Abstract
Activation of the T cell–mediated immune response has been associated with changes in the expression of specific microRNAs (miRNAs). However, the role of miRNAs in the development of an effective immune response is just beginning to be explored. This study focuses on the functional role of miR-146a in T lymphocyte–mediated immune response and provides interesting clues on the transcriptional regulation of miR-146a during T-cell activation. We show that miR-146a is low in human naive T cells and is abundantly expressed in human memory T cells; consistently, miR-146a is induced in human primary T lymphocytes upon T-cell receptor (TCR) stimulation. Moreover, we identified NF-kB and c-ETS binding sites as required for the induction of miR-146a transcription upon TCR engagement. Our results demonstrate that several signaling pathways, other than inflammation, are influenced by miR-146a. In particular, we provide experimental evidence that miR-146a modulates activation-induced cell death (AICD), acting as an antiapoptotic factor, and that Fas-associated death domain (FADD) is a target of miR-146a. Furthermore, miR-146a enforced expression impairs both activator protein 1 (AP-1) activity and interleukin-2 (IL-2) production induced by TCR engagement, thus suggesting a role of this miRNA in the modulation of adaptive immunity.
Introduction
During adaptive immune response T-cell receptor (TCR) engagement by the antigen triggers a signal cascade, which leads to the activation of 3 main transcription factors: AP-1, NF-kB, and NFAT, critically involved in cytokine production. In particular, these transcription factors regulate the expression of early cytokines, especially interleukin-2 (IL-2), that mediate the lymphocytic “clonal expansion” phase.1 Once the foreign threat has been overcome and T lymphocytes have served their effector functions, they must be removed. The death of activated lymphocytes serves to limit the immune response by killing cells that are no longer needed or cells that may have developed the potential to recognize and generate a response to self-antigens. Deregulation of apoptotic pathways in T cells may lead to a spectrum of diseases, including autoimmune diseases and proliferative disorders.2,3
MicroRNAs (miRNAs) recently came into focus as a novel class of posttranscriptional regulatory elements. They are small endogenous noncoding RNAs that repress mRNA translation by base pairing to 3′ untranslated region (3′UTR) of the target genes. In particular, miRNAs are powerful tools that can be promptly expressed by the cell and have the potential to coordinately regulate a large number of different target genes. This suggests that they can be optimal candidates for the control of immune response, a process involving large regulative networks and fine prompt modulation.
The first indication that miRNAs are involved in immunity has emerged by studies showing the selective expression of miR-223 in bone marrow and the involvement of miR-223, miR-155, and miR-146a in the differentiation of myeloid cells.4-7 A growing number of reports have been focused on miR-146a as an important modulator of immune function. In particular, miR-146a has been shown to be involved in innate immunity.8 It was also demonstrated that Toll/Il-1 receptor–mediated miR-146a expression is predominantly driven by NF-κB and it has been speculated that miR-146a might “fine-tune” negative feedback regulation of inflammation through down-regulation of IRAK1 and tumor necrosis factor receptor–associated factor 6 (TRAF6), two proteins involved in Toll/Il-1 receptor signaling.8
Interestingly, miR-146a disregulation has been observed in autoimmune diseases. In particular it has been shown that miR-146 levels are higher in human rheumatoid arthritis synovial tissue, compared with healthy controls.9,10 On the contrary, miR-146a is underepressed in peripheral blood lymphocytes of patients with systemic lupus erythematosus and this low expression has been demonstrated to be associated with the overactivation of the type I interferon pathway.11
Furthermore, expression-profiling studies in mice have demonstrated distinct patterns of miR-146a expression in various T-cell subtypes. For example, miR-146a expression is higher in murine T helper type 1 cells but lower in T helper type 2 and naive T cells.12 In addition, miR-146a is among the most highly expressed miRNAs in murine regulatory T cells, thus suggesting a possible role for miR-146a in maintaining differentiated T-cell lineages.13
Notably, despite the great number of reports showing a selective miR-146a expression in differentiated immune cells, the role of miR-146a in the development of an adequate immune response is just beginning to be explored.
This study focuses on the functional role of miR-146a in T lymphocyte–mediated immune response and provides interesting clues on the transcriptional regulation of miR-146a during T-cell activation. We show that miR-146a is low in human naive T cells and is abundantly expressed in memory T cells; consistently, miR-146a is induced in human primary T lymphocytes upon TCR stimulation. Moreover, we identified NF-kB and c-ETS binding sites as required for the modulation of miR-146a transcription upon TCR engagement.
Our results also demonstrate that several signaling pathways, other than inflammation, are influenced by miR-146a. In particular, we provide experimental evidence that miR-146a modulates activation-induced cell death (AICD), acting as an antiapoptotic factor and that Fas-associated death domain (FADD) is a target of miR-146a. Furthermore, we show that miR-146a overexpression impairs both activator protein 1 (AP-1) activity and IL-2 expression induced by TCR engagement, thus suggesting a role of miR-146a in the modulation of adaptive immunity.
Methods
Purification of naive and differentiated CD4+ and CD8+ T cells
Human primary CD4+ and CD8+ T lymphocytes were purified from healthy donor peripheral blood mononuclear cells using T-cell isolation kit (Miltenyi Biotec), as previously described.14 The purity of the CD4+CD45RA+, CD8+CD45RA+, CD4+CD45RA−CD27+ T, and CD8+CD45RA−CD27+ T cells was more than 98%, as determined by flow cytometry.
Cell cultures and stimulation condition
Human primary naive T lymphocytes were grown in RPMI 1640 (Gibco) supplemented with recombinant IL-2 (50 U/mL; Sigma-Aldrich), 10% heat-inactivated fetal bovine serum (FBS; Sigma-Aldrich), penicillin/streptomycin (Gibco), and l-glutamine (l-glut; Gibco) at 37°C in 5% CO2. The human leukemic T-cell line Jurkat (DSMZ) was grown in RPMI 1640 (Gibco) supplemented with 10% heat-inactivated FBS (Sigma-Aldrich), penicillin/streptomycin (Gibco), and l-glut (Gibco) at 37°C in 5% CO2. 293T cells were grown in Dulbecco modified Eagle medium (Gibco) supplemented with 10% FBS, penicillin/streptomycin, and l-glut at 37°C in 5% CO2. For treatment with pharmacologic inhibitors, Jurkat cells (5 × 105 cells/mL) were preincubated with cyclosporine A (CsA; 0.5 μg/mL; Sigma-Aldrich) or PD98059 (20μM; Sigma-Aldrich) or SP600125 (6μM; Sigma-Aldrich) for 30 minutes before stimulation with anti-CD3 (5 μg/mL; clone OKT3) and anti-CD28 (5 μg/mL) antibodies (Abs). TCR stimulation of human primary CD4+CD45RA+ T lymphocytes and of Jurkat cells was performed using either phorbol myristate acetate (PMA, 50 ng/mL; Sigma-Aldrich) and ionomycin (1 μg/mL; Sigma-Aldrich) or anti-CD3 (5 μg/mL; clone OKT-3) and anti-CD28 (5 μg/mL) Abs.
RNA extraction and analysis of miRNA expression
Total cellular RNA was extracted using the TRIzol reagent (Invitrogen), following the manufacturer's instructions. For Northern blot analysis 20 μg of total RNA was separated by electrophoresis in a 15% acrylamide gel and then transferred to Gene Screen Plus Hybridization Transfer membrane (Perkin Elmer). After ultraviolet cross-linking, membrane hybridization was carried out overnight at 50°C with miR-146a probe labeled with [γ-32P] deoxyoligonucleotide antisense to miR-146a, using T4 PNK system (New England Biolabs). Membranes were washed at 37°C, 3 times with 2× saline sodium citrate/0.1% sodium dodecyl sulfate for 20 minutes and autoradiographed on x-ray films (Kodak) using an intensifying screen at −80°C. After autoradiography the blot was reprobed with a probe for U6 RNA as a loading control. Quantitative reverse-transcription–polymerase chain reaction (qRT-PCR) was performed as described.15 The first-strand synthesis reaction for qRT-PCR was performed using the Superscript III RNase-H− Reverse Transcriptase (Invitrogen) for 30 minutes at 50°C. qPCR was performed with iCycler (BioRad) using iQ-SYBR-green supermix (BioRad) as described.15 U6 RNA was measured with the same method and used for normalization. All oligonucleotides used (Sigma-Aldrich) are listed in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Expression constructs and stable transduction
MiR-146a lentiviral construct was obtained as follows: a region of 668 nucleotides (nts) containing pre-miR-146a was amplified from Jurkat cell genomic DNA and cloned into the BamHI/XhoI sites of a pcDNA4TO vector (Invitrogen). The fragment containing the cytomegalovirus (CMV)–TO promoter and the pre-miRNA coding region, was PCR amplified, inserting BstB1/XhoI sites, and cloned into pSuperior-Puro (Oligoengine) in the same sites. The XmaI-XhoI fragment was finally cloned into the AgeI-XhoI sites in pRRLSIN.cPPT.PGK-GFP.WPRE (plasmid no. 12252; Addgene), replacing the phosphoglycerate kinase (PGK) promoter and the green fluorescent protein (GFP) coding sequence. The vector obtained was named pRRLSIN.cPPT.PURO.CMV-TO-miR-146a.WPRE, herein referred to as pRRL-146a. Similarly, we prepared a control lentiviral construct: pRRLSIN.cPPT.PURO.CMV-TO-ct.WPRE (pRRL-ct), obtained by substituting the miR-146a precursor fragment (BamHI-XhoI) with a sequence encoding for a hairpin yielding a 22-mer RNA, designed to lack homology to any human gene. Viral particles were produced by calcium phosphate–mediated transfection of 293T cells as described.16 Lentiviral supernatants were collected 48 hours after transfection, passed through a 0.45-μm filter, and purified by ultracentrifugation for 2 hours at 175 000g. Jurkat cells were transduced overnight at a multiplicity of infection of 15 in the presence of 4 μg/mL of polybrene (Sigma-Aldrich). Thereafter cells were selected with 3 μg/mL puromycin for 24 hours.
Cloning of reporter constructs, transient transfection, and luciferase assays
The pMLuc-1 AccepTor plasmid (Novagen) was used for generation of miR-146a promoter reporter constructs. Briefly, the 1100-bp promoter region was amplified by PCR from genomic DNA. PCR products were ligated into the pMLuc-1 AccepTor plasmid according to the manufacturer's instructions. Deletions were introduced using the site Quick-Change II Site Directed Mutagenesis kit (Stratagene). Jurkat cells (5 × 105 cells/mL) were transfected (Lipofectamine 2000; Invitrogen) with pMLuc-1 AccepTor plasmid containing the promoter fragment and pGL3-tk for normalization of transfection efficiency. After 24-hour recovery period, transfected cells were either left untreated or induced for 24 hours with either anti-CD3/CD28 Abs or with PMA and ionomycin. Cells were then lysed and analyzed for firefly and renilla luciferase activity using the Dual-Luciferase assay (Promega). For miR-146a target validation, 293T cells (105 cells/500 μL) were grown in 24-well culture plates and 24 hours later transfected with a pGL-3-luc vector containing the 3′UTR of interest, a miR-146a mimic or a scramble mimic and with a pRL-tk-luc vector for normalization of transfection efficiency (Promega). The luciferase activity was assayed using the Dual-Luciferase assay kit (Promega) according to manufacturer's instructions.
Bioinformatics analysis of hsa-pri-miRNA-146a promoter
A bioinformatics analysis covering 3000-bp upstream of the pri-miRNA coding region was performed using the MULAN software (National Center for Biotechnology Information, http://mulan.dcode.org) to scan for transcription factor binding sites conserved between human and mouse.
IL-2 ELISA
pRRL-ct and pRRL-146a Jurkat cells (5 × 105 cells/mL) were stimulated with anti-CD3/CD28 Abs and the supernatants were collected 4 and 24 hours after stimulation. Human IL-2 ELISA Ready-SET-Go kit (eBioscience) was used according to the manufacturer's instructions. Plates were read at 405 nm using an enzyme-linked immunosorbent assay (ELISA) reader (Multiskan-Labsystem).
AP-1 reporter luciferase assay
pRRL-ct and pRRL-146a Jurkat cells (5 × 105 cells/mL) were transiently transfected using Lipofectamine 2000 (Invitrogen) with (AP-1)-luc reporter plasmid, containing 7 copies of the AP-1 enhancer 5′-TGACT-3′ connected to a TATA box located upstream of the firefly luciferase gene or with a control vector, (AP-1)del-luc reporter plasmid, in which the 7 copies of the AP-1 enhancer were removed. The renilla luciferase vector pRL-tk-luc (Promega) was cotransfected to normalize for transfection efficiency, and 24 hours after transfection cells were stimulated with anti-CD3/CD28 Abs. After 24-hour stimulation the luciferase activity was assayed.
Induction and analysis of apoptosis
pRRL-ct and pRRL-146a Jurkat T cells (5 × 105 cells/mL) were grown in RPMI with 3% FBS and were stimulated with anti-CD3 Ab (10 μg/mL, clone OKT3) or anti-Fas Ab (100 ng/mL, clone CH11; Upstate) for the indicated time. Apoptotic cell fractions were determined by annexin V–fluorescein isothiocyanate (FITC) and propidium iodide double staining according to the manufacturer's instructions. Cells were analyzed on a FACSCalibur flow cytometer and data were analyzed with CellQuest software (Becton Dickinson).
Intracellular staining
pRRL-ct, pRRL-146a Jurkat cells and human primary T lymphocytes (5 × 105 cells) were fixed and permeabilized with Cytofix/Cytoperm buffer (100 μL; BD Biosciences) and incubated for 20 minutes at 4°C. Cells were washed twice with saponin 0.1% (Sigma-Aldrich) and then incubated for 1 hour at 4°C with primary Ab diluted in saponin 0.5%. Anti-FADD mAb (BD Pharmingen) was used at dilution of 1:10. Subsequently, cells were washed twice and were incubated for 45 minutes at 4°C in the dark with FITC-conjugated secondary Ab (Sigma-Aldrich) at a dilution of 1:500. Cells were washed with 0.1% saponin and resuspended in phosphate-buffered saline before flow cytometry analysis.
Results
MiR-146a is up-regulated during T-cell differentiation
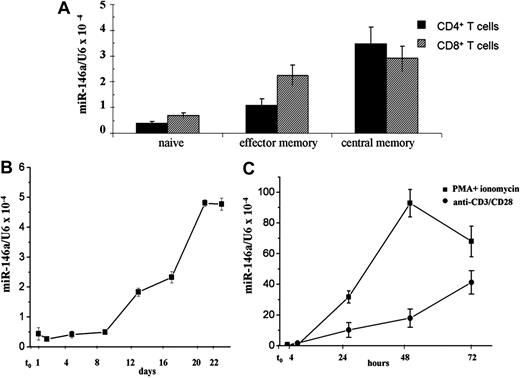
To identify candidate miRNAs whose expression is associated with T-cell differentiation, we analyzed miRNA expression profiles of human naive and central memory CD4+ and CD8+ T lymphocytes, using a miRNA microarray platform developed by LC Sciences. Among the miRNAs that were differentially expressed in these distinct subsets of human primary T cells, we identified miR-146a, a member of miR-146 family composed by miR-146a and miR-146b (supplemental Figure 1). The data obtained by microarray analysis were further validated by quantitative RT-PCR (qRT-PCR), using primers that specifically recognized the mature forms of miR-146a/b, and the analysis was extended to CD4+ and CD8+ effector memory T lymphocytes. We observed that miR-146a expression is low in CD4+ naive T cells and increases approximately 3-fold and 9-fold in CD4+ memory effectors and CD4+ central memory T cells, respectively. Similarly, levels of miR-146a were 3-fold higher in both CD8+ memory effectors and CD8+ central memory T lymphocytes, compared with CD8+ naive T lymphocytes (Figure 1A). Because by qRT-PCR no differences of expression were detected for miR-146b either in CD4+ or in CD8+ memory T cells compared with naive T cells (data not shown), we focused our analysis on miR-146a.
MiR-146a is expressed in T lymphocytes. (A) MiR-146a levels were measured in human purified primary CD4+ and CD8+ cells by qRT-PCR, using U6 levels to normalize expression. (B) CD4+CD45RA+ human naive T lymphocytes were stimulated with PMA and ionomycin and miR-146a levels were measured at different times by qRT-PCR. (C) Jurkat cells were stimulated with anti-CD3/CD28 Abs or with PMA and ionomycin and MiR-146a levels were measured at different times by qRT-PCR. Each bar is the mean of 3 independent experiments.
MiR-146a is expressed in T lymphocytes. (A) MiR-146a levels were measured in human purified primary CD4+ and CD8+ cells by qRT-PCR, using U6 levels to normalize expression. (B) CD4+CD45RA+ human naive T lymphocytes were stimulated with PMA and ionomycin and miR-146a levels were measured at different times by qRT-PCR. (C) Jurkat cells were stimulated with anti-CD3/CD28 Abs or with PMA and ionomycin and MiR-146a levels were measured at different times by qRT-PCR. Each bar is the mean of 3 independent experiments.
MiR-146a expression is induced upon TCR stimulation
The observed up-regulation of miR-146a in memory T lymphocytes led us to study the effects of TCR stimulation on miR-146a expression. To investigate the role of TCR engagement on miR-146a expression, CD4+CD45RA+ naive T lymphocytes, freshly purified from healthy donors, were stimulated with PMA and ionomycin and miR-146a levels were measured by qRT-PCR at different times. As shown in Figure 1B, miR-146a levels increase after 8 days of stimulation, reaching a plateau around 20 days. Interestingly, this kinetic reflected the expression changes of markers commonly used to indicate the transition to a memory cell phenotype, such as CD45RA and CD45RO (supplemental Table 2).
To better define the role of miR-146a on T-cell gene expression, we used Jurkat T cells, a well-established model for in vitro study of TCR signaling. Jurkat cells were treated with either PMA and ionomycin or anti-CD3/CD28 Abs and miR-146a levels were measured at different times by qRT-PCR. As shown in Figure 1C, miR-146a expression, low in nonstimulated Jurkat cells, progressively increased upon PMA and ionomycin treatment, reaching a plateau after 48 hours. When Jurkat cells were stimulated with anti-CD3/CD28 Abs, to resemble a more physiologic stimulation of TCR, we observed the same kinetic of induction, but lower miR-146a levels (Figure 1C). The increase of miR-146a expression in Jurkat cells, stimulated with anti-CD3/CD28 Abs, was confirmed by Northern blot analysis (supplemental Figure 2).
This experimental evidence, demonstrating that miR-146a is induced upon TCR stimulation, prompted us to further investigate the expression of miR-146a in T cell–mediated immune response.
Identification of TCR signaling pathways affecting miR-146a expression
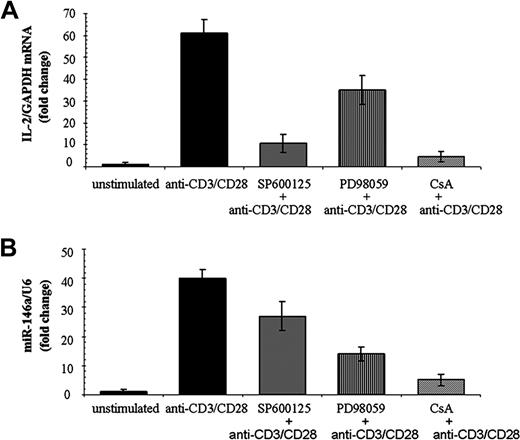
Given that multiple signaling pathways are activated downstream of TCR engagement, we used specific inhibitors to analyze the contribution of different pathways to the expression of miR-146a after TCR stimulation in Jurkat cells. A specific inhibitor of the Janus kinase (JNK) pathway, SP600125,17 caused a drop of IL-2 mRNA level but no significant reduction of miR-146a expression upon TCR stimulation with anti-CD3/CD28 Abs (Figure 2), thus indicating that the JNK pathway is not involved in the transcriptional modulation of miR-146a in Jurkat cells.
TCR signaling pathways affecting miR-146a expression. Jurkat cells were treated for 30 minutes with the JNK inhibitor SP600125, the MEK/ERK inhibitor PD98059, or CsA. Thereafter, cells were stimulated for 24 hours with anti-CD3/CD28 Abs. (A) Relative IL-2 mRNA levels were measured by qRT-PCR using GAPDH levels to normalize expression. Relative IL-2 mRNA levels in stimulated cells are compared with, which are set to 1. Each bar is the mean of 3 independent experiments. (B) MiR-146a levels were measured by qRT-PCR and U6 levels were used to normalize expression. Relative miR-146a expression in stimulated cells is compared with that in nonstimulated cells, which set to 1. Each bar is the mean of 3 independent experiments.
TCR signaling pathways affecting miR-146a expression. Jurkat cells were treated for 30 minutes with the JNK inhibitor SP600125, the MEK/ERK inhibitor PD98059, or CsA. Thereafter, cells were stimulated for 24 hours with anti-CD3/CD28 Abs. (A) Relative IL-2 mRNA levels were measured by qRT-PCR using GAPDH levels to normalize expression. Relative IL-2 mRNA levels in stimulated cells are compared with, which are set to 1. Each bar is the mean of 3 independent experiments. (B) MiR-146a levels were measured by qRT-PCR and U6 levels were used to normalize expression. Relative miR-146a expression in stimulated cells is compared with that in nonstimulated cells, which set to 1. Each bar is the mean of 3 independent experiments.
On the contrary, using PD98059, an inhibitor of mitogen-activated protein kinase (MEK)/extracellular signal-related kinase (ERK) pathway, we observed 50% inhibition of IL-2 expression, as expected,17 and 75% inhibition of miR-146a expression, 24 hours after TCR stimulation (Figure 2). These results indicated that the activation of MEK/ERK pathway could play a role on miR-146a expression, but also suggested that additional signaling pathways, responsive to anti-CD3/CD28 stimulation, could regulate miR-146a transcription.
Finally, Jurkat cells were treated with cyclosporine (CsA), a specific calcineurin inhibitor, able to suppress the production of cytokines during T-cell activation. In particular CsA directly inhibits calcineurin-mediated activation of NFAT family of transcription factors,18 but it also influences the function of other transcriptional regulators, such as AP-1 and NF-kB.19,20
Treatment with CsA caused the expected strong reduction of IL-2 expression and markedly decreased miR-146a expression (Figure 2), thus suggesting that at least one of the CsA-sensitive transcriptional regulators (NFAT, AP-1, and NF-kB) could control miR-146a expression.
Analysis of miR-146a promoter region upon TCR stimulation
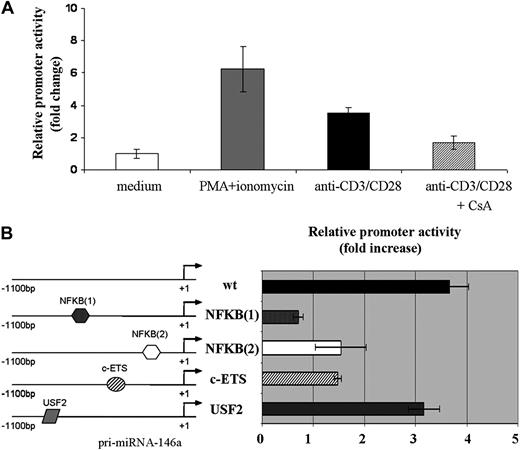
Because 5′ genomic sequences proximal to transcription start sites often act as critical cis-regulatory elements for gene transcription, we performed a comparative bioinformatics analysis covering 3000-bp upstream of the pri-miR146a coding region. By comparing human and mouse genome we identified a region consisting of 1100 bp, upstream of the start site, which contains several putative conserved cis-regulatory elements. In addition to the 2 NF-kB binding sites previously described,8,21 we identified 2 other conserved binding sites, c-ETS and USF-2, potentially relevant for miR-146a expression in T lymphocytes. It should be noted that in this 3000-bp region we could not find putative binding sites for either AP-1 or NFAT.
The 1100-bp region, located upstream of the pri-miR-146a coding region, was cloned in pMluc luciferase reporter and the relative promoter activity was tested after transient transfection in Jurkat cells. As shown in Figure 3A, the 1100-bp region was able to drive the expression of the reporter gene, in response to the same stimuli leading to miR-146a expression. Indeed, stimulation with either PMA and ionomycin or anti-CD3/CD28 Abs induced the relative reporter activity by approximately 8-fold and 4-fold, respectively. Moreover, the treatment of Jurkat cells with CsA caused a sharp decrease of the relative miR-146a promoter activity induced by anti-CD3/CD28 Abs (Figure 3A).
A region of 1100-nt upstream of pri-miR-146a is able to induce expression of a reporter gene upon TCR stimulation. Jurkat cells were cotransfected with pMluc luciferase reporter containing the 1100-bp miR-146a promoter region and with pGL3-tk-luc, used for normalization of transfection. A renilla/firefly Dual-Luciferase assay was performed. Relative promoter activity in stimulated cells is compared with that in nonstimulated cells, which is set to 1. (A) Cells were stimulated for 24 hours with either PMA and ionomycin or anti-CD3/CD28 Abs. Where indicated, Jurkat cells were pretreated for 30 minutes with CsA. Each bar is the mean of 4 independent experiments (P < .05). (B) Jurkat cells were cotransfected with the wild-type or with the mutagenized 1100-bp promoter constructs: NFKB(1); NFKB(2); c-ETS; or USF-2 together with pGL3-tk-luc, used for normalization of transfection. Cells were stimulated with anti-CD3/CD28 Abs for 24 hours. Each bar is the mean of 4 independent experiments (P < .05).
A region of 1100-nt upstream of pri-miR-146a is able to induce expression of a reporter gene upon TCR stimulation. Jurkat cells were cotransfected with pMluc luciferase reporter containing the 1100-bp miR-146a promoter region and with pGL3-tk-luc, used for normalization of transfection. A renilla/firefly Dual-Luciferase assay was performed. Relative promoter activity in stimulated cells is compared with that in nonstimulated cells, which is set to 1. (A) Cells were stimulated for 24 hours with either PMA and ionomycin or anti-CD3/CD28 Abs. Where indicated, Jurkat cells were pretreated for 30 minutes with CsA. Each bar is the mean of 4 independent experiments (P < .05). (B) Jurkat cells were cotransfected with the wild-type or with the mutagenized 1100-bp promoter constructs: NFKB(1); NFKB(2); c-ETS; or USF-2 together with pGL3-tk-luc, used for normalization of transfection. Cells were stimulated with anti-CD3/CD28 Abs for 24 hours. Each bar is the mean of 4 independent experiments (P < .05).
Finally, mutations of both NF-kB sites as well as of the c-ETS site resulted in a significant decrease of miR-146a promoter activity after TCR stimulation with anti-CD3/CD28 Abs (Figure 3B), thus indicating that these binding sites are involved in the transcriptional regulation of miR-146a gene in response to TCR engagement. By contrast, USF-2 site mutation did not significantly affect miR-146a promoter activity. Similar results were obtained after stimulation of Jurkat cells with PMA and ionomycin (data not shown).
MiR-146a ectopic expression impairs IL-2 production and AP-1 activity
To gain insight into the role of miR-146a induction in cellular responses, we generated a miR-146a lentiviral-based expression vector (pRRL-146a) and transduced it in Jurkat cells. To assess the specificity of miR-146a effects we used as control a lentiviral construct (pRRL-ct), encoding for a hairpin yielding a 22-mer RNA, designed to lack homology to any human gene. Northern blot analysis of pRRL-146a Jurkat cells revealed a stable expression of the mature form of miR-146a at levels comparable with those observed after stimulation of Jurkat cells with anti-CD3/CD28 Abs (supplemental Figure 2). On the contrary, in both unstimulated Jurkat cells and in pRRL-ct Jurkat cells, miR-146a expression was undetectable by Northern blot.
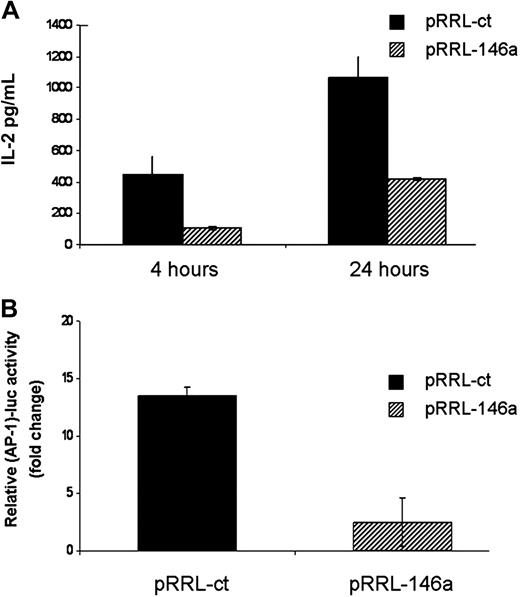
Because miR-146a is induced upon TCR stimulation and TCR engagement triggers the signaling events that lead to IL-2 expression and clonal expansion, we first analyzed IL-2 production in pRRL-ct and pRRL-146a Jurkat cells upon anti-CD3/CD28 Ab stimulation. As shown in Figure 4A, IL-2 level expressed by pRRL-146a Jurkat cells was 3-fold lower compared with that of pRRL-ct Jurkat cells, suggesting that miR-146a plays a role in the timing of different T-cell activation phases by impairing the IL-2 signal.
MiR-146a impairs IL-2 production and AP-1 activity. (A) pRRL-ct and pRRL-146a Jurkat cells were stimulated with anti-CD3/CD28 Abs for 4 and 24 hours and IL-2 levels were determined by ELISA assay. Each bar is the mean of 3 independent experiments. (B) Jurkat cells were cotransfected with the (AP-1)-luc reporter plasmid together with pGL3-tk-luc used for normalization of transfection. Cells were stimulated with anti-CD3/CD28 Abs for 24 hours and the relative promoter activity was measured by renilla/firefly Dual-Luciferase assay. Relative promoter activity in stimulated cells is compared with that in nonstimulated cells, which is set to 1. Each bar is the mean of 3 independent experiments (P < .05).
MiR-146a impairs IL-2 production and AP-1 activity. (A) pRRL-ct and pRRL-146a Jurkat cells were stimulated with anti-CD3/CD28 Abs for 4 and 24 hours and IL-2 levels were determined by ELISA assay. Each bar is the mean of 3 independent experiments. (B) Jurkat cells were cotransfected with the (AP-1)-luc reporter plasmid together with pGL3-tk-luc used for normalization of transfection. Cells were stimulated with anti-CD3/CD28 Abs for 24 hours and the relative promoter activity was measured by renilla/firefly Dual-Luciferase assay. Relative promoter activity in stimulated cells is compared with that in nonstimulated cells, which is set to 1. Each bar is the mean of 3 independent experiments (P < .05).
Given that IL-2 expression is driven by AP-1 transcription factor,22 we asked whether overexpression of miR-146a influenced AP-1 activity. The AP-1 activity was measured in both pRRL-ct and pRRL-146a Jurkat cells using an AP-1 luciferase reporter, (AP-1)-luc. As shown in Figure 4B, the enforced expression of miR-146a in pRRL-146a Jurkat cells dramatically reduces the AP-1 reporter activity observed in pRRL-ct Jurkat cells after 24-hour stimulation with anti-CD3/CD28 Abs. To demonstrate that miR-146a specifically inhibited AP-1 activity, we used as a negative control the same AP-1 responsive reporter lacking in the AP-1 binding sites. As expected, the luciferase activity of this construct did not change upon stimulation in either pRRL-ct or pRRL-146a Jurkat cells.
Overall, these data suggested that miR-146a impairs IL-2 production by affecting AP-1 transcriptional activity.
MiR-146a ectopic expression protects Jurkat cells from AICD
In vivo, upon repeated TCR triggering, activated T cells undergo apoptosis through a mechanism called activation-induced cell death (AICD), which is mediated mainly via the FAS/FAS ligand system. AICD plays a critical role in the termination of the immune response and induction of peripheral tolerance to self-antigens.
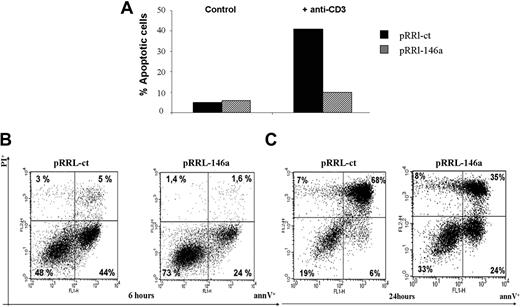
In vitro, the AICD process can be mimicked either by cross-linking the TCR complex with anti-CD3 Ab or treating cells with FAS agonistic Ab. To address whether enforced expression of miR-146a influenced the response to apoptotic signals, the pRRL-146a Jurkat cells were stimulated with anti-CD3 Ab. As shown in Figure 5A, 24 hours after TCR stimulation with anti-CD3 Ab the fraction of apoptotic cells in pRRL-146a Jurkat was 10%, almost the same level (7%) of the unstimulated cells. Instead the level of apoptosis was 40% in pRRL-ct Jurkat cells, thus indicating that miR-146a enforced expression protects Jurkat cells from apoptosis.
MiR-146a overexpression inhibits the AICD of Jurkat T cells. (A) pRRL-ct and pRRL-146a Jurkat cells, plated in RPMI medium with 3% FBS, were left untreated (control) or stimulated with anti-CD3 (OKT3; 10 μg/mL) Ab. After 24 hours the apoptotic cells fraction was measured by annexin V staining. Results shown are 1 representative of 3 independent experiments. (B) pRRL-ct and pRRL-146a Jurkat cells, plated in RPMI medium with 3% FBS, were left untreated (control) or stimulated with Fas-agonistic Ab (CH11 clone; 100 ng/mL) for 6 hours or (C) for 24 hours. The apoptotic cells fraction was measured by annexin V and propidium iodide double staining. Results shown are 1 representative of 3 independent experiments.
MiR-146a overexpression inhibits the AICD of Jurkat T cells. (A) pRRL-ct and pRRL-146a Jurkat cells, plated in RPMI medium with 3% FBS, were left untreated (control) or stimulated with anti-CD3 (OKT3; 10 μg/mL) Ab. After 24 hours the apoptotic cells fraction was measured by annexin V staining. Results shown are 1 representative of 3 independent experiments. (B) pRRL-ct and pRRL-146a Jurkat cells, plated in RPMI medium with 3% FBS, were left untreated (control) or stimulated with Fas-agonistic Ab (CH11 clone; 100 ng/mL) for 6 hours or (C) for 24 hours. The apoptotic cells fraction was measured by annexin V and propidium iodide double staining. Results shown are 1 representative of 3 independent experiments.
Furthermore, we found that in pRRL-146a Jurkat cells, the early apoptosis, induced by treatment with Fas agonist antibody CH11, was markedly reduced; indeed after 6 hours the annexin V–positive cells were only 24%, compared with 44% apoptotic cells observed in pRRL-ct Jurkat cells (Figure 5B). Of note, 24 hours after induction, the rate of late apoptosis was still lower in pRRL-146a Jurkat cells (35%) compared with pRRL-ct Jurkat cells (68%; Figure 5C).
Taken together, these findings pointed to miR-146a as a negative regulator of Fas-mediated apoptosis.
Validation of predicted target genes
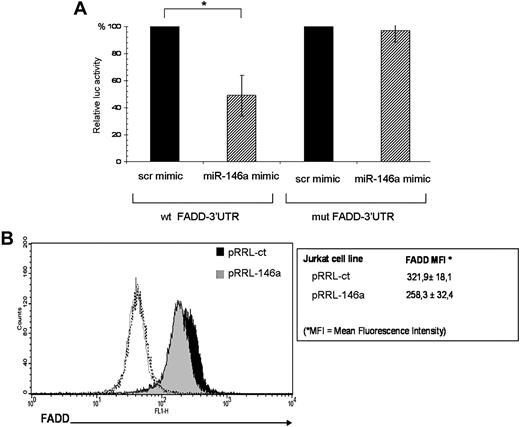
To explore the biologic relevance of miR-146a induction upon TCR stimulation, we used bioinformatics to identify miR-146a potential targets. The miRGen database (http://www.diana.pcbi.upenn.edu/miRGen/v3/miRGen.html),23 which integrates analysis from TargetScan,24 PicTar,25 and MiRanda,26 generated a list of approximately 900 predicted miR-146a targets. Focusing on genes relevant for T-lymphocyte biology, implicated in T-cell activation, apoptosis, and proliferation, we selected approximately 20 putative target genes (supplemental Table 3). To validate them, we first performed luciferase reporter assays on constructs containing the firefly luciferase gene, fused to 500 bp of the 3′UTR of each selected miR-146a putative target. The reporter constructs were transiently transfected in 293T cells together with miR-146a mimic or with a scramble mimic, as negative control. We observed that overexpression of miR-146a significantly decreased luciferase activity of a reporter gene containing the FADD 3′UTR. Notably, the deletion of 6 nucleotides in the 3′UTR seed match sequence restored luciferase levels, thus indicating that the observed down-regulation was specific for the predicted miR-146a target site (Figure 6A).
FADD is a target of miR-146a. (A) The FADD-3′UTR luciferase construct was cotransfected in 293T cells with miR-146a mimic or with a negative control mimic (scr mimic). Results are expressed as mean (±SD, n = 3) of the ratio between renilla luciferase and firefly control luciferase activities (*P < .01). (B) Intracellular staining and FACS analysis for FADD protein. FADD protein levels were measured as mean fluorescence intensity (MFI). Values of MFI are expressed as mean (±SD, n = 7). Black histogram represents pRRL-ct Jurkat cells stained with FADD Ab, gray histogram represents pRRL-146a Jurkat cells stained with FADD Ab, and dashed open line represents pRRL-ct and pRRL-146a Jurkat cells stained with anti–rabbit fluorescein isothiocyanate (FITC) secondary antibody alone.
FADD is a target of miR-146a. (A) The FADD-3′UTR luciferase construct was cotransfected in 293T cells with miR-146a mimic or with a negative control mimic (scr mimic). Results are expressed as mean (±SD, n = 3) of the ratio between renilla luciferase and firefly control luciferase activities (*P < .01). (B) Intracellular staining and FACS analysis for FADD protein. FADD protein levels were measured as mean fluorescence intensity (MFI). Values of MFI are expressed as mean (±SD, n = 7). Black histogram represents pRRL-ct Jurkat cells stained with FADD Ab, gray histogram represents pRRL-146a Jurkat cells stained with FADD Ab, and dashed open line represents pRRL-ct and pRRL-146a Jurkat cells stained with anti–rabbit fluorescein isothiocyanate (FITC) secondary antibody alone.
To directly confirm that FADD is a target of miR-146a, we determined the endogenous levels of FADD protein in pRRL-146a and pRRL-ct Jurkat cells by intracellular staining. Notably, we found that FADD protein levels, expressed as mean fluorescence intensity (MFI), were 20% lower in pRRL-146a Jurkat cells compared with the control pRRL-ct Jurkat cells (Figure 6B). Meanwhile, Fas expression was the same in pRRL-ct and pRRL-146a Jurkat cells (data not shown).
In keeping with the observations that miR-146a levels increase upon TCR stimulation and that FADD is a target of this miRNA, we asked whether FADD protein expression was modulated during in vitro stimulation of naive T lymphocytes.
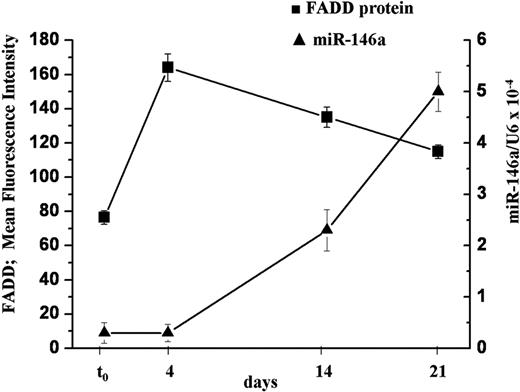
To this aim, freshly isolated human naive T lymphocytes were stimulated with PMA and ionomycin. The levels of FADD protein and miR-146a were measured at different times, together with the markers of differentiation CD45RA and CD45RO. We found that 4 days after TCR stimulation, FADD protein levels were 2-fold higher than in naive T cells (t0), whereas miR-146a levels were still low. Fourteen days after stimulation, concomitant to the augmentation of miR-146a level (5-fold), FADD protein expression decreased 20%. Furthermore, after 21 days, along with higher levels of miR-146a, we observed further lower FADD levels (Figure 7), comparable with that measured in freshly isolated memory T lymphocytes (data not shown). In parallel to the changes in FADD and miR-146a expression we observed a decrease of CD45RA+ cells and an increase of CD45RO+ cells, indicating a transition from naive to memory phenotype as already shown in supplemental Table 2.
FADD protein level negatively correlates with miR-146a expression in human primary T cells. Human primary naive CD4+ T lymphocytes were stimulated with PMA and ionomycin and samples were analyzed to quantify FADD protein and miR-146a levels at different times. FADD levels were measured by intracellular staining and expressed as mean fluorescence intensity (MFI). MiR-146a expression was measured by qRT-PCR and U6 levels were used for normalization. Each bar is the mean of 3 independent experiments.
FADD protein level negatively correlates with miR-146a expression in human primary T cells. Human primary naive CD4+ T lymphocytes were stimulated with PMA and ionomycin and samples were analyzed to quantify FADD protein and miR-146a levels at different times. FADD levels were measured by intracellular staining and expressed as mean fluorescence intensity (MFI). MiR-146a expression was measured by qRT-PCR and U6 levels were used for normalization. Each bar is the mean of 3 independent experiments.
These findings highlight a negative correlation (r = −0.97) between endogenous miR-146a levels and endogenous FADD protein in stimulated primary CD4+ T lymphocytes, thus supporting our previous results that indicate FADD as a target of miR-146a.
Discussion
Activation of the T cell–mediated immune response has been associated with changes in the expression of specific miRNAs. In mice, miR-181a has been demonstrated to be an intrinsic modulator of T-cell sensitivity to the antigen27 and miR-155 has been indicated as a positive regulator of cytokine production and T-cell lineage choice by an uncharacterized mechanism.28,29 However, the capability of TCR stimulation to modulate miRNA expression and, more importantly, the role of miRNAs in the development of an effective immune response are just beginning to be explored. In contrast to the emerging role of miR-146a in the innate immunity, a role of miR-146a in the adaptive immune response remains to be investigated.
The major contribution of our work is to demonstrate that in T lymphocytes miR-146a expression is induced upon TCR engagement and that miR-146a enforced expression impairs IL-2 production and protects T cells from AICD, possibly contributing to the modulation of the adaptive immune response.
Our finding that miR-146a levels are higher in human memory CD4+ and CD8+ T lymphocytes compared with naive T lymphocytes led us to speculate that miR-146a could be induced upon TCR stimulation. By stimulating primary T lymphocytes with PMA and ionomycin we observed that miR-146a levels gradually increase after 8 days, parallel to the increase of CD45RO, a marker of transition to a memory phenotype. These results suggest that miR-146a may contribute to the cell changes triggered by TCR engagement. Because TCR stimulation in Jurkat cells induced a similar increase of miR-146a levels, we chose this well-established cellular model to further investigate the functional role of miR-146a in T-cell response.
One of the first detectable consequences of TCR triggering is the nuclear localization of AP-1, NFAT, and NF-kB transcription factors, all known to bind to the IL-2 promoter region and to induce IL-2 expression.22,30-32 Furthermore, MEK/ERK pathway has a well-established role in propagating TCR signals, leading to the activation of c-ETS transcription factors.33 Interestingly, CsA, an inhibitor of AP-1, NFAT, and NF-κB, and the MEK/ERK inhibitor PD98059 markedly decreased miR-146a expression upon TCR stimulation, thus suggesting that transcription factors sensitive to these inhibitors modulate miR-146a transcription.
Notably, a bioinformatics analysis excluded the presence of any AP-1 or NFAT conserved binding sites 3000-bp upstream of the pre-miR-146a coding region and confirmed the presence of the 2 putative NF-κB binding sites, already described by Taganov et al.8 Therefore, the impairment of miR-146a promoter activity, observed either in the presence of CsA or after mutagenesis of NF-κB binding sites, points to a role of NF-κB in the modulation of miR-146a expression not only in monocytes, in response to proinflammatory stimuli,8 but also in T lymphocytes, upon TCR stimulation.
Furthermore, the down-modulation of miR-146a expression after treatment of Jurkat cells with the MEK/ERK inhibitor, together with the reduced miR-146a promoter activity, observed when the c-ETS binding site was mutated, suggests the involvement of c-ETS family of transcription factors in the regulation of miR-146a expression. Interestingly, members of c-ETS family have been demonstrated to play a key role in T-cell development and maturation in fetal thymus,34 as well as in T-cell activation in adults.35
From a functional point of view, the induction of miR-146a expression appears to be relevant for the modulation of T-lymphocyte differentiation phases, after TCR activation. Indeed, we demonstrated that miR-146a overexpression in Jurkat cells impairs IL-2 production, which is consistent with the reduced activity of AP-1 transcription factor. These findings suggest that miR-146a plays a role in the timing of different T-cell activation steps, by contributing to switch off the IL-2 signal. Moreover, we found that miR-146a enforced expression renders Jurkat cells resistant to AICD induced by anti-CD3 Ab, as a consequence of a decrease in their sensitivity to Fas-induced apoptosis.
Examination of the miRGen database and the luciferase reporter assays indicated 2 attractive putative miR-146a targets: FADD, the adaptor protein that, once recruited by FAS, binds to procaspase 8 resulting in the activation of the apoptotic protease cascade, and c-FOS, a component of AP-1 transcription factor. Notably, the decreased levels of FADD protein observed in pRRL-146a Jurkat cells confirmed the results obtained by luciferase assays in 293T cells, indicating that FADD is a target of miR-146a. Consistent with these observations, we also demonstrated a negative correlation between FADD protein levels and miR-146a expression in stimulated primary T lymphocytes, suggesting that miR-146a may down-modulate FADD expression also in human primary T cells.
Taken together these findings highlight FADD as a target of miR-146a. This is consistent with the antiapoptotic role of miR-146a observed in pRRL-146a Jurkat cells, even though it is plausible that the protection from apoptosis mediated by miR-146a may be consequent to the concurrent down-regulation of more than one protein involved in the apoptotic pathway.
Conversely, despite the results obtained by c-FOS 3′UTR reporter in 293T cells (supplemental Figure 3), we did not succeed in demonstrating a decrease of c-FOS protein levels in pRRL-146a Jurkat cells. The reduction observed in some experimental replicates was not always reproduced. Thus, to date, we are not able to indicate c-FOS as a target of miR-146a and to define to what degree the down-modulation of c-FOS protein directly contributes to the observed phenotypes: AP-1 activity impairment and IL-2 reduction. Further studies are needed and additional miR-146a targets may be taken into account to fully explain miR-146a mechanism of action.
To the best of our knowledge, all work published thus far aimed at demonstrating a role for miR-146a in modulating inflammation in cellular contexts related to innate immunity.8-11 Here we show that other signaling pathways, apart from inflammation, are influenced by miR-146a modulation and function. Indeed, we have provided experimental evidence that supports a role for miR-146a as a modulator of IL-2 expression and as an antiapoptotic factor, protecting T lymphocytes from Fas-mediated apoptosis.
The up-regulation of miR-146a in memory T lymphocytes and the reduced apoptosis observed in the presence of high levels of miR-146a are consistent with a possible role of this miRNA in the accumulation and/or maintenance of memory T-cell “pool.” Memory T lymphocytes are long-lasting cells, less susceptible to apoptosis, thus miR-146a could be one of the antiapoptotic factors up-regulated in memory T cells.
Under normal conditions homeostasis and immune response are tightly regulated, being very sensitive to defects at critical checkpoints of cell death pathways and reduced cell death is often associated with autoimmunity and cancer.36-39 Our finding that miR-146a enforced expression protects T lymphocytes from Fas-mediated apoptosis is consistent with the observation that miR-146a is up-regulated in rheumatoid arthritis synovial tissue9,10,40 and that synovial inflammation in rheumatoid arthritis is often characterized by a unique resistance to Fas-related apoptosis.41
Understanding miR-146a functions in the complex molecular networks that orchestrate T-cell immune response will contribute not only to a better comprehension of immune homeostasis, but also to identification of novel therapeutic targets for the treatment of autoimmune disorders and cancer.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We offer special thanks to G. Ruberti for technical help, as well as for discussions and sharing ideas. A special thanks also to Teresa Colombo for help with the bioinformatics analysis. pRRLSIN.cPPT.PGK-GFP.WPRE was kindly provided by Didier Trono through Addgene.
This work was supported by the SIROCCO project of the European Union (G.M.), the Italian Association for Cancer Research (AIRC; G.M. and V.B.), and Istituto Pasteur-Fondazione Cenci Bolognetti (V.B.).
Authorship
Contribution: G.C. designed research, generated and analyzed miR-146a promoter reporter constructs, performed the experiments with pharmacologic inhibitors, designed and generated pRRL-146a Jurkat cell line, designed miR-146a target identification strategy, generated miR-146a target reporter constructs and performed luciferase assays, designed and performed apoptotic assays and intracellular staining, analyzed data, and wrote the paper; F.C. analyzed and interpreted data and drafted the paper; C.C. designed and generated pRRL-ct and pRRL-146a Jurkat cells; M.G. measured by qRT-PCR miR-146a in human primary T lymphocytes and tested the pharmacologic inhibitors; N.C. performed ELISA assays; V.F. performed bioinformatics analysis; D.F. and F.M. purified primary T-lymphocyte populations; and V.B. and G.M. drafted the paper and supervised research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Franca Citarella, Dipartimento di Biotecnologie Cellulari ed Ematologia, Sezione di Genetica Molecolare, Sapienza Università di Roma, Viale Regina Elena 324, 00161 Rome, Italy; e-mail: citarella@bce.uniroma1.it.