Abstract
Chromosomal abnormalities are increasingly used to risk stratify adults with acute lymphoblastic leukemia. Published data describing the age-specific incidence of chromosomal abnormalities and their prognostic relevance are largely derived from clinical trials. Trials frequently have age restrictions and low recruitment rates. Thus we investigated these factors in a population-based cohort of 349 patients diagnosed during the course of 19 years in the northern part of England. The incidence of most chromosomal abnormalities varied significantly with age. The incidence of t(9;22)(q34;q11) increased in each successive decade, up to 24% among 40- to 49-year-old subjects. Thereafter the incidence reached a plateau. t(4;11)(q21;q23) and t(1;19)(q23;p13) were a rare occurrence among patients older than 60 years of age. In contrast, the frequency of t(8;14)(q24;q32) and t(14;18)(q32;q21) increased with age. High hyperdiploidy occurred in 13% of patients younger than 20 years of age but in only 5% of older patients. The incidence of low hypodiploidy/near-triploidy and complex karyotype increased with age from 4% (15-29 years) to 16% (≥ 60 years). Overall survival varied significantly by age and cytogenetics. Older patients and those with t(9;22), t(4;11), low hypodiploidy/near-triploidy, or complex karyotype had a significantly inferior outcome. These population-based results demonstrate the cytogenetic heterogeneity of adult acute lymphoblastic leukemia. These data will inform the delivery of routine clinical services and the design of new age-focused clinical trials.
Introduction
Recurrent and clonal chromosomal abnormalities in the leukemic cells of patients with acute lymphoblastic leukemia (ALL) are the hallmark of the disease and are now routinely used in the pediatric setting to assist patient management, particularly in terms of diagnosis, disease monitoring, prognosis, and risk stratification.1 The clinical utility of cytogenetics in adult ALL is an emerging topic, and more studies are urgently required.2,3 To date the majority of cytogenetic studies (pediatric and adult) have been based on patients enrolled in local, national, or international clinical trials. In pediatric ALL these studies can be considered representative because recruitment rates in this age group are very high. However, trial recruitment is much lower among adolescents and adults.4 Moreover, adults older than 60 years of age are rarely eligible for clinical trials.5,6
Population-based studies of adult ALL are rare. The lack of studies describing the age-specific incidence of chromosomal abnormalities and their prognostic relevance makes planning the delivery of routine clinical care services and clinical trials difficult. There are several current issues that would benefit significantly from such data. First, the treatment of adolescents and young adults is controversial, with many clinical trial groups opting to treat them with the use of pediatric protocols.7-10 However, the distinct genetic profile of these patients has not been taken into consideration.11 Second, there is renewed interest in developing treatment protocols for older (ie, > 60 years) adults.12 Finally, the authors of recent studies2,3 have demonstrated that cytogenetics is highly predictive of outcome in adult ALL, but the effectiveness of these markers has not been tested outside clinical trials. In this report we present population-based cytogenetic, clinical, and outcome data from 349 adult ALL patients diagnosed during a 19-year period in a single region in the northern part of England.
Methods
Study area and population data
The study area comprised the region served by the National Health Service (NHS) North East Strategic Health Authority during this period. The total population of this region is 3.1 million and includes 2.5 million people ages 15 years or older (Office for National Statistics; http://www.statistics.gov.uk/). Age-specific incidence rates were calculated by the use of the average of the population determined by the 1991 and 2001 United Kingdom censuses (Office for National Statistics). Since October 1982, the consultant hematologists in this region have collaborated to keep a register of all newly diagnosed patients with ALL.5 Ethical approval to conduct this audit was granted by the institutional review boards of all relevant treatment centers.
Diagnosis
All patients diagnosed with ALL between January 1, 1983, and December, 31, 2001, who were 15 years or older were included in this study. In all cases, the diagnosis was confirmed by morphology and appropriate cytochemical staining. Immunophenotyping was performed where possible by the use of standard methodologies, and cases were retrospectively classified into 1 of 5 subgroups (pro-B, B-cell precursor ALL, mature-B, T-lineage [T]-ALL, and biphenotypic) by the use of the original immunologic classification.
Cytogenetics
Cytogenetic studies were performed at 1 of 3 laboratories in the region (see Acknowledgments) but collated by the NHS Northern Genetics Service. Analysis was performed on pretreatment bone marrow or peripheral blood samples by the use of standard G-banding techniques. Screening for the major chromosomal translocations, ie, t(9;22)/BCR-ABL1, MLL/11q23 rearrangements, by the use of fluorescence in situ hybridization (FISH) and/or reverse-transcriptase polymerase chain reaction (RT-PCR) assays became routine practice at the beginning of 1997. Chromosomal abnormalities were defined and recorded according to the International System for Human Cytogenetic Nomenclature.13 None of the patients presented in this report was included in the previous cytogenetic study of patients treated on Medical Research Council (MRC) UKALLXII/Eastern Cooperative Oncology Group (ECOG) 2993.2
Treatment
Patients younger than 55 years of age with ALL (B-cell precursor, T-cell, and mature-B) or acute biphenotypic leukemia who were treated with curative intent were mostly treated according to regional North East ALL protocols (NE-ALL III-VI).14,15 Occasionally, patients were treated according to, but not registered on, national protocols (UKALLXII, UKALLXA).16,17 Acute biphenotypic leukemia was defined as terminal deoxynucleotidyl transferase (TdT)–positive patients who also expressed some myeloid antigens. In this era TdT positivity was considered the key marker indicating involvement of the lymphoid lineage and hence ALL therapy. The multiagent protocol NE-ALL III15 was introduced in 1982 and correlates with the start of this study. In 1984, routine standard maintenance was discontinued for patients younger than 55 years of age and instead these patients were offered, in first remission, an allogeneic transplant or, in the absence of a family donor, an autologous transplant.14 Patients younger than 45 years of age underwent a preconditioning regimen of cyclophosphamide (60 mg/kg) and total-body irradiation (1200 cGy), and those aged 45 to 55 years received melphalan (3 mg/kg), total-body irradiation (1050 cGy), and noncryopreserved marrow rescue.
Between 1988 and 1994 (NE-ALL IV-V), the use of idarubicin replaced doxorubicin in induction because of improved penetration into the central nervous system, and prednisolone was replaced by dexamethasone. Cranial irradiation was used throughout NE-ALL III-V unless the patient received irradiation as part of his or her conditioning for a transplantation. The last NE-ALL protocol in the series, NE-ALL VI, started in 1994. Cranial irradiation was abandoned and replaced by a consolidation phase, in which 2 courses of high-dose methotrexate were alternated with 2 courses of high-dose iphosphamide, epirubicin, and etoposide. The transplant policy in NE-ALL VI was the same as NE-ALL IV-V and was applied consistently until 2001. Patients aged 55 years or older were treated as previously described.5
Statistics
Categorical variables were compared by the use of the χ2 or Fisher exact tests. The Mann-Whitney rank test was used to compare the age distribution of different cytogenetic subgroups. Complete remission (CR) was defined morphologically as fewer than 5% blasts in the bone marrow, immunologically as TdT/CD10 negativity (before 1995), or by molecular assessment of immunoglobulin and T-cell gene rearrangements (after 1995). Survival analysis was restricted to those patients who were treated by an intention-to-cure protocol (n = 250). Patients who received only palliative care, those who presented with advanced organ failure, and those who died before a CR could be assessed were deemed not to have received treatment with curative intent (n = 99) and, therefore, were not included in the survival analysis.
Overall survival (OS) was defined as the time from diagnosis to death or last contact. Survival estimates, life tables, and curves were constructed using the Kaplan-Meier method. The variables age and cytogenetic risk group initially were analyzed in isolation by the use of univariate Cox regression models. Multivariate Cox regression on these variables was then performed via a stepwise modeling process on the basis of the difference between successive models calculated by use of the log likelihood. At this stage of analysis there was a further reduction of the number of patients analyzed as the result of missing cytogenetics (n = 73), dates (n = 13), or because of statistical concerns over their excessive influence on the model (n = 2). Thus a total of 162 patients were included in the multivariate model. Because of the exploratory nature of this analysis, all calculations were considered at the 5% significance level.
Results
Descriptive epidemiology
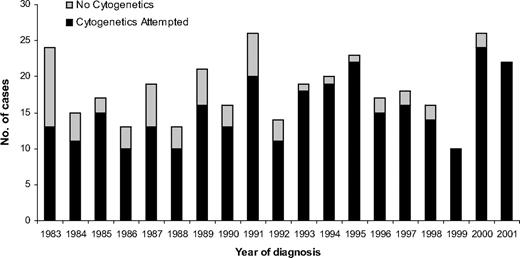
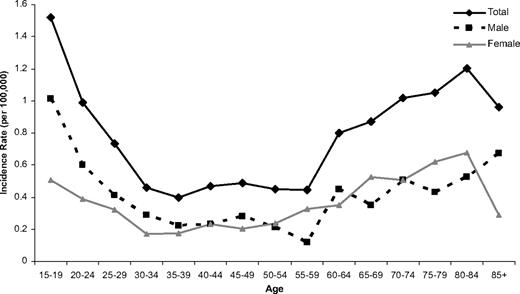
A total of 349 adults aged 15 years or older were diagnosed with ALL during the study period. Although the number of patients diagnosed each year varied, there was no trend toward either an increase or decrease in the incidence of the disease during the study period (Figure 1). The incidence of ALL varied by age, and a bimodal distribution was observed, with the younger (15-19 years old) and older (≥ 70 years) adults having the greatest incidence (Figure 2). Approximately one-third of the patients were 60 years or older (Table 1).
Number of patients diagnosed in the northern part of England with adult ALL between 1983 and 2001 subdivided by cytogenetic analysis at diagnosis.
Number of patients diagnosed in the northern part of England with adult ALL between 1983 and 2001 subdivided by cytogenetic analysis at diagnosis.
Age-specific incidence of adults with ALL in the northern part of England by sex.
Age-specific incidence of adults with ALL in the northern part of England by sex.
Demographic, clinical, and cytogenetic features of 349 adults with ALL
| . | Age, y . | Total . | ||
|---|---|---|---|---|
| 15 to 29 . | 30 to 59 . | 60 or older . | ||
| No. of patients | 115 | 110 | 124 | 349 |
| Sex ratio, M:F | 1:0.6 | 1:1.1 | 1:1.1 | 1:0.9 |
| WCC × 109/L, n (%)* | ||||
| < 10 | 55 (48) | 40 (38) | 42 (41) | 137 (43) |
| 10-49.9 | 35 (30) | 33 (32) | 46 (45) | 114 (36) |
| ≥ 50 | 24 (21) | 31 (30) | 14 (14) | 69 (21) |
| Immunophenotype, n (%)† | ||||
| Pro-B | 7 (7) | 15 (15) | 11 (12) | 33 (10) |
| B-cell precursor | 71 (68) | 60 (59) | 44 (49) | 175 (55) |
| Mature B | 1 (< 1) | 10 (10) | 21 (23) | 32 (10) |
| T-ALL | 26 (25) | 11 (11) | 5 (6) | 42 (13) |
| Biphenotypic | 0 | 6 (6) | 9 (10) | 15 (5) |
| Cytogenetic investigations | ||||
| Attempted | 98 (85) | 100 (91) | 94 (73) | 292 (84) |
| Successful | 77 (79) | 79 (79) | 80 (85) | 236 (81) |
| Normal | 25 (32) | 19 (24) | 18 (23) | 62 (26) |
| Abnormal | 52 (68) | 60 (76) | 62 (77) | 174 (74) |
| Cytogenetic investigations before January 1993, n (%) | ||||
| Attempted | 44 (76) | 51 (86) | 37 (61) | 132 (74) |
| Successful | 32 (73) | 38 (75) | 30 (81) | 100 (76) |
| Normal | 12 (38) | 8 (21) | 11 (37) | 31 (31) |
| Abnormal | 20 (63) | 30 (79) | 19 (63) | 69 (69) |
| Cytogenetic investigations after January 1993, n (%) | ||||
| Attempted | 54 (95) | 49 (96) | 57 (90) | 160 (94) |
| Successful | 45 (88) | 41 (84) | 50 (88) | 136 (85) |
| Normal | 13 (29) | 11 (27) | 7 (14) | 31 (23) |
| Abnormal | 32 (71) | 30 (73) | 43 (86) | 105 (77) |
| Chromosomal abnormality, n (%)‡ | ||||
| Total | 77 | 79 | 80 | 236 |
| t(9;22)(q34;q11) | 6 (8) | 16 (20) | 14 (18) | 36 (15) |
| t(4;11)(q21;q23) | 4 (5) | 5 (6) | 1 (1) | 10 (4) |
| t(1;19)(q23;p13) | 4 (5) | 2 (3) | 0 | 6 (3) |
| t(8;14)(q24;q32) | 0 | 7 (9) | 10 (13) | 17 (7) |
| t(14;18)(q32;q21)§ | 0 | 2 (3) | 8 (10) | 10 (4) |
| HeH‖ | 10 (13) | 4 (5) | 3 (4) | 17 (7) |
| HoTr¶ | 1 (1) | 2 (3) | 4 (5) | 7 (3) |
| Complex# | 2 (3) | 4 (6) | 9 (11) | 15 (7) |
| Other | 25 (32) | 18 (23) | 16 (20) | 59 (25) |
| Normal** | 25 (32) | 19 (24) | 18 (23) | 62 (26) |
| CR rate, % (n)†† | 89 (99/111) | 77 (75/97) | 51 (20/39) | 79 (194/247) |
| OS estimate, 5 y (95% confidence interval), %‡‡ | 45 (35-55) | 19 (11-28) | 12 (4-27) | 30 (24-36) |
| . | Age, y . | Total . | ||
|---|---|---|---|---|
| 15 to 29 . | 30 to 59 . | 60 or older . | ||
| No. of patients | 115 | 110 | 124 | 349 |
| Sex ratio, M:F | 1:0.6 | 1:1.1 | 1:1.1 | 1:0.9 |
| WCC × 109/L, n (%)* | ||||
| < 10 | 55 (48) | 40 (38) | 42 (41) | 137 (43) |
| 10-49.9 | 35 (30) | 33 (32) | 46 (45) | 114 (36) |
| ≥ 50 | 24 (21) | 31 (30) | 14 (14) | 69 (21) |
| Immunophenotype, n (%)† | ||||
| Pro-B | 7 (7) | 15 (15) | 11 (12) | 33 (10) |
| B-cell precursor | 71 (68) | 60 (59) | 44 (49) | 175 (55) |
| Mature B | 1 (< 1) | 10 (10) | 21 (23) | 32 (10) |
| T-ALL | 26 (25) | 11 (11) | 5 (6) | 42 (13) |
| Biphenotypic | 0 | 6 (6) | 9 (10) | 15 (5) |
| Cytogenetic investigations | ||||
| Attempted | 98 (85) | 100 (91) | 94 (73) | 292 (84) |
| Successful | 77 (79) | 79 (79) | 80 (85) | 236 (81) |
| Normal | 25 (32) | 19 (24) | 18 (23) | 62 (26) |
| Abnormal | 52 (68) | 60 (76) | 62 (77) | 174 (74) |
| Cytogenetic investigations before January 1993, n (%) | ||||
| Attempted | 44 (76) | 51 (86) | 37 (61) | 132 (74) |
| Successful | 32 (73) | 38 (75) | 30 (81) | 100 (76) |
| Normal | 12 (38) | 8 (21) | 11 (37) | 31 (31) |
| Abnormal | 20 (63) | 30 (79) | 19 (63) | 69 (69) |
| Cytogenetic investigations after January 1993, n (%) | ||||
| Attempted | 54 (95) | 49 (96) | 57 (90) | 160 (94) |
| Successful | 45 (88) | 41 (84) | 50 (88) | 136 (85) |
| Normal | 13 (29) | 11 (27) | 7 (14) | 31 (23) |
| Abnormal | 32 (71) | 30 (73) | 43 (86) | 105 (77) |
| Chromosomal abnormality, n (%)‡ | ||||
| Total | 77 | 79 | 80 | 236 |
| t(9;22)(q34;q11) | 6 (8) | 16 (20) | 14 (18) | 36 (15) |
| t(4;11)(q21;q23) | 4 (5) | 5 (6) | 1 (1) | 10 (4) |
| t(1;19)(q23;p13) | 4 (5) | 2 (3) | 0 | 6 (3) |
| t(8;14)(q24;q32) | 0 | 7 (9) | 10 (13) | 17 (7) |
| t(14;18)(q32;q21)§ | 0 | 2 (3) | 8 (10) | 10 (4) |
| HeH‖ | 10 (13) | 4 (5) | 3 (4) | 17 (7) |
| HoTr¶ | 1 (1) | 2 (3) | 4 (5) | 7 (3) |
| Complex# | 2 (3) | 4 (6) | 9 (11) | 15 (7) |
| Other | 25 (32) | 18 (23) | 16 (20) | 59 (25) |
| Normal** | 25 (32) | 19 (24) | 18 (23) | 62 (26) |
| CR rate, % (n)†† | 89 (99/111) | 77 (75/97) | 51 (20/39) | 79 (194/247) |
| OS estimate, 5 y (95% confidence interval), %‡‡ | 45 (35-55) | 19 (11-28) | 12 (4-27) | 30 (24-36) |
ALL indicates acute lymphoblastic leukemia; CR, complete remission; HeH, high hyperdiploidy; HoTr, low hypodiploidy/near triploidy; OS, overall survival; T-ALL, T-lineage acute lymphoblastic leukemia; and WCC, white cell count.
Information only available for 320 patients.
Information only available for 297 patients.
Mutually exclusive.
This chromosomal abnormality was expressed in conjunction with the t(8;14) in 3 patients.
High hyperdiploidy (51-65 chromosomes).
Low hyperdiploidy (30-39 chromosomes)/near triploidy (60-78 chromosomes).
Complex karyotype defined as 5 or more chromosomal abnormalities in the absence of an established cytogenetic subgroup.
A total of 20 or more normal metaphases in the absence of any clonal chromosomal abnormality.
CR only available for 247 patients.
OS rate available for 234 patients as the result of missing data.
Overall there were more male than female subjects (1.2:1 M:F), but this ratio varied significantly with age (P < .03; Table 1; Figure 2). The excess of male patients was confined to younger adults, where the ratio was 1.8:1 M:F. In addition, the excess of male patients was more pronounced among T-ALL patients, where a ratio of 2.2:1 M:F was observed. T-ALL patients were also younger (see next paragraph). Figure 2 shows the age-specific incidence of ALL for male and female patients separately, demonstrating that the change in the sex ratio with increasing age is not simply a function of the increased life expectancy enjoyed by females.
The majority of patients had a B-lineage ALL (240, 81%), whereas 42 (14%) had T-ALL and 15 (5%) were described as biphenotypic (see “Methods”). However, this distribution differed significantly with age (P < .001; Table 1). Patients with mature-B ALL, which comprised 11% of patients overall, were significantly older, with a median age of 63 years (interquartile range [IQR], 53-71 years) versus 42 years (IQR, 23-66 years; P < .001). In contrast, T-ALL patients were significantly younger, with a median age of 25 years (IQR, 19-42 years) versus 48 years (IQR, 26-69 years; P < .001).
White cell count (WCC) was strongly correlated with immunophenotype. Patients with T-ALL were significantly more likely to have a WCC of greater than 50 × 109/L compared with B-lineage patients: 17 (40%) of 42 versus 47 (19%) of 247 (P = .001). Given that patients with T-ALL were also significantly younger (see previous paragraph), this finding translated into older patients appearing to have a lower WCC (Table 1). However, no such effect was observed if the T-ALL patients were excluded (data not shown).
Diagnostic cytogenetic analysis
Cytogenetic analysis was attempted in 292 (84%) of 349 patients but was significantly more prevalent after 1993: 160 (94%) of 171 versus 132 (74%) of 178 (P < .001; Table 1; Figure 1). This finding was true across all age groups (data not shown). A total of 236 (81%) of 292 had a successful cytogenetic result, and a clonal chromosomal abnormality was detected in 174 (74%) of 236 patients. These rates did not vary during the study period (P > .05) or by age (P > .05). Overall cytogenetics revealed a normal karyotype in 63 (27%) patients, but this finding was more prevalent among T-ALL patients (14/30, 47%) compared with B-lineage patients (38/176, 22%; P = .006). Because T-ALL patients are younger, this explains the greater incidence of normal karyotype in adolescents (15-19 years).
The most prevalent specific chromosomal abnormality was the Philadelphia chromosome, t(9;22)(q34;q11), BCR-ABL1, which was present in 36 (15%) of 236 patients. There was no difference in the incidence before and after the introduction of routine FISH/RT-PCR at the beginning of 1997: 24 (15%) of 158 versus 12 (13%) of 92 (P > .1). The prevalence increased with age up to, but not beyond, the fourth decade: 15 to 19 years, 2 (5%) of 42; 20 to 29 years, 4 (11%) of 35; 30 to 39 years 4 (15%) of 26; 40 to 49 years, 7 (24%) of 29; 50 to 59 years 5 (21%) of 24; 60 to 69 years, 8 (21%) of 38; 70 to 79 years, 4 (13%) of 30; 80 years or older, 2 (17%) of 12. Thus, although there was a significant difference in the incidence of t(9;22) between patients ages 15 and 30 years (7%) and those older than 30 years of age (24%; P = .02), there was no difference between those ages 30 to 60 years (22%) and older than 60 years of age (16%; P = .4). The majority of t(9;22) patients had B-cell precursor–ALL (32/35, 91%) but 3 patients were described as having a biphenotypic immunophenotype. Just more than one-half the t(9;22) patients (19/34, 56%) had a WCC less than 50 × 109/L, whereas the remaining 15 patients had a WCC of more than 50 × 109/L, including 10 patients with a WCC of more than 100 × 109/L.
Other established chromosomal translocations, such as t(4;11), t(1;19), t(8;14), and t(14;18), each occurred in less than 10% of patients but showed strong correlations with age. Both t(4;11) and t(1;19) occurred more often in younger adults and t(8;14) and t(14;18) in older adults (≥ 60 years; Table 2). The 2 ploidy subgroups, high hyperdiploidy (51-65 chromosomes, HeH) and low hypodiploidy/near triploidy (30-39 and 60-78 chromosomes, HoTr), were observed in 7% and 3% of patients, respectively. None of these patients had T-ALL or mature-B ALL, and they were associated with younger and older age, respectively (Table 2). Patients without one of the aforementioned established chromosomal abnormalities or tetraploidy were classified as having a complex karyotype if 5 or more clonal chromosomal abnormalities were observed. This subgroup accounted for 7% of patients overall but was more prevalent among patients older than 60 years of age. Collectively, Philadelphia-negative patients with high-risk cytogenetics, defined as t(4;11), t(8;14), HoTr, and complex karyotype,2 were significantly more prevalent among patients older than 60 years of age: 24 (36%) of 66 versus 25 (19%) of 134 (P = .006).
Demographic, clinical, and survival data for 349 adults with ALL
| . | Total, n (%) . | Sex ratio, M/F . | Age . | WCC* . | Outcome† . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Treated, n (%) . | CR rate, n (%)‡ . | Deaths, n (%)§ . | OS at 5 years‖ . | ||||||||
| Mean . | Older than 60 y, % . | Median . | More than 50, % . | OS, % . | 95% CI, % . | ||||||
| Total (full dataset) | 349 | 1:0.9 | 46.4 | 36 | 13.6 | 20 | 250 (72) | 194 (70) | 177 (73) | 30 | (24- 36) |
| Successful cytogenetics | |||||||||||
| Total | 236 | 1:0.8 | 45.4 | 34 | 14.6 | 20 | 177 (75) | 133 (75) | 126 (71) | 29 | (22- 37) |
| t(9;22)(q34;q11) | 36 (15) | 1:0.6 | 50.6 | 39 | 30.1 | 42 | 26 (72) | 17 (65) | 24 (92) | 9 | (2- 26) |
| t(4;11)(q21;q23) | 10 (4) | 1:1.5 | 39.1 | 10 | 43.6 | 50 | 9 (90) | 7 (78) | 6 (67) | 33 (at 3 years) | (8-62) |
| t(1;19)(q23;p13) | 6 (3) | 1:5 | 33.2 | 21.5 | 16 | 6 (100) | 6 (100) | 3 (50) | 60 | (13- 88) | |
| t(8;14)(q24;q32) | 17 (7) | 1:0.9 | 60.8 | 59 | 15.7 | 6 | 10 (59) | 4 (44) | 8 (80) | 20 | (3%- 48%) |
| t(14;18)(q32;q21)¶ | 10 (4) | 1:0.8 | 60.0 | 67 | 20.0 | 16 | 5 (50) | 2 (40) | 4 (80) | 33 (at 1 year) | (1-77) |
| HeH# | 17 (7) | 1:0.5 | 33.9 | 18 | 3.1 | 6 | 15 (88) | 15 (100) | 5 (36) | 77 | (45-92) |
| HoTr‡‡ | 7 (3) | 1:1.3 | 55.1 | 86 | 2.6 | — | 6 (86) | 4 (67) | 6 (100) | 0 | — |
| Complex†† | 15 (7) | 1:0.5 | 58.9 | 60 | 13.0 | — | 5 (33) | 4 (80) | 4 (80) | 20 | (1- 58) |
| Other | 59 (25) | 1:0.8 | 42.2 | 29 | 16.3 | 24 | 48 (85) | 39 (81) | 34 (76) | 27 | (14-41) |
| Normal‡‡ | 62 (26) | 1:0.7 | 40.8 | 29 | 10.0 | 16 | 49 (79) | 35 (71) | 34 (69) | 32 | (18-46) |
| Cytogenetic risk group | |||||||||||
| t(9;22) | 36 (15) | 1:0.6 | 50.6 | 39 | 30.1 | 42 | 26 (72) | 17 (65) | 24 92% | 9 | (2-26) |
| Poor | 56 (24) | 1:1.3 | 55.8 | 52 | 15.2 | 13 | 33 (59) | 21 (66) | 26 79% | 19 | (7-36) |
| Standard | 144 (61) | 1:0.8 | 40.0 | 26 | 11.0 | 18 | 118 (82) | 95 (82) | 76 67% | 37 | (27-46) |
| . | Total, n (%) . | Sex ratio, M/F . | Age . | WCC* . | Outcome† . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Treated, n (%) . | CR rate, n (%)‡ . | Deaths, n (%)§ . | OS at 5 years‖ . | ||||||||
| Mean . | Older than 60 y, % . | Median . | More than 50, % . | OS, % . | 95% CI, % . | ||||||
| Total (full dataset) | 349 | 1:0.9 | 46.4 | 36 | 13.6 | 20 | 250 (72) | 194 (70) | 177 (73) | 30 | (24- 36) |
| Successful cytogenetics | |||||||||||
| Total | 236 | 1:0.8 | 45.4 | 34 | 14.6 | 20 | 177 (75) | 133 (75) | 126 (71) | 29 | (22- 37) |
| t(9;22)(q34;q11) | 36 (15) | 1:0.6 | 50.6 | 39 | 30.1 | 42 | 26 (72) | 17 (65) | 24 (92) | 9 | (2- 26) |
| t(4;11)(q21;q23) | 10 (4) | 1:1.5 | 39.1 | 10 | 43.6 | 50 | 9 (90) | 7 (78) | 6 (67) | 33 (at 3 years) | (8-62) |
| t(1;19)(q23;p13) | 6 (3) | 1:5 | 33.2 | 21.5 | 16 | 6 (100) | 6 (100) | 3 (50) | 60 | (13- 88) | |
| t(8;14)(q24;q32) | 17 (7) | 1:0.9 | 60.8 | 59 | 15.7 | 6 | 10 (59) | 4 (44) | 8 (80) | 20 | (3%- 48%) |
| t(14;18)(q32;q21)¶ | 10 (4) | 1:0.8 | 60.0 | 67 | 20.0 | 16 | 5 (50) | 2 (40) | 4 (80) | 33 (at 1 year) | (1-77) |
| HeH# | 17 (7) | 1:0.5 | 33.9 | 18 | 3.1 | 6 | 15 (88) | 15 (100) | 5 (36) | 77 | (45-92) |
| HoTr‡‡ | 7 (3) | 1:1.3 | 55.1 | 86 | 2.6 | — | 6 (86) | 4 (67) | 6 (100) | 0 | — |
| Complex†† | 15 (7) | 1:0.5 | 58.9 | 60 | 13.0 | — | 5 (33) | 4 (80) | 4 (80) | 20 | (1- 58) |
| Other | 59 (25) | 1:0.8 | 42.2 | 29 | 16.3 | 24 | 48 (85) | 39 (81) | 34 (76) | 27 | (14-41) |
| Normal‡‡ | 62 (26) | 1:0.7 | 40.8 | 29 | 10.0 | 16 | 49 (79) | 35 (71) | 34 (69) | 32 | (18-46) |
| Cytogenetic risk group | |||||||||||
| t(9;22) | 36 (15) | 1:0.6 | 50.6 | 39 | 30.1 | 42 | 26 (72) | 17 (65) | 24 92% | 9 | (2-26) |
| Poor | 56 (24) | 1:1.3 | 55.8 | 52 | 15.2 | 13 | 33 (59) | 21 (66) | 26 79% | 19 | (7-36) |
| Standard | 144 (61) | 1:0.8 | 40.0 | 26 | 11.0 | 18 | 118 (82) | 95 (82) | 76 67% | 37 | (27-46) |
ALL indicates acute lymphoblastic leukemia; CI, confidence interval; CR, complete remission; HeH, high hyperdiploidy; HoTr, low hypodiploidy/near triploidy; OS, overall survival; and WCC, white cell count.
Information only available for 320 patients.
This only includes those 250 patients treated with curative intent.
CR only available for 247 patients.
Status unavailable for 16 patients.
OS information available for 234 patients as the result of missing data.
This chromosomal abnormality was expressed in conjunction with the t(8;14) in 3 patients
High hyperdiploidy (51-65 chromosomes).
Low hyperdiploidy (30-39 chromosomes)/near triploidy (60-78 chromosomes).
Complex karyotype defined as 5 or more chromosomal abnormalities in the absence of an established cytogenetic subgroup.
A total of 20 or more normal metaphases in the absence of any clonal chromosomal abnormality.
The most prevalent cytogenetic subgroups were the other abnormal group and those with a normal karyotype, which together accounted for more than 50% of patients with successful cytogenetics. Among the 59 patients classified as “other abnormal,” some known chromosomal abnormalities were observed but were too infrequent to be analyzed separately. Examples included t(10;14)(q24;q11), n = 2, and t(11;14)(p13;q11), n = 1. Known secondary chromosomal abnormalities, such as deletions of 6q (n = 9) and 9p (n = 6), also were observed both in combination with established abnormalities and within the “other abnormal” and complex karyotype groups but at too low a frequency to be analyzed separately.
Outcome and prognostic relevance of chromosomal abnormalities
Among the 349 patients in this cohort, 250 (72%) were treated with curative intent. This number included 211 (94%) of 225 patients younger than 60 years of age but only 39 (31%) of 124 of those older than 60 years of age. Only the outcome of patients who were actively treated has been considered in this section. CR was achieved in 194 (78%) of 247 patients who were actively treated. However, this rate varied by age and cytogenetics (Tables 1–2). Significantly fewer patients older than 60 years of age achieved a CR compared with those younger than 60 years: 20 (51%) of 39 versus 174 (84%) of 208 (P < .001). In addition, fewer patients with t(9;22) or other high-risk cytogenetics achieved a CR (Tables 1–2).
Among the 250 actively treated patients, 100 (40%) received an autologous (n = 66), allogeneic (n = 29), or matched unrelated donor (n = 6) bone marrow transplant. All but 6 of the transplants were performed in first CR. None of the patients older than 60 years of age received a transplant, compared with 59 (51%) of 115 patients 15 to 29 years old and 41 (37%) of 110 patients 30 to 60 years old. The remaining 99 patients received chemotherapy alone.
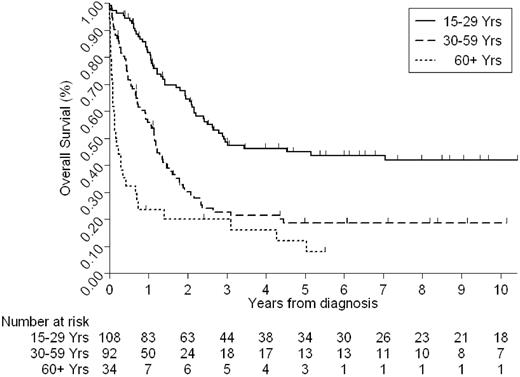
Given the duration of the study and the heterogeneity of treatments received, we have only considered OS. Recent data suggest that in adult ALL, outcome after relapse is extremely poor18 ; hence this is a suitable end point to be considered by this study. The median follow-up time for this cohort was 6.25 years. The 5-year OS rate for the whole cohort was 30% (95% confidence interval [CI], 24%-36%). However, this figure varied considerably by age and cytogenetics (Tables 1–2; Figures 3–4). Estimates of the 5-year OS rates by each decade were as follows: 15 to 19 years, 47% (95% CI, 33%-61%); 20 to 29 years, 43% (95% CI, 29%-57%); 30 to 39 years, 16% (95% CI, 6%-32%); 40 to 49 years, 20% (95% CI, 8%-36%); 50 to 59 years, 19% (95% CI, 7%-36%); 60 years or older, 12% (3%-27%).
Older patients and those with poor-risk cytogenetics, as defined by the MRC UKALLXII/ECOG 2993 trial, had a significantly worse outcome (Tables 1–2; Figures 3–4). We have already demonstrated (Tables 1–2) that cytogenetics and age are closely related. The frequency of poor-risk cytogenetics, for example, t(9;22), t(8;14), HoTr, and complex karyotype, was greater among older patients, and fewer older patients had HeH. Little change was noted in the outcome of patients during the study period. Although there was a marginally increase in the OS at 5 years for patients ages 30 years and older, the difference was not statistically significant (data not shown).
To determine whether the adverse affect of cytogenetics was independent of age we performed multivariate analysis. Because of the relatively small number of patients with both cytogenetic and outcome data (n = 162), we were able to consider only 3 cytogenetic risk groups: t(9;22), all patients with t(9;22)/BCR-ABL; poor, patients with t(4;11), t(8;14), t(14;18), HoTr, and complex karyotype; and standard, all other patients. A Cox proportional hazards model containing just age and cytogenetic risk group indicated a statistical interaction between age and cytogenetics. When we added an interaction term to the model, it revealed that both age and cytogenetics were contributing significantly to outcome. Patients with t(9;22) had a 12.5-fold increase risk of dying (hazard ratio 12.50, 95% CI, 2.69-58.07, P = .001) whereas those with poor-risk cytogenetics had a 3.5-fold increased risk (hazard ratio 3.47, 95% CI, 1.45-8.37, P = .007), both in comparison with patients with standard-risk cytogenetics. The statistical interaction between cytogenetics and age indicates that the effect of cytogenetics might not be the same at all ages. However, given the size of the cohort, the relatively crude measure of outcome used, and the heterogeneity of treatment, it was not possible to investigate this further.
Incidence of major chromosomal abnormalities in this study compared with previously published data from clinical trials
We compared the incidence of the major chromosomal abnormalities in this study to that observed in 6 major clinical trials (Table 3). Because researchers of most clinical trials impose age limits on potential subjects, we restricted the comparison with those patients in this study between 15 and 59 years of age. Aside from the variation in the incidence of normal karyotype and t(9;22) there was little difference. However, it should be noted that several of the abnormalities were only classified in a few studies. The overall frequency of t(9;22) in this population-based study was 15%, which compares well with that reported by the 2 recent United Kingdom–based trials: MRC UKALLXA (11%)17 and UKALLXII (16%).2 In contrast, the overall frequency of t(9;22) ALL reported by other trial based studies is much greater: ECOG2993 (25%),2 GMALL (36%),19 GIMEMA0496 (23%-31%),20 GFCH (29%),21 SWOG9400 (27%),3 and CALGB (26%)22 (Table 4). These differences are likely to be attributable to a combination of factors, including the age profile of the underlying populations, recruitment bias, and detection method. Although there are exceptions, generally studies with a lower frequency of t(9;22) comprise a greater proportion of younger patients and vice versa.
Incidence of major chromosomal abnormalities in adult ALL observed in this study compared with previously published clinical trials
| Study . | Reference . | Patients tested, n . | t(9;22) % . | t(4;11) % . | t(1;19) % . | t(8;14) % . | t(14;18) % . | HeH, %* . | HoTr, %† . | Complex, %‡ . | Normal, %§ . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| This study (15- to 59-year-old patients only) | NA | 236 | 15 | 6 | 4 | 4 | 1 | 9 | 2 | 4 | 29 |
| MRC UKALLXA | 17 | 350 | 11 | 3 | 3 | NA | NA | 8 | NA | NA | 34 |
| UKALLXII/ECOG2993 | 2 | 1373 | 19 | 4 | 2 | 1 | NA | 6 | 2 | 3 | 14 |
| CALGB | 22 | 256 | 26 | 5 | NA | NA | NA | NA | NA | NA | 25 |
| GIMEMA 0496 | 20 | 386 | 26 | 7 | 2 | NA | NA | NA | NA | NA | 26 |
| SWOG 9400 | 3 | 140 | 26 | 5 | 5 | NA | NA | NA | NA | NA | 22 |
| GFCH | 21 | 443 | 29 | 4 | 3 | 5 | NA | 7 | NA | NA | 15 |
| Study . | Reference . | Patients tested, n . | t(9;22) % . | t(4;11) % . | t(1;19) % . | t(8;14) % . | t(14;18) % . | HeH, %* . | HoTr, %† . | Complex, %‡ . | Normal, %§ . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| This study (15- to 59-year-old patients only) | NA | 236 | 15 | 6 | 4 | 4 | 1 | 9 | 2 | 4 | 29 |
| MRC UKALLXA | 17 | 350 | 11 | 3 | 3 | NA | NA | 8 | NA | NA | 34 |
| UKALLXII/ECOG2993 | 2 | 1373 | 19 | 4 | 2 | 1 | NA | 6 | 2 | 3 | 14 |
| CALGB | 22 | 256 | 26 | 5 | NA | NA | NA | NA | NA | NA | 25 |
| GIMEMA 0496 | 20 | 386 | 26 | 7 | 2 | NA | NA | NA | NA | NA | 26 |
| SWOG 9400 | 3 | 140 | 26 | 5 | 5 | NA | NA | NA | NA | NA | 22 |
| GFCH | 21 | 443 | 29 | 4 | 3 | 5 | NA | 7 | NA | NA | 15 |
ALL indicates acute lymphoblastic leukemia; CALGB, Cancer and Leukemia Group B; GFCH, Français de Cytogénétique Hématologique; GIMEMA, Gruppo Italiano Malattie Ematologiche Ddell'Adulto; HeH, high hyperdiploidy; HoTr, low hypodiploidy/near triploidy; MRC, Medical Research Council; NA, not available; SWOG, Southwest Oncology Group.
High hyperdiploidy (51-65 chromosomes).
Low hyperdiploidy (30-39 chromosomes)/near triploidy (60-78 chromosomes).
Complex karyotype defined as 5 or more chromosomal abnormalities in the absence of an established cytogenetic subgroup.
A total of 20 or more normal metaphases in the absence of any clonal chromosomal abnormality.
The incidence of t(9;22) in adult ALL in different studies by age
| Study . | Reference . | No. of patients tested . | t(9;22), n (%) . | Detection method . | Age range, y . | Mean age, y . | Less than 20 y, n (%) . | Less than 30 y, n (%) . |
|---|---|---|---|---|---|---|---|---|
| MRC UKALLXA | 17 | 350 | 40 (11) | Cytogenetics | 15-60+ | NA | 115 (33) | 186 (53) |
| This study | NA | 236 | 36 (15) | Cytogenetics, plus some FISH and RT-PCR | 15-91 | 45 | 42 (18) | 77 (33) |
| UKALLXII | 2 | 872 | 142 (16) | Cytogenetics, FISH, and RT-PCR | 15-55 | 31 | 207 (24) | 453 (52) |
| ECOG2993 | 2 | 501 | 125 (25) | Cytogenetics, FISH, and RT-PCR | 15-65 | 36 | 61 (12) | 190 (38) |
| CALGB | 22 | 256 | 67 (26) | Cytogenetics | 15-60+ | 33 | NA | 109 (43) |
| GIMEMA 0496 | 20 | 386 | 102 (26) | Cytogenetics and RT-PCR | 15-60 | 31 | NA | 148 (46) |
| SWOG 9400 | 3 | 140 | 36 (26) | Cytogenetics and RT-PCR | 15-65 | 32 | NA | 86 (43) based on all 200 patients |
| GFCH | 21 | 443 | 127 (29) | Cytogenetics | 15-60+ | 37 | 90 (20) | 169 (38) |
| GMALL | 19 | 2498 | 904 (36) | RT-PCR | 15-74 | NA | 481 (19) < 25 y | 902 (36) < 35 y |
| Study . | Reference . | No. of patients tested . | t(9;22), n (%) . | Detection method . | Age range, y . | Mean age, y . | Less than 20 y, n (%) . | Less than 30 y, n (%) . |
|---|---|---|---|---|---|---|---|---|
| MRC UKALLXA | 17 | 350 | 40 (11) | Cytogenetics | 15-60+ | NA | 115 (33) | 186 (53) |
| This study | NA | 236 | 36 (15) | Cytogenetics, plus some FISH and RT-PCR | 15-91 | 45 | 42 (18) | 77 (33) |
| UKALLXII | 2 | 872 | 142 (16) | Cytogenetics, FISH, and RT-PCR | 15-55 | 31 | 207 (24) | 453 (52) |
| ECOG2993 | 2 | 501 | 125 (25) | Cytogenetics, FISH, and RT-PCR | 15-65 | 36 | 61 (12) | 190 (38) |
| CALGB | 22 | 256 | 67 (26) | Cytogenetics | 15-60+ | 33 | NA | 109 (43) |
| GIMEMA 0496 | 20 | 386 | 102 (26) | Cytogenetics and RT-PCR | 15-60 | 31 | NA | 148 (46) |
| SWOG 9400 | 3 | 140 | 36 (26) | Cytogenetics and RT-PCR | 15-65 | 32 | NA | 86 (43) based on all 200 patients |
| GFCH | 21 | 443 | 127 (29) | Cytogenetics | 15-60+ | 37 | 90 (20) | 169 (38) |
| GMALL | 19 | 2498 | 904 (36) | RT-PCR | 15-74 | NA | 481 (19) < 25 y | 902 (36) < 35 y |
ALL indicates acute lymphoblastic leukemia; CALGB, Cancer and Leukemia Group B; ECOG, Eastern Cooperative Oncology Group; FISH, fluorescence in situ hybridization; GFCH, Français de Cytogénétique Hématologique; GIMEMA, Gruppo Italiano Malattie Ematologiche Ddell'Adulto; GMALL, German Multicenter Acute Lymphoblastic Leukemia; MRC, Medical Research Council; NA, not available; RT-PCR, reverse-transcription polymerase chain reaction; and SWOG, Southwest Oncology Group.
Discussion
We have reported the largest population-based cytogenetic study of adults with ALL. The overall incidence rate and pattern of incidence by age and sex was similar to that observed in other areas of the United Kingdom.23 This was an appropriate region in which to examine the relationship between age and cytogenetics. First, there was no overlap with the previous publication on the basis of the MRC UKALLXII/ECOG 2993 cohort from which we derived our cytogenetic risk criteria. Although some patients (< 10%) were treated according to the same protocol, they were not officially registered on the trial. Second, the age-specific OS rates observed in this region were similar to that observed in other regions of the United Kingdom.4,24 The results clearly demonstrate that the incidence of cytogenetic and immunophenotypic subgroups varies markedly with the age. Although it is known that the incidence of specific chromosomal abnormalities differs between children and adults, the age-specific frequency of such lesions within adult ALL, especially among those older than 60 years of age, was hitherto unknown or poorly studied.
The results of recent clinical trials of adult ALL have highlighted the importance of cytogenetics in predicting the risk of relapse.2,3 Such studies support the development of cytogenetic-based risk stratification of adults in future trials. One of the limitations of these studies was that they were based on a relatively small proportion of the total available patients because most adult patients with ALL are not recruited to clinical trials. One of the key questions in this field of research is whether the specific chromosomal abnormalities identified as being indicators of poor outcome in the MRC UKALLXII/ECOG2993 trial retained their prognostic relevance outside the context of a clinical trial. Our analyses suggest that both age and cytogenetics are important predictors of outcome in adult ALL. The relatively small number of patients with individual poor-risk chromosomal abnormalities prevented an in-depth analysis. Overall the data in this study support our previous observations that patients with t(4;11), t(8;14), HoTr, and complex karyotype have an inferior outcome. However, it should be noted that (1) there was evidence to suggest that the effect of cytogenetics varied with age, and (2) the treatment received by this cohort was during a 19-year period and hence heterogeneous.
We confirmed the findings of Burmeister et al,19 who reported that the frequency of t(9;22)/BCR-ABL–positive adult ALL does not continue to increase beyond the fourth decade of life. These results are in contrast to the popularly held belief that the incidence of t(9;22)-positive ALL continues to increase with age. The overall frequency of t(9;22) ALL in this study and other United Kingdom trial studies was considerably lower than that observed in series from other countries (Table 4). Although differences in the age profile of the underlying populations are likely to explain some of the variation, the picture appears more complicated and may reflect geographical heterogeneity.25 Patients with t(9;22) now usually are treated with imatinib mesylate or another tyrosine kinase inhibitor in combination with multiagent chemotherapy. These data indicate that such therapy will be suitable for a smaller fraction of patients older than 60 years of age than had been anticipated.
Although this study is the largest and most comprehensive of its kind to date, it does have limitations. First, the number of patients was still quite small, and hence, the number with specific chromosomal abnormalities was too few to undertake detailed survival analysis. Second, rarer and secondary chromosomal abnormalities could not be considered. Third, although most treatment occurred in a single center and with the same ethos, this study spanned nearly 20 years. Finally, the global genomic analysis of childhood ALL has revealed a wide spectrum of copy number alterations beyond the resolution of cytogenetics.26 Many are microdeletions targeting B-cell development genes (eg, IKZF1, PAX5, VpreB1), and most correlate with established cytogenetic subgroups.26,27 Initial investigations in adult ALL by Paulsson et al28 suggest that similar copy number alterations are present. Given that approximately one-half of the patients in this study could not be assigned to a clinically relevant cytogenetic subgroup, future studies will need to incorporate high-quality cytogenetic, FISH and genomic data to fully characterize patients. Moreover, such studies will need to be population based to efficiently plan services, design clinical trials, and fully use the prognostic relevance of genetic markers.
The authors of several recent studies7-10 have reported that adolescents and young adults (AYAs) have a better outcome when treated with pediatric protocols. Several groups worldwide10,29 are now treating AYAs up to the age of 29 years with the use of pediatric protocols. Because AYAs are much less likely to enroll in clinical trials than children,4 this study is the most informative to date as to their cytogenetic profile. Interestingly it is markedly different from both the classical pediatric profile and the adult profile.1 The frequency of HeH is much lower than observed among childhood cohorts (13% vs ∼ 30%), whereas the frequency of t(9;22) and other “adult” cytogenetic abnormalities such as HoTr and complex karyotype is much lower. Unfortunately, our patients have not been screened for ETV6-RUNX1 fusion. However, our previous study30 suggested that this would be present at a much lower frequency than in childhood ALL. The frequency of T-ALL is this age group is noteworthy; at 25%, it is greater than in both childhood ALL and adults aged older than 30 years of age.
One of the major strengths of this study is the unselected nature of the cohort, and we were able to estimate the incidence of ALL at all ages, and by sex and year of diagnosis. There was no evidence to suggest that the incidence of this disease was increasing with time. However, the observation that the incidence continues to increase with age among both male and female patients suggests that the absolute number of patients is likely to increase in the context of an aging population. Thus “elderly” ALL is likely to become an increasing health burden during the next 2 decades, especially as other authors6 have reported that there has been no improvement in outcome for these patients. Hence, new clinical trials targeting this age group are urgently required.
We were able to examine the cytogenetic profile of 124 patients older than 60 years of age. This age group is largely underreported because the upper age limit of most clinical trials is 55 to 65 years. Although diagnostic cytogenetic investigations were less frequently requested in this age group, when they were attempted the success and abnormality rates were similar, if not better, compared with younger patients. These results suggest that pretreatment cytogenetic analysis for adults with ALL should be performed irrespective of age. Interestingly the biologic and cytogenetic profile of these “older” patients was quite different compared with younger patients. Mature-B ALL, plus t(8;14)/t(14;18), was more prevalent, whereas T-ALL was markedly less prevalent. Such information is important for planning the delivery of routine clinical care services and designing clinical trials. Patients with mature-B ALL will now routinely receive treatment on lymphoma protocols.31 The translocations t(4;11) and t(1;19) were rare among patients older than 60 years of age, as was HeH. In contrast, the recently established poor-risk groups—HoTr and complex karyotype—were more prevalent in this older subgroup.
Because older adults (≥ 60 years) were less likely to receive curative treatment, the proportion of such patients included in the multivariate model was lower. Hence, it is difficult to assess the effect of cytogenetics in this age group. Recent data from the Surveillance, Epidemiology and End Results Program showed that this age group was the only one that has not benefited from recent improvements in survival.6 Therefore, further investigation into the cytogenetics of this age group is urgently required to maximize the usefulness of future clinical trials aimed at this age group.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Penny R. A. Taylor for compiling and maintaining the Northern Region adult acute lymphoblastic register. We thank all consultant hematologists in the Northern Region for their consistent support in provision of samples. We would also like to acknowledge the contribution of Dr Janet Tawn (Geoffrey Schofield Cytogenetics Laboratory) and Dr Connie Clarke (Department of Cytogenetics, Middlesbrough General Hospital), who performed cytogenetic analyses on local samples until 1995 and 1997, respectively.
Funding for the registry during the period of study was provided by The Tyneside Leukaemia Research Fund and the Bone Marrow Transplant 2000 Fund. This research was funded in part by Leukemia Research.
Authorship
Contribution: A.V.M. and S.J.P. initiated and designed the study; L.C. and J.W. collected and analyzed the data; H.M.E. performed survival analyses; N.B. provided cytogenetic data; A.V.M. and L.C. wrote the manuscript; and all authors critically reviewed the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Anthony V. Moorman, Leukaemia Research Cytogenetics Group, Northern Institute for Cancer Research, Level 5, Sir James Spence Institute, Royal Victoria Infirmary, Queen Victoria Rd, Newcastle upon Tyne, NE1 4LP United Kingdom; e-mail: anthony.moorman@ncl.ac.uk.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal