMAP protein expression in B- and T-cell development. BM cells (A), splenic B cells (B), and thymocytes (C) from C57BL/6 mice were FACS-purified into subsets as described in “Fluorescence-activated cell sorting.” Expression of GIMAP4, GIMAP5, GIMAP1, GIMAP6, and GIMAP9 was examined by Western blot, with β-actin as control. *Parentheses surround nonspecific bands caused by the secondary anti–rat IgG reagent.
Analysis of GIMAP protein expression in B- and T-cell development. BM cells (A), splenic B cells (B), and thymocytes (C) from C57BL/6 mice were FACS-purified into subsets as described in “Fluorescence-activated cell sorting.” Expression of GIMAP4, GIMAP5, GIMAP1, GIMAP6, and GIMAP9 was examined by Western blot, with β-actin as control. *Parentheses surround nonspecific bands caused by the secondary anti–rat IgG reagent.
These data suggest 2 hypotheses: (1) that the GIMAP proteins are under a degree of coordinate regulation bringing about high expression at the critical stages of the late thymocyte and the transitional B cell, and their subsequent entry into the mature peripheral T- or B-lymphocyte pools; and (2) that, in the thymus (but not the BM), GIMAP1 may have an ongoing function relevant at all developmental stages.
Discussion
Investigations of the GIMAP GTPases have led to the proposal that they are important regulators of T-lymphocyte development and survival.1 The data presented here, however, crucially extend the scope of interest in the GIMAPs to include corresponding processes in the B-cell lineage, in which the phenotype of GIMAP1/CD2-cKO mice diverges dramatically from its GIMAP5-deficient forerunners (Figures 2–3,5). The T-lineage phenotype displayed by the GIMAP1/CD2-cKO mice described here is similar to that seen previously in GIMAP5-deficient rats and mice. Features in common include (1) severe peripheral T lymphopenia in combination with a thymus that is normal to initial inspection, and (2) effects on cells at the later stages of intrathymic development, including reductions in the number of SP cells and failure of these cells to acquire a long-lived phenotype measurable in in vitro survival assays (Figures 2,4).12 GIMAP1/CD2-cKO mice, however, exhibit, in addition, severe reductions in all types of mature B cells studied: follicular, mature recirculating, MZ, peritoneal B1a and B1b. No deficit of B cells has ever been reported in the GIMAP5-mutant rat model, and only MZ B cells showed a deficit in the GIMAP5 knockout mouse.13 Paralleling the effects in the T-cell lineage, the earlier stages in B-cell development in the BM of GIMAP1/CD2-cKO mice were apparently unaffected (Figure 3).
Our analysis of the expression patterns of various GIMAP proteins during lymphocyte development raised the interesting possibility that the predominant role of GIMAP family proteins may be to act in a coordinated fashion to regulate lymphocyte survival from a late intrathymic (or intra-BM) stage onward. The analysis of intrathymic GIMAP expression also highlighted the expression of family members in DN thymocytes, a phenomenon first noticed for GIMAP4.33 An effect on the cellular events after β-selection would explain the reported effect of GIMAP5 loss on DP thymocyte numbers/production.1
It is unclear why inactivation of the GIMAP1 gene should have more extensive effects (ie, including all mature B cells) than observed for inactivation of its close relative GIMAP5. It seems possible from our data that the T-cell phenotype in GIMAP1/CD2-cKO mice is more severe than that of GIMAP5-mutant mice or rats (bearing in mind that a proportion of the remaining peripheral lymphocytes are cells in which Cre-mediated deletion of GIMAP1 may have failed to occur). This might indicate that GIMAP1 is more “central” to the survival processes regulated by this GTPase family (and that its loss may therefore be capable of tilting the balance of survival signals in B cells as well as T cells). Note here that germline GIMAP1 loss appears to be lethal soon after birth (data not shown), which is, again, a more severe phenotype than that described previously for germline loss of GIMAP5 in mice.13 Peripheral T lymphopenia in GIMAP5-mutant rats is due to the deleterious effect of GIMAP5 loss on the thymic SP compartment, resulting in reduced thymic T-cell output and reduced survival of thymic emigrants. Thus, the peripheral T-cell pool consists largely of recent thymic emigrants and additional T cells of unusual “activated” phenotype(s) with a paucity of recognizable resting, naive T cells and a defect in survival in the overall population. It is probable that a similar general description will apply to the GIMAP1/CD2-cKO phenotype.
The mechanism by which GIMAP5 promotes T-lymphocyte survival has been the subject of diverse proposals,1,26,34-37 including hypotheses relating to mitochondrial permeability, mitogen-activated protein kinase and nuclear factor κB (NF-κB) signaling, interaction with Bcl-2 family members, regulation of calcium signaling and the endoplasmic reticulum stress pathway. A weakness intrinsic to a number of these conclusions is their basis in data from the GIMAP5-mutant rat, in which the few residual peripheral T cells are intrinsically short-lived and of unusual phenotype. Conditional models for GIMAP gene deletion should allow comparisons of normal lymphocytes before and after elective GIMAP gene deletion and hence better controlled investigations of the mechanism(s) by which GIMAP GTPases regulate lymphocyte survival.
In principle, there are 2 classes of survival process in which GIMAP proteins may participate. The first is protection from an endogenous mediator or process that is potentially cytotoxic/proapoptotic but normally present at subthreshold levels. Endogenous molecules known to have cytotoxic potential include inflammatory mediators such as tumor necrosis factor38 and nicotinamide adenine dinucleotide.39 Cell-autonomous processes such as the endoplasmic reticulum stress response and atrophy-apoptosis modulation would also fit into this category.
The second class of survival process is constitutive survival signaling. Continuing survival of recirculating lymphocytes requires stimulation via their antigen receptors40,41 as well as through specific cytokine receptors (BAFF-R for B cells; IL-7R for T cells).32,42 Appropriate stimulation through these receptors activates a nexus of downstream pathways which maintains cell growth and restrains apoptosis. A central role is played by the canonical and noncanonical NF-κB pathways, alongside the PI3-kinase/Akt/mTOR pathway, the Pim2 pathway, and the Erk pathway, which may themselves contain targets of NF-κB transcriptional control.43,44 The phenotypes of several spontaneous or engineered mouse mutants in components of these pathways (eg, NEMO, IKK, TAK1, NIK, Pim2) attest to their importance for establishing and maintaining the mature lymphocyte repertoire.45 That the phenotypes of GIMAP1- and GIMAP5-deficient rodents are focused around the same critical processes is highly suggestive. The fact that GIMAP1 loss effects both B and T lineages tends to favor common pathways, such as NF-κB signaling, against lineage-specific pathways (BAFF-R, IL-7R), and this is supported by our demonstration of intact IL-7 signaling and responsiveness (Figure 6).
Nitta et al1 have reported that GIMAPs 3, 4, and 5 are capable of interacting with members of the bcl-2 protein family; a role for the GIMAPs in the modulation of survival signaling might provide a layer of fine control to the regulation of apoptosis in lymphoid lineages. In work to be submitted elsewhere, we have established that GIMAP1 and GIMAP5 occupy separate intracellular locations, indicating that the processes in which these GTPases participate span at least 2 cellular compartments. This result may also serve to explain the absence of redundancy between these 2 closely related proteins.
In conclusion, our findings have shown a major, nonredundant, role for GIMAP1 in the development of mature lymphocytes. This has uncovered an unexpected involvement of this GTPase family in B-cell development.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr N. Copeland for “recombineering” plasmids; and Ross Miller, Margaret Graham, Ted Saunders, Martin George, and Martyn Cooke for contributions to this work.
This work was supported by the United Kingdom Biotechnology and Biological Sciences Research Council. A.S. received a BBSRC Research Studentship.
Authorship
Contribution: A.S. and L.M.C.W. designed and conducted research, analyzed data, and wrote the paper; M.L.J. and C.C. designed and conducted research; A.H., J.P., N.P., and G.M. conducted research; M.T. designed research; and G.W.B. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: G. W. Butcher, The Babraham Institute, Cambridge, CB22 3AT, United Kingdom; e-mail: geoff.butcher@bbsrc.ac.uk.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal