Abstract
The transcription factor PU.1 is essential for myeloid development. Targeted disruption of an upstream regulatory element (URE) decreases PU.1 expression by 80% and leads to acute myeloid leukemia (AML) in mice. Here, we sequenced the URE sequences of PU.1 in 120 AML patients. Four polymorphisms (single nucleotide polymorphisms [SNPs]) in the URE were observed, with homozygosity in all SNPs in 37 patients. Among them, we compared samples at diagnosis and remission, and one patient with cytogenetically normal acute myeloid leukemia M2 was identified with heterozygosity in 3 of the SNPs in the URE at remission. Loss of heterozygosity was further found in this patient at 2 polymorphic sites in the 5′ promoter region and in 2 intronic sites flanking exon 4, thus suggesting loss of heterozygosity covering at least 40 kb of the PU.1 locus. Consistently, PU.1 expression in this patient was markedly reduced. Our study suggests that heterozygous deletion of the PU.1 locus can be associated with human AML.
Introduction
The lineage-specific transcription factor PU.1 is essential for myeloid development, and its disruption leads to a block of myeloid and B-cell development.1-4 Consequently, PU.1 expression is tightly regulated.5-9 Several studies have indicated that PU.1 function is suppressed in subsets of acute myeloid leukemia (AML) patients, such as in the presence of the leukemic fusion proteins PML-RARA or AML1-ETO.10,11 In addition, genomic mutations of PU.1 in AML patients have been reported.11-16
A highly conserved distal upstream regulatory element (URE) is mediating transcriptional control of Pu.1 expression. Targeted disruption of this element suppresses Pu.1 expression to 80% and induces AML in mice.17-20 Here, we hypothesized that mutations within the URE could cause dysregulation of PU.1 and thus contribute to leukemogenesis in human AML. We therefore analyzed the URE in AML patients by direct sequencing. We identified one AML patient with heterozygous loss of the entire URE sequences. Our data suggest that heterozygous deletion of the PU.1 locus can be associated with human AML.
Methods
Patients
DNA was extracted from blasts of 120 AML patients at diagnosis. Characteristics of the patients are depicted in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). In addition, DNA was studied from peripheral blood of 141 healthy volunteers, with informed consent of patients and volunteers in accordance with the Declaration of Helsinki. The studies were approved by the cantonal ethics committee of Bern, Switzerland.
Sequencing
The primers for distal regulatory unit (DRU) were (607 bp): 5′-AGAGGAAACTGAGGCCAAGTG-3′ and 5′-TGGCAGTCCTCACTGAGGCCATTG-3; and for PRU (634 bp) 5′-CAATGGCCTCAGTGAGGACTGCCA-3′ and 5′-TCTTGGCGGAAGCTGTTAGGGAAG-3′. Sequencing reactions were performed on an Applied Biosystems 3730 Analyzer (Applied Biosystems).
Quantitative real-time PCR for PU.1 expression
The primers were as follows: 5′-TGTTACAGGCGTGCAAAATGGAAGG-3′, 5′-CTCGTGCGTTTGGCGTTGGTATAGA-3′; and 5′-FAM-CCTCGTCCCCCCTCCATCAGAAGACCTGG-TAMRA-3′. Expression levels were obtained using standard curves, and normalized to ABL expression.
FISH
For locus-specific fluorescence in situ hybridization (FISH), 2 probes were used: a polymerase chain reaction (PCR)-generated 1.4-kb fragment covering the entire URE sequences (red signal) and a probe directed against the centromer of chromosome 11 (green signal). Labeling, probe preparation, and hybridization of slides were performed as previously described.21,22
Results and discussion
We analyzed the DRU and proximal regulatory unit (PRU) of the URE of the hematopoietic master transcription factor PU.1 for genomic mutations by direct sequencing (Figure 1). We screened DNA from malignant cells of 120 AML patients and peripheral blood of 141 healthy volunteers, and no mutations were detected. This suggests that genomic mutation in the URE of PU.1 is a rare event in human AML.
Location of the SNPs in the URE and the coding region of PU.1. (A) PU.1 is located in the centromeric part of chromosome 11p (band 11p11.2). (B) Organization of the PU.1 locus: The PU.1 gene consists of 5 exons (E1-E5), spanning 25 kb. The URE is located 17 kb upstream of the transcription start site. One SNP was identified within the DRU (DRU1), 3 SNPs are located within (PRU3) or close to the PRU (PRU1 and PRU2), 4 SNPs (PROM1 to 4) exist within the 625 bp of the PU.1 promoter just upstream of the translation start site, and 2 SNPs in intron 3 (SNPi3) and intron 4 (SNPi4) are just flanking exon 4. The ruler depicts distance from the PU.1 transcription start site. The location of each SNP is encoded with the last 4 digits of their position in the reference sequence with gene accession number cNT009237.17; eg, DHR1 is located at position 46194424. Previously described SNPs are marked with their accession numbers. (C) Allelic distribution is indicated for each SNP, with the observed incidence of the wild-type sequence (bottom part of the box) and the polymorphic sequence (top part of the box) indicated for each SNP.
Location of the SNPs in the URE and the coding region of PU.1. (A) PU.1 is located in the centromeric part of chromosome 11p (band 11p11.2). (B) Organization of the PU.1 locus: The PU.1 gene consists of 5 exons (E1-E5), spanning 25 kb. The URE is located 17 kb upstream of the transcription start site. One SNP was identified within the DRU (DRU1), 3 SNPs are located within (PRU3) or close to the PRU (PRU1 and PRU2), 4 SNPs (PROM1 to 4) exist within the 625 bp of the PU.1 promoter just upstream of the translation start site, and 2 SNPs in intron 3 (SNPi3) and intron 4 (SNPi4) are just flanking exon 4. The ruler depicts distance from the PU.1 transcription start site. The location of each SNP is encoded with the last 4 digits of their position in the reference sequence with gene accession number cNT009237.17; eg, DHR1 is located at position 46194424. Previously described SNPs are marked with their accession numbers. (C) Allelic distribution is indicated for each SNP, with the observed incidence of the wild-type sequence (bottom part of the box) and the polymorphic sequence (top part of the box) indicated for each SNP.
We observed one single nucleotide polymorphism (SNP) in the DRU (DRU1) and 3 SNPs in the PRU (PRU1-3) of the URE sequences as illustrated in Figure 1. DRU1 and PRU3 correspond to the 2 SNPs described by Steidl et al.21 We could not detect a higher incidence of the SNP DRU1 in AML patients with complex abnormalities (n = 24) as previously reported,20 possibly because of the small size of this group.
In mice, targeted disruption of the entire URE sequences suppresses Pu.1 mRNA by 80% and induces AML.19 We therefore hypothesized that loss of heterozygosity (LOH) of the URE might be observed in AML patients. Accordingly, we focused on AML patients with noninformative SNPs in all 4 locations of the URE. We identified homozygosity for all SNPs in 37 of 120 patients (30.8%) and in 47 of 141 healthy volunteers (33.3%).
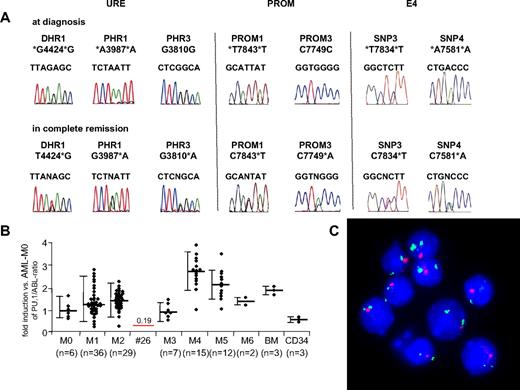
Next, we analyzed samples obtained at diagnosis and at remission in the patients with noninformative SNPs in the URE. Appropriate material at remission was not available in 14 of the 37 patients because of early death (8 patients), treatment failure and thus unavailability of remission material (5 patients), or loss during follow-up (1 patient). In 22 patients, identical results were observed at diagnosis and remission. However, 1 patient showed a heterozygous pattern in the SNPs DRU1, PRU1, and PRU3 at remission. This result was verified in a second remission sample obtained at a later time point (Figure 2). These data indicate LOH of the URE sequences in leukemic cells at diagnosis.
Heterozygous loss of the PU.1 locus in human AML. (A) Comparison of chromatograms obtained at diagnosis and in first remission in an AML-M2 patient (patient 26). Three informative SNPs within the URE, 2 SNPs within the promoter region (PROM), and 2 SNPs flanking exon 4 (1 each in intron 3, SNPi3; and in intron 4, SNPi4) showed heterozygosity at remission but LOH at diagnosis. The other SNPs (PRU2, PROM2, and PROM4) were not informative. (B) PU.1 mRNA expression of 107 (of 120) AML patients. Material was not available from 13 patients. The mean value for each AML subtype is presented as PU.1/ABL-ratio, with the value of AML-M0 patients being arbitrarily selected as 100%. Bars represent SD. The single line represents PU.1 expression of patient 26 with LOH in the PU.1 locus. BM indicates unsorted bone marrow of healthy persons; CD34, CD34+ selected cells of AML patients in remission. (C) FISH analysis of bone marrow at diagnosis of patient 26 with LOH in the PU.1 locus. The single red signal obtained from a probe directed against the URE sequences represents LOH of the URE, and the 2 green signals obtained from a probe against the centromeric region of chromosome 11 represent that indeed 2 chromosome 11 alleles are present.
Heterozygous loss of the PU.1 locus in human AML. (A) Comparison of chromatograms obtained at diagnosis and in first remission in an AML-M2 patient (patient 26). Three informative SNPs within the URE, 2 SNPs within the promoter region (PROM), and 2 SNPs flanking exon 4 (1 each in intron 3, SNPi3; and in intron 4, SNPi4) showed heterozygosity at remission but LOH at diagnosis. The other SNPs (PRU2, PROM2, and PROM4) were not informative. (B) PU.1 mRNA expression of 107 (of 120) AML patients. Material was not available from 13 patients. The mean value for each AML subtype is presented as PU.1/ABL-ratio, with the value of AML-M0 patients being arbitrarily selected as 100%. Bars represent SD. The single line represents PU.1 expression of patient 26 with LOH in the PU.1 locus. BM indicates unsorted bone marrow of healthy persons; CD34, CD34+ selected cells of AML patients in remission. (C) FISH analysis of bone marrow at diagnosis of patient 26 with LOH in the PU.1 locus. The single red signal obtained from a probe directed against the URE sequences represents LOH of the URE, and the 2 green signals obtained from a probe against the centromeric region of chromosome 11 represent that indeed 2 chromosome 11 alleles are present.
The cytogenetic analysis of 20 metaphases in this patient indicated a normal karyotype at diagnosis. No mutations were detected in the genes encoding CEBPA, FLT3, and NPM1. The patient was diagnosed at the age of 32 years with AML-M2, the blasts were positive for CD11b, CD13, CD33, CD34, and HLA-DR, and leukocytes at diagnosis were 2.3 g/L, with 3% blasts. Hemoglobin was 92 g/L, platelets 72 g/L, and lactate dehydrogenase 398 U/mL. The bone marrow indicated 80% blast infiltration. The patient was treated with 3 cycles of chemotherapy and enjoys ongoing complete remission.
Because the karyotype was normal, LOH detected in SNP of the URE suggests a submicroscopic deletion in band 11p11.2. However, the size of the deletion remained to be determined. We therefore investigated whether the PU.1 coding region located 17 kb downstream of the URE might also be affected. We sequenced 625 basepairs of the 5′ PU.1 promoter region (PROM) starting immediately upstream of the translation start site in all 120 AML patients and in the healthy volunteers. Again, no mutations were detected. Remarkably, the patient with LOH in the URE sequences also showed homozygosity in all 4 promoter SNPs at diagnosis, whereas heterozygosity was found for PROM1 and PROM3 at remission. We also tested 2 additional SNPs located in intron 3 (SNPi3) and intron 4 (SNPi4) both just flanking exon 4. Both SNPs were homozygous at diagnosis but heterozygous at remission. This indicates that the deletion spans at least from the URE to exon 4, thus covering 40 kb.
Additional SNP analysis within the telomeric region of chromosome 11p and the centromeric region of chromosome 11 indicated conserved heterozygosity of both regions at diagnosis (supplemental Figure 1). Furthermore, SNPs were analyzed located 20, 50, and 100 kb upstream and downstream of the PU.1 translation start site. Interestingly, heterozygosity was conserved at diagnosis in an SNP 20 kb upstream, narrowing down the upstream breakpoint of the deletion between 20 kb and 17 kb (location of the URE), whereas the downstream distant SNPs flanking the LOH of the PU.1 locus were not informative.
We ultimately confirmed the deletion by FISH analysis using a probe that corresponded to the URE region (Figure 2). The probe showed loss of one signal in leukemic cells at diagnosis, whereas 2 signals were detectable using a probe against the centromeric region of chromosome 11.
Finally, we assessed PU.1 mRNA expression by real-time PCR available in 107 AML patients (Figure 2). We observed that the AML-M2 patient with the heterozygous PU.1 deletion expressed significantly lower PU.1 mRNA compared with our collection of AML-M2 patients, and the expression was 19% of the mean expression observed in AML-M0 patients, which served as a reference (100%).
Our data indicate that heterozygous deletion of the PU.1 locus can be associated with AML; however, this appears to be a rare event. Submicroscopic deletions spanning several kilobases as identified in our patient will necessarily escape the detection by conventional sequencing strategies. Our findings may therefore contribute to further clarify divergent reports on the incidence of PU.1 mutations in AML patients.11-15,23
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Andreas Steege for help with the SNP analysis, Ludwig Wilkens for technical assistance with the FISH analyses, and Marianne Eyholzer for help with sample preparation.
This work was supported by the Swiss National Science Foundation (SF 310000-113761; B.U.M.) and the Swiss Cancer League (OCS 01731082005; B.U.M.).
Authorship
Contribution: N.B. performed research; T.P. analyzed data and wrote the paper; and B.U.M. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Beatrice U. Mueller, Department of Internal Medicine, University Hospital, 3010 Bern, Switzerland; e-mail: beatrice.mueller@insel.ch.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal