Abstract
The B6.SJL-Ptprc(d)Pep3(b)/BoyJ (B6.SJL) congenic mouse strain, a valuable and widely used tool in murine bone marrow transplantation studies, has long been considered equivalent to the parental C57B/L6 (B6) strain with the exception of a small congenic interval on chromosome 1 harboring an alternative CD45/Ly-5 alloantigen (Ly-5.1). In this study we compared functional properties of stem and stromal cells between the strains, and delineated the boundary of the B6.SJL congenic interval. We identified a 25% reduction in homing efficiency, 3.8-fold reduction in transplantable long-term hematopoietic stem cells (LT-HSCs), a 5-fold reduction in LT-HSCs capable of 24-hour homing, and a cell-intrinsic engraftment defect of 30% to 50% in B6.SJL-derived bone marrow cells relative to B6-derived cells. These functional differences were independent of stem cell number, cycling, or apoptosis. Genotypic analysis revealed a 42.1-mbp congenic interval in B6.SJL including 306 genes, and at least 124 genetic polymorphisms. Moreover, expression profiling revealed 288 genes differentially expressed between nonhematopoietic stromal cells of the 2 strains. These results indicate that polymorphisms between the B6 and SJL genotype within the B6.SJL congenic interval influence HSC engraftment and result in transcriptional variation within bone marrow stroma.
Introduction
Efforts to identify unique surface antigens on functional subsets of T lymphocytes led to the discovery of several classes of lymphocyte (Ly) antigen groups.1-3 The discovery of the Ly-5, later termed CD45, antigens in 1975 also revealed that this antigen exists as 2 allotypes, Ly-5.1 and Ly-5.2, in different mouse strains.4 Soon after the identification of Ly-5 antigens on T lymphocytes, Ly-5 was found to be a panhematopoietic marker on the surface of primitive hematopoietic cells and differentiated cells with the exception of the erythroid lineage.5-7 By the 1980s investigators at the Sloan-Kettering institute had used the Ly-5.1–bearing SJL strain from The Jackson Laboratory as a donor strain and the C57BL/6 (B6) strain as the recipient to generate congenic mice carrying the Ly-5.1 alloantigen on a B6 background.8 Two studies in the late 1980s using B6 and Ly-5 congenic mice set the stage for extensive use of this strain combination in hematopoietic stem cell (HSC) and bone marrow transplantation research. In the first study, the Ly-5 cell surface markers were used in the characterization of a purified HSC population capable of reconstituting lethally irradiated hosts.9 This technique was given further credence when a later paper described no alloreactivity for mismatched Ly-5 alloantigens in bone marrow transplantation or skin grafts.10
Despite successful use in HSC transplantation, there have been several subsequent demonstrations of mild alloreactivity in this system. In 1998, Chen et al demonstrated that B6 and B6.SJL congenic mice could be immunized against a small peptide corresponding to part of the polymorphic region of Ly-5 from the opposite strain.11 Ly-5 immunogenicity was also cited as a mechanism for decreased engraftment of Ly-5.1–bearing cells into Ly-5.2–bearing recipients relative to Gpi-congenic transplants.12 The authors of this study advised the use of high-dose (> 6 Gy) total body irradiation in Ly-5–mismatched transplants to ensure efficient engraftment, because this was sufficient to equalize engraftment efficiency between the 2 models. Another study specifically identified αβ T-cell receptor–positive, CD4+, and CD8+ T cells as mediators of an anti–Ly-5.1 immune response.13 Interestingly, a more recent publication investigating mobilized peripheral blood stem cells demonstrated no immunogenicity toward a mismatched Ly-5 alloantigen when Ly-5.2 cells were transplanted into Ly-5.1 hosts, the converse of the transplant strategy used in the earlier studies citing immunogenicity.14
To identify potential engraftment differences, we compared stem cells of both strains in recipients of both strains. We also compared stem cell number, cell cycling status, apoptosis, stromal cell gene expression, and genetic polymorphisms between B6 and B6.SJL. Our results demonstrate a 25% reduction in 24-hour homing efficiency of B6.SJL-derived cells transplanted in B6 recipients, an approximate 4- to 5-fold reduction in the absolute numbers of transplantable stem cells, and a unidirectional engraftment deficit of 30% to 50%. Genome sequence variations and stromal transcriptome variation may underlie these functional variations.
Methods
Animals
Young (6- to 12-week-old) female C57B/L6J (B6) and B6.SJL mice purchased from The Jackson Laboratory were used in all experiments. Mice were maintained in the Department of Laboratory Animal Resources at the University of Kentucky Chandler Medical center under pathogen-free conditions and given food and acidified water ad libitum. Transplant recipients received sulfamethoxazole and trimethoprim (Actavis) diluted in sterile water to a final concentration of 143 μg/mL sulfamethoxazole and 29 μg/mL trimethoprim as a prophylaxis to opportunistic bacterial infection. Quarterly analyses of sentinel animals have revealed no pathogens present in our mouse colony. Approval was obtained from the University of Kentucky Institutional Animal Care and Use Committee for these studies.
Conditioning and bone marrow transplant
A dose (9-Gy) of total body gamma radiation was administered in a single dose at 0.0289 gray per second from a 137Cs source in a J. L. Shepherd Mark I Irradiator (J. L. Shepherd and Associates). Anesthetized recipients received a transplant of specified numbers of whole bone marrow cells via the retro-orbital venous plexus.
Competitive repopulation
In each of 2 independent experiments, 4 B6 and 4 B6.SJL mice (8 total mice per strain) received a transplant of 4 × 106 freshly isolated bone marrow cells. Transplanted cells were harvested and admixed from B6 and B6.SJL donors (2 × 106 cells from each strain). Recipients were evaluated for Ly-5 chimerism in peripheral blood from 5 to 30 weeks after transplant, and in bone marrow and spleen at 30 weeks after transplant.
Determination of chimerism in hematopoietic tissues
Peripheral blood obtained from the retro-orbital plexus of isoflurane-anesthetized recipients was depleted of red cells by hypotonic lysis and stained for Ly-5.1/Ly-5.2 chimerism in total peripheral blood and within the macrophage/granulocyte, B-cell, and T-cell populations. Antigens were identified by the following antibodies: phycoerythrin (PE)– or fluorescein isothiocyanate–conjugated CD45.1(A20) and CD45.2(104), and PE-conjugated Ly-6G/Ly6C(Gr-1/)(RB6-8C5), CD11b(integrinαM chain, Mac-1 alpha chain)(M1/70), CD45R/B220(RA3-6B2), and CD90.2(Thy1.2)(30-H-12). All antibodies were purchased from Becton Dickinson (BD) Pharmingen. Stained mononucleated blood cells were incubated in media containing propidium iodide (PI)15 to determine cell viability by PI exclusion and analyzed on a FACScan flow cytometry instrument (BD). Thirty weeks after transplant, bone marrow and spleen were harvested from recipients and stained as described.
Quantification of hematopoietic stem cells
The competitive repopulating unit (CRU) assay was used to quantify stem cell numbers in B6 and B6.SJL mice in vivo. Graded numbers of B6 or B6.SJL bone marrow cells, ranging from 2000 to 60 000, were combined with 200 000 freshly harvested competitor cells of the opposite strain and transplanted intravenously into B6 or B6.SJL hosts. Peripheral blood was screened at 5, 10 to 12, and 15 to 17 weeks after transplant for the presence of donor-derived cells. Animals were considered positive for CRU if greater than 5% of peripheral blood leukocytes were of donor origin. The absolute frequency CRU was calculated by limiting dilution analysis using the L-Calc software package from StemCell Technologies. We performed simultaneous analysis of B6 CRUs in B6.SJL recipients and B6.SJL CRUs in B6 recipients. Statistical comparisons between the 2 transplantation schemes were performed by comparing the 2 sets of limiting dilution data using the L-Calc software package. Four independent CRU assays were conducted for B6.SJL cells transplanted into B6 recipients and 3 independent CRU assays were performed with B6 cells transplanted into B6.SJL recipients.
The cobblestone area forming cell (CAFC) assay was used to quantify stem and progenitor cell number in B6 and B6.SJL mice in vitro as we have described previously.16-18 The frequency of HSCs was determined by cobblestone areas counted on days 28 and 35 in 3 replicate experiments using marrow pooled from 2 to 3 mice per strain, and analyzed by limiting dilution analysis using the L-Calc software package.
Homing efficiency
Homing efficiency of B6-derived and B6.SJL-derived CRUs was measured using a modification of previously described competitive repopulation and homing assays.19 Briefly, lethally irradiated B6 or B6.SJL mice received a transplant of 2 × 106 B6.SJL or B6 donor cells, respectively, from the same inoculum used in the CRU assay described and were killed 24 hours after transplant to obtain a single-cell suspension of bone marrow cells from pooled femora and tibiae. Based on the assumption that 2 femora and 2 tibiae represent 25% of the mouse marrow,20 cells were resuspended at a concentration of 1 primary bone marrow equivalent (4 recipient mice) per milliliter. To measure the number of CRUs in this suspension of “homed” cells, graded numbers of cells were transplanted into 6 groups of secondary, lethally irradiated B6 or B6.SJL recipients that received 0.5% to 20% homed bone marrow (BM) equivalents per mouse along with 200 000 freshly harvested B6 or B6.SJL competitor cells. Peripheral blood was screened as described. The frequency of homed CRUs per primary bone marrow fraction was calculated by limiting dilution analysis using L-Calc software package, and homing efficiency was defined as the proportion of homed relative to transplanted CRUs. Four independent homing assays were conducted for B6.SJL cells transplanted into B6 recipients and 3 independent homing assays were performed with B6 cells transplanted into B6.SJL recipients. Limiting dilution data from the 2 transplantation schemes were compared using the L-Calc software package.
Cell cycling and apoptosis
Cell cycle and apoptosis analysis was performed using a BrdU Flow Kit (BD Biosciences) according to the manufacturer's instructions. Briefly, intraperitoneal injections of 100 μL of a 10-mg/mL solution of 5-bromo-2-deoxyuridine (BrdU) were administered to 3 mice of each strain in 2 independent experiments. One hour after BrdU injection, mice were killed and whole bone marrow was harvested. Red blood cells were depleted by hypotonic lysis and the remaining bone marrow cells were labeled with monoclonal antibodies specific to either differentiated or primitive HSCs. Lineage-specific cells were labeled by a cocktail of biotinylated antibodies directed against the following antigens: CD5(Ly-1)(53-7.3) (T cells), CD45R/B220(RA3-6B2) (B cells), CD11b-(integrinαMchain, Mac-1 αchain)(M1/70) (macrophages), CD8a(Ly-2)(53-6.7) (T cells), Ly-6G and Ly-6C(RB6-8C5) (granulocytes), and TER-119(Ly-76) (erythrocytes), and labeled with streptavidin–allophycocyanin–cyanin 7. Hematopoietic stem and progenitor cells were identified with antibodies to the Kit receptor (allophycocyanin anti–mouse CD117/c-Kit [2B8]) and stem cell antigen-1 (PE anti–mouse Ly6A/E/Sca-1 [E13-161.7]). Subsequent colabeling with BrdU and 7-aminoactinomycin D enabled the determination of cell cycle stage and DNA content in whole bone marrow, lineage-negative, and stem cell (lineage-negative, Sca-1+, and c-Kit+ or “LSK”) populations. All antibodies used in this assay were purchased from BD.
Data analysis
CRU frequency, frequency of homed CRUs, and CRU-frequency comparisons between strains were performed using the L-Calc limiting dilution software analysis program. Homing efficiency was measured by dividing the number of CRUs recovered 24 hours after transplant by the absolute number of CRUs contained in the original 2 × 106 cells transplanted into primary recipients.
To assess differences in Ly-5 chimerism in the peripheral blood of B6 and B6.SJL recipients at each time point, we used a paired t test with a Bonferroni correction for multiple testing. To assess changes in the contribution of Ly-5.1 and Ly-5.2 cells over time in each group of recipients, we performed mixed model analyses. To assess changes in lymphoid and myeloid cells within the Ly-5.1 population over time, we applied mixed model analysis to the percentages of Mac-1/Gr-1, B220, and Thy-1.2 cells that were also Ly-5.1 positive. To compare the Mac-1/Gr-1, B220, and Thy1.2 lineages between B6 and B6.SJL recipients, we used an unpaired t test with a Tukey-Kramer adjustment for multiple testing.
Statistical comparisons of hematopoietic chimerism at 30 weeks after transplant, cell cycling, and apoptosis were performed using the 2-tailed, unpaired Student t test assuming unequal variance, with a significance threshold of P = .05. Statistical comparisons in limiting dilution experiments (CAFC, CRU, and homing assays) were made using L-Calc software. Significance was determined by a P value of less than .05 in all comparisons.
Genotype analysis
Tail biopsies from 2 female B6.SJL mice were sent to the Cincinnati Children's Hospital Medical Center Genotyping Core Facility for DNA isolation and genotyping on a fee-for-service basis. The Illumina GoldenGate Genotyping Assay protocol (Illumina Inc) was used with the mouse medium density linkage panel consisting of 1449 loci across the mouse genome. Samples were run on the 16-sample Sentrix Universal BeadChip (Illumina Inc). The B6.SJL genotype was compared with the genotype of B6 and SJL using an interactive analysis tool developed by Bala Abiramikuma available through the Genotyping Core Facility website (http://dna.chmcc.org/bala/MOUSE_SNP). Gene lists within this interval were generated in January 2008 using NCBI Mapviewer Build 37.1.21 Lists of single nucleotide polymorphisms (SNPs) between B6 and SJL strains within this interval were generated in January 2008 using the Wellcome Trust Centre for Human Genetics Mouse SNP Selector, Build 37.22
Isolation of bone marrow stromal cells for microarray analysis
Nonhematopoietic marrow-associated stromal cells were isolated from femora and tibiae by flushing marrow into Hanks balanced salt solution (GIBCO) with 2% serum. Bone-associated nonhematopoietic stromal cells were subsequently harvested from the same bones by crushing and enzymatic digestion in Dulbecco modified Eagle medium (GIBCO) containing 1 mg/mL collagenase P (Roche), 10% serum, 3mM CaCl2 (Sigma), 40 μg/mL DNase 1 (Sigma), and 0.01M N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (GIBCO). After a 40-minute incubation at 37°C while shaking, bone-associated stromal cells were separated from debris by washing and Ficoll-gradient centrifugation. Ficoll-enriched bone marrow–derived cells were combined and stained with a cocktail of biotinlyated, lineage-specific antibodies as described for cell cycling and apoptosis assays and subsequently stained with streptavidin–fluorescein isothiocyanate and PE-conjugated anti-CD45.1(A20) or anti-CD45.2(104). All antibodies used in this assay were purchased from BD. Viable, nonhematopoietic cells were selected from this population by cell sorting on a FACSVantage (BD) on the basis of PI exclusion and negative selection for lineage markers and CD45 antigens. Each sample was pooled from 5 individual mice, and 2 samples were collected per strain.
Microarray analysis
Total RNA was extracted from bone marrow stromal cells using a QIAGEN RNeasy Mini Kit according to the manufacturer's instructions. Duplicate RNA samples were isolated independently from 81 000 to 326 000 cells of B6 and B6.SJL mice. Each sample was reverse transcribed, purified, labeled, and hybridized to individual arrays on an Illumina Sentrix-Mouse 6 Whole Genome Expression Beadchip (Illumina Inc). Illumina expression arrays were processed on a fee-for-service basis at the Cincinnati Children's Hospital Medical Center Microarray Core Facility. Expression was quantified with an Illumina Bead Scanner. Gene expression analysis was performed using Illumina BeadStudio V3. Average expression values were obtained from the duplicate samples from each group and normalized by the rank invariant method. Differences in gene expression between strains were calculated using the Illumina Custom differential expression algorithm. Reproducibility between independent samples of the same strain was confirmed by correlation analysis, in which r2 ≥ 0.96. All microarray data have been submitted to ArrayExpress under accession number E-TABM-829.23
URLs
Website addresses are as follows: Cincinnati Children's Hospital Medical Center Genotyping Core Facility Analysis tool http://dna.chmcc.org/bala/MOUSE_SNP24 ; NCBI Mapviewer http://www.ncbi.nlm.nih.gov/mapview/maps.cgi?org=mouse&chr=1; Wellcome Trust Centre for Human Genetics Mouse SNP Selector http://gscan.well.ox.ac.uk/gs/strains.cgi.
Results
Competitive repopulation
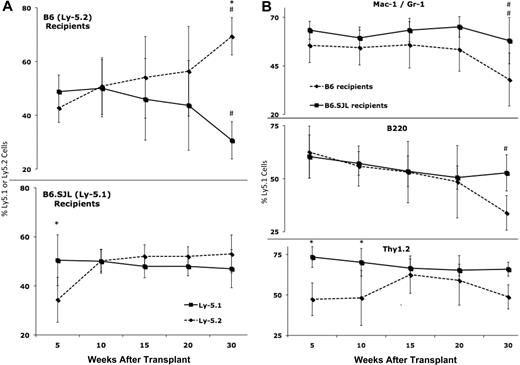
In light of disparate results that have been observed in reciprocal Ly-5 transplants,12-14 and to account for the possible influence of donor microenvironment on recipient engraftment, we admixed and transplanted equal numbers (2 × 106) of hematopoietic cells from B6 and B6.SJL strains (4 × 106 total cells) into myeloablated recipients of both genotypes. Results of peripheral blood analysis at 5, 10, 15, 20, and 30 weeks after transplant are displayed in Figure 1A and B. Thirty weeks after transplant, B6 recipients had an average of 69% Ly-5.2 cells and 31% Ly-5.1 cells (*P = .001), constituting a 38% relative engraftment defect. Mixed model analyses revealed significant changes in Ly-5 chimerism over time including a drop in Ly-5.1 cells (#P = .001) and an increase in Ly-5.2 cells (#P < .0001) in B6 recipients. There were corresponding significant decreases in the Mac-1/Gr-1 lineage in both strains (# #P = .009) and the B220 lineage in B6 recipients (#P = .03). Thy-1.2 cells also decreased over time in both strains (m = −0.069) but not significantly (P = .6). The fact that reductions in the Ly-5.1 contribution to B6 peripheral blood were evident in both the lymphoid and myeloid lineages demonstrates a stem cell origin for the decrease (Figure 1B).
B6-derived Ly-5.2 cells exhibit competitive advantage in B6 recipients. (A top panel) Ly-5.2 cells increase significantly (#P < .0001) and Ly-5.1 cells decrease significantly (#P = .001) from 5 to 30 weeks after transplant in B6 recipients. Within the Ly-5+ population, the percentage of Ly-5.2 (mean ± SD) cells is significantly higher (*P = .001) than the percentage of Ly-5.1 (mean ± SD) cells at 30 weeks after transplant in B6 recipients. (Bottom panel) Ly-5.2 cells increase significantly (#P = .002) in B6.SJL recipients from 5 to 30 weeks after transplant. Within the Ly-5+ population, the percentage of Ly-5.1 cells (mean ± SD) is significantly higher (*P = .001) than the percentage of Ly-5.2 cells (mean ± SD) at 5 weeks after transplant in B6 recipients. (B) Within the Ly-5.1 population, the percentage of Mac-1/Gr-1 cells (mean ± SD) decreased significantly (##P = .009) in B6 and B6.SJL recipients, the percentage of B220 cells (mean ± SD) decreased significantly in B6 (#P = .03) but not B6.SJL (P = .5) recipients, and the percentage of Thy-1.2 cells dropped slightly (slope = −0.069) but not significantly (P = .6) in both strains. Cross-strain comparisons revealed a significant difference in Thy-1.2 cells at 5 (*P = .03) and 10 (*P = .02) weeks after transplant. Data were pooled from a total of 3 to 8 mice per strain at each time point.
B6-derived Ly-5.2 cells exhibit competitive advantage in B6 recipients. (A top panel) Ly-5.2 cells increase significantly (#P < .0001) and Ly-5.1 cells decrease significantly (#P = .001) from 5 to 30 weeks after transplant in B6 recipients. Within the Ly-5+ population, the percentage of Ly-5.2 (mean ± SD) cells is significantly higher (*P = .001) than the percentage of Ly-5.1 (mean ± SD) cells at 30 weeks after transplant in B6 recipients. (Bottom panel) Ly-5.2 cells increase significantly (#P = .002) in B6.SJL recipients from 5 to 30 weeks after transplant. Within the Ly-5+ population, the percentage of Ly-5.1 cells (mean ± SD) is significantly higher (*P = .001) than the percentage of Ly-5.2 cells (mean ± SD) at 5 weeks after transplant in B6 recipients. (B) Within the Ly-5.1 population, the percentage of Mac-1/Gr-1 cells (mean ± SD) decreased significantly (##P = .009) in B6 and B6.SJL recipients, the percentage of B220 cells (mean ± SD) decreased significantly in B6 (#P = .03) but not B6.SJL (P = .5) recipients, and the percentage of Thy-1.2 cells dropped slightly (slope = −0.069) but not significantly (P = .6) in both strains. Cross-strain comparisons revealed a significant difference in Thy-1.2 cells at 5 (*P = .03) and 10 (*P = .02) weeks after transplant. Data were pooled from a total of 3 to 8 mice per strain at each time point.
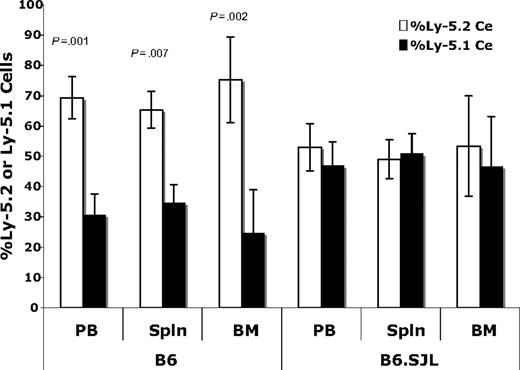
We also measured Ly-5 chimerism in the spleen and bone marrow of the same mice at 30 weeks after transplant (Figure 2). In B6 recipients, there was a significantly larger percentage of Ly-5.2 cells in all hematopoietic organs (38% in blood, 31% in spleen, and 51% in bone marrow), whereas chimerism was balanced and stable in B6.SJL recipients, providing evidence for a unidirectional engraftment defect. These data (Figures 1–2) reveal that engraftment differs between the 2 transplantation schemes even when recipients are lethally irradiated and receive a transplant of 4 × 106 cells (2 × 106 cells from B6 and 2 × 106 cells from B6.SJL donors).
Ly-5.1 cells are underrepresented in all hematopoietic organs of B6 recipients. (A) In Ly-5–positive cells, the percentage of Ly-5.1 cells (mean ± SD) was significantly lower than the percentage of Ly-5.2 cells (mean ± SD) in the peripheral blood (P = .001), spleen (P = .007), and bone marrow (P = .002) of B6 mice 30 weeks after transplant.
Ly-5.1 cells are underrepresented in all hematopoietic organs of B6 recipients. (A) In Ly-5–positive cells, the percentage of Ly-5.1 cells (mean ± SD) was significantly lower than the percentage of Ly-5.2 cells (mean ± SD) in the peripheral blood (P = .001), spleen (P = .007), and bone marrow (P = .002) of B6 mice 30 weeks after transplant.
In vivo quantification of hematopoietic stem cell numbers and homing efficiency
We quantified HSCs in vivo and measured HSC homing efficiency using a modification of the method of Szilvassy et al.19,25 We found that B6.SJL mice possess 3.8-fold fewer in vivo–defined long-term (LT)–HSCs, termed competitive repopulating units (CRUs) than B6 at 15 to 17 weeks after transplant (Tables 1–2). Importantly, this assay revealed that the homing efficiency of less primitive CRUs (measured at 5 and 10-12 weeks after transplant) did not differ significantly between the strains. This suggests that the reason for the engraftment defect detected only at late time points (30 weeks after transplant) in competitive repopulation studies results, at least in part, from a diminished ability of LT-HSCs to home to the bone marrow microenvironment.
Determination of CRU frequency and homing efficiency of B6.SJL-derived cells transplanted into B6 recipients
| . | Proportion of positive mice* . | ||
|---|---|---|---|
| 5 weeks . | 10-12 weeks . | 15-17 weeks . | |
| No. of 1° BM cells injected per mouse | |||
| 2 × 103 | 0 of 16 | 2 of 15 | 0 of 12 |
| 6 × 103 | 1 of 14 | 2 of 14 | 3 of 14 |
| 2 × 104 | 9 of 16 | 7 of 15 | 5 of 15 |
| 6 × 104 | 10 of 13 | 10 of 12 | 9 of 11 |
| 1/CRU frequency,† ± SE | 36 838 (29 084-46 660) | 31 335 (24 723-39 715) | 39 318 (30 509-50 670) |
| No. of CRUs injected,‡ ± SE | 54.3 (42.9-68.8) | 63.8 (50.4-80.9) | 50.9 (39.5-65.6) |
| Proportion of 1° BM injected per mouse | |||
| 0.005 | 0 of 15 | 3 of 11 | 0 of 8 |
| 0.014 | 1 of 14 | 2 of 12 | 0 of 12 |
| 0.042 | 0 of 12 | 2 of 11 | 1 of 11 |
| 0.125 | 2 of 13 | 4 of 12 | 1 of 9 |
| 0.16 | 4 of 13 | 5 of 12 | 3 of 12 |
| 0.2 | 3 of 11 | 3 of 9 | 5 of 9 |
| 1/CRU frequency per primary BM fraction,† ± SE | 0.59 (0.43-0.81) | 0.3 (0.2-0.3) | 0.5 (0.3-0.6) |
| CRUs recovered per primary BM,‡ ± SE | 1.7 (1.2-2.3) | 3.3 (3.3-5) | 2 (1.7-3.3) |
| Homing efficiency,§ % (CRUs recovered/CRUs injected × 100) | 3.10 | 5.20 | 3.90 |
| . | Proportion of positive mice* . | ||
|---|---|---|---|
| 5 weeks . | 10-12 weeks . | 15-17 weeks . | |
| No. of 1° BM cells injected per mouse | |||
| 2 × 103 | 0 of 16 | 2 of 15 | 0 of 12 |
| 6 × 103 | 1 of 14 | 2 of 14 | 3 of 14 |
| 2 × 104 | 9 of 16 | 7 of 15 | 5 of 15 |
| 6 × 104 | 10 of 13 | 10 of 12 | 9 of 11 |
| 1/CRU frequency,† ± SE | 36 838 (29 084-46 660) | 31 335 (24 723-39 715) | 39 318 (30 509-50 670) |
| No. of CRUs injected,‡ ± SE | 54.3 (42.9-68.8) | 63.8 (50.4-80.9) | 50.9 (39.5-65.6) |
| Proportion of 1° BM injected per mouse | |||
| 0.005 | 0 of 15 | 3 of 11 | 0 of 8 |
| 0.014 | 1 of 14 | 2 of 12 | 0 of 12 |
| 0.042 | 0 of 12 | 2 of 11 | 1 of 11 |
| 0.125 | 2 of 13 | 4 of 12 | 1 of 9 |
| 0.16 | 4 of 13 | 5 of 12 | 3 of 12 |
| 0.2 | 3 of 11 | 3 of 9 | 5 of 9 |
| 1/CRU frequency per primary BM fraction,† ± SE | 0.59 (0.43-0.81) | 0.3 (0.2-0.3) | 0.5 (0.3-0.6) |
| CRUs recovered per primary BM,‡ ± SE | 1.7 (1.2-2.3) | 3.3 (3.3-5) | 2 (1.7-3.3) |
| Homing efficiency,§ % (CRUs recovered/CRUs injected × 100) | 3.10 | 5.20 | 3.90 |
CRU indicates competitive repopulating unit; BM, bone marrow; and 1°, primary.
Recipients were considered positive if at least 5% of red cell-depleted peripheral blood cells were of donor origin (Ly-5.1).
CRU frequency is the number of BM cells containing 1 CRU.
Absolute numbers of CRUs were determined by multiplying 1 CRU frequency by the number of cells injected.
Homing efficiency is the ratio of CRUs detected in secondary recipients over the number of CRUs transplanted into the primary recipients multiplied by 100.
Determination of CRU frequency and homing efficiency of B6 derived cells transplanted into B6.SJL recipients
| . | Proportion of positive mice* . | ||
|---|---|---|---|
| 5 weeks . | 10-12 weeks . | 15-17 weeks . | |
| No. of 1° BM cells injected per mouse | |||
| 2 × 103 | 1 of 11 | 1 of 10 | 6 of 9 |
| 6 × 103 | 3 of 11 | 4 of 10 | 4 of 8 |
| 2 × 104 | 2 of 9 | 5 of 7 | 6 of 7 |
| 6 × 104 | 5 of 10 | 8 of 10 | 9 of 10 |
| 1/CRU frequency,† ± SE | 60 203 (43 649-82 949) | 23 081 (17 363-30 681) | 10 429 (7 655-14 208) |
| No. of CRUs injected,‡ ± SE | 33.2 (24.1-45.8) | 86.7 (65.2-115.2) | 191.8 (140.8-261.3) |
| Proportion of 1° BM injected per mouse | |||
| 0.005 | 0 of 8 | 0 of 5 | 2 of 5 |
| 0.014 | 0 of 10 | 2 of 9 | 1 of 8 |
| 0.042 | 0 of 10 | 2 of 9 | 5 of 8 |
| 0.125 | 2 of 10 | 1 of 8 | 4 of 7 |
| 0.16 | 1 of 10 | 5 of 10 | 6 of 10 |
| 0.2 | 0 of 10 | 2 of 7 | 3 of 6 |
| 1/CRU frequency per primary BM fraction,† ± SE | 1.7 (1-3.1) | 0.3 (0.2-0.4) | 0.1 (0.1-0.2) |
| CRUs recovered per primary BM,‡ ± SE | 0.59 (0.32-1) | 3.3 (2.5-5) | 10 (5-10) |
| Homing efficiency,§ % (CRU recovered/CRU injected × 100) | 1.80 | 3.80 | 5.20 |
| Comparison of the ratio of proportions, Table 1 versus Table 2 | |||
| P for primary CRUs | .22 | .41 | .001 |
| P for homed CRUs | .07 | .57 | .002 |
| . | Proportion of positive mice* . | ||
|---|---|---|---|
| 5 weeks . | 10-12 weeks . | 15-17 weeks . | |
| No. of 1° BM cells injected per mouse | |||
| 2 × 103 | 1 of 11 | 1 of 10 | 6 of 9 |
| 6 × 103 | 3 of 11 | 4 of 10 | 4 of 8 |
| 2 × 104 | 2 of 9 | 5 of 7 | 6 of 7 |
| 6 × 104 | 5 of 10 | 8 of 10 | 9 of 10 |
| 1/CRU frequency,† ± SE | 60 203 (43 649-82 949) | 23 081 (17 363-30 681) | 10 429 (7 655-14 208) |
| No. of CRUs injected,‡ ± SE | 33.2 (24.1-45.8) | 86.7 (65.2-115.2) | 191.8 (140.8-261.3) |
| Proportion of 1° BM injected per mouse | |||
| 0.005 | 0 of 8 | 0 of 5 | 2 of 5 |
| 0.014 | 0 of 10 | 2 of 9 | 1 of 8 |
| 0.042 | 0 of 10 | 2 of 9 | 5 of 8 |
| 0.125 | 2 of 10 | 1 of 8 | 4 of 7 |
| 0.16 | 1 of 10 | 5 of 10 | 6 of 10 |
| 0.2 | 0 of 10 | 2 of 7 | 3 of 6 |
| 1/CRU frequency per primary BM fraction,† ± SE | 1.7 (1-3.1) | 0.3 (0.2-0.4) | 0.1 (0.1-0.2) |
| CRUs recovered per primary BM,‡ ± SE | 0.59 (0.32-1) | 3.3 (2.5-5) | 10 (5-10) |
| Homing efficiency,§ % (CRU recovered/CRU injected × 100) | 1.80 | 3.80 | 5.20 |
| Comparison of the ratio of proportions, Table 1 versus Table 2 | |||
| P for primary CRUs | .22 | .41 | .001 |
| P for homed CRUs | .07 | .57 | .002 |
CRU indicates competitive repopulating unit; BM, bone marrow; and 1°, primary.
Recipients were considered positive if at least 5% of red cell-depleted peripheral blood cells were of donor origin (Ly-5.1).
CRU frequency is the number of BM cells containing 1 CRU.
Absolute numbers of CRUs were determined by multiplying 1 CRU frequency by the number of cells injected.
Homing efficiency is the ratio of CRUs detected in secondary recipients over the number of CRUs transplanted into the primary recipients multiplied by 100.
In 4 independent experiments, lethally irradiated B6 recipients were injected with 2 × 106 BM cells from B6.SJL mice and killed 24 hours later to obtain homed cells for secondary transplantation. Reciprocal transplants (B6 into B6.SJL) were performed in 3 independent experiments. The frequency of CRU in the original 2 × 106 cells was determined by transplanting graded numbers of primary cells from the same innoculumn and homed cells into separate groups of B6 or B6.SJL recipients. Recipients of primary and homed BM cells were evaluated for donor engraftment at the indicated times after transplantation.
Cell cycling and apoptosis
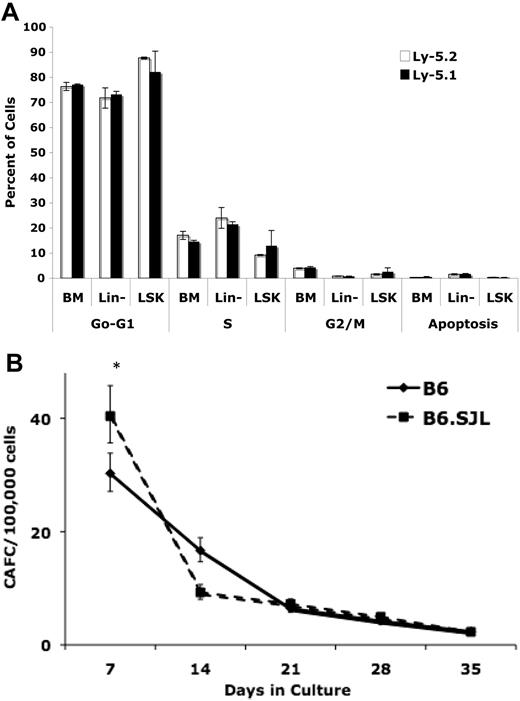
To address the possibility that Ly-5 or other polymorphic passenger loci may influence engraftment and/or subsequent self-renewal or differentiation by influencing cell cycling dynamics, we measured the percentage of whole bone marrow, lineage-negative, and LSK cells in various stages of the cell cycle or apoptosis by colabeling with BrdU and 7-aminoactinomycin D. Although the experimental design and variation in the data would have enabled us to detect differences in the percentage of cells in S-phase between the strains of 2.7% in BM, 4.8% in Lin− cells, and 6.1% in LSK cells as statistically significant at 80% power, no statistically significant differences were identified in this or any other phase of the cell cycle (detectable effects ranging from 0.2%-9%) or apoptosis (detectable effects ranging from 0.2%-1.5%) between the strains (Figure 3A).
B6 and B6.SJL strains possess equivalent patterns of cell cycling, apoptosis, and CAFCs. (A) There were no significant differences between B6 and B6.SJL cells in any phase of the cell cycle or apoptosis in any bone marrow population tested. Values represent the mean ± SD calculated from 3 independent experiments each using 2 mice per strain. Statistical comparisons were made using the unpaired Student t test assuming unequal variance. (B) There were no significant differences in in vitro defined bone marrow HSC number (CAFCd28 and CAFCd35) between B6 and B6.SJL mice (*P < .05). Data were combined from 3 independent CAFC assays each using marrow pooled from 2 to 3 mice per strain.
B6 and B6.SJL strains possess equivalent patterns of cell cycling, apoptosis, and CAFCs. (A) There were no significant differences between B6 and B6.SJL cells in any phase of the cell cycle or apoptosis in any bone marrow population tested. Values represent the mean ± SD calculated from 3 independent experiments each using 2 mice per strain. Statistical comparisons were made using the unpaired Student t test assuming unequal variance. (B) There were no significant differences in in vitro defined bone marrow HSC number (CAFCd28 and CAFCd35) between B6 and B6.SJL mice (*P < .05). Data were combined from 3 independent CAFC assays each using marrow pooled from 2 to 3 mice per strain.
In vitro stem cell quantification
The fact that B6.SJL marrow contains 3.8-fold fewer HSCs than B6 as determined by CRU analysis suggests that the mice differ in absolute stem cell numbers, yet because this assay incorporates homing efficiency into the readout of HSC number, and we also detected a 25% homing defect in B6.SJL cells relative to B6, we also quantified HSCs in vitro using the cobblestone area forming cell (CAFC) assay. The results of the CAFC assay indicate that mice of both strains contain approximately 2 to 5 stem cells (CAFCs on days 28 and 35) per every 105 total mononucleated bone marrow cells (Figure 3B). In 3 independent experiments, any apparent variation between strains was irreproducible and not statistically significant.
Delineation of the B6.SJL congenic interval
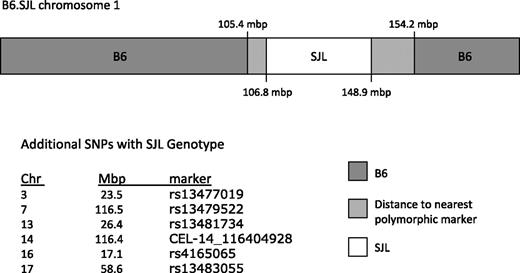
Because the differences in competitive repopulating ability, CRU number, and homing efficiency could not be explained by differences in total stem cell number (measured by CAFC), or percentages of cycling or apoptotic cells, we believe that the engraftment differences between the 2 transplantation strategies may be related to homing and stem cell–microenvironment interactions. Allotypic variation in the Ly-5 gene has never been linked to HSC engraftment in lethally irradiated recipients, and the congenic interval of B6.SJL has never been defined with dense markers. Therefore, to determine whether polymorphisms in Ly-5 and/or additional polymorphic passenger genes in the interval could influence engraftment, we compared the genotype of B6.SJL mice to the B6 and SJL inbred strains using the Illumina GoldenGate SNP Genotyping Assay. This assay allowed us to identify, for the first time, the boundaries of the B6.SJL congenic interval, which range from at least 106 831 730 to 148 913 234 bases on chromosome 1 (Figure 4, supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article). The nearest flanking markers detecting the B6 genotype are at the 105 423 103 and 154 187 910-bp position on chromosome 1 (Figure 4, supplemental Table 1). It is, therefore, possible that the SJL congenic interval extends further toward these B6 boundaries. Because genome sequence analysis proximal and distal from the SJL SNPs would be required to confirm this, we chose the most conservative description, the SJL SNP marker coordinates, to define the congenic interval (Figure 4, supplemental Table 1). In addition to this congenic interval, isolated SNPs were also identified on chromosomes 3, 7, 13, 14, 16, and 17 (Figure 4, supplemental Table 1). A query of the congenic interval (106 to 148 mbp) using NCBI Mapviewer Build 37.1 returned 306 known or predicted genes. Included in this list are genes with known functions in the hematopoietic system such as CXCR-4. Of greater importance is the fact that 45 of these genes, including CXCR-4, contain 1 or more polymorphisms between B6 and SJL according to the Wellcome Trust Centre for Human Genetics Mouse SNP Selector. A comparison of the set of genes within this interval using NCBI Build 37 and the Build 37 SNP database available through the SNP Selector revealed that at least 45 known or predicted genes span 1 or more SNPs (Table 3).
B6.SJL congenic interval spans 106 to 148 mbp on mouse chromosome 1. Schematic representation of B6.SJL chromosome 1: the SJL donor-interval is designated in white, the distance to the nearest informative B6 marker is light gray, and the B6 genotype is represented in darker gray.
B6.SJL congenic interval spans 106 to 148 mbp on mouse chromosome 1. Schematic representation of B6.SJL chromosome 1: the SJL donor-interval is designated in white, the distance to the nearest informative B6 marker is light gray, and the B6 genotype is represented in darker gray.
B6.SJL congenic interval contains at least 45 genes with or one or more SNPs between the B6 and SJL strains
| . | Gene symbol . | Gene start . | Gene stop . | SNP . | SNP position . | B6 genotype . | SJL genotype . |
|---|---|---|---|---|---|---|---|
| 1 | Rnf152 | 107 176 508 | 107 253 247 | rs3707424 | 107 216 898 | CC | TT |
| rs3696200 | 107 241 903 | TT | AA | ||||
| 2 | Pign | 107 417 716 | 107 560 239 | CEL-1_105566001 | 107 541 004 | AA | GG |
| 3 | Serpinb8 | 109 486 648 | 109 505 486 | rs13476039 | 109 503 687 | CC | TT |
| 4 | Cdh19 | 112 785 772 | 112 873 949 | rs3710646 | 112 847 342 | AA | CC |
| 5 | Cntnap5a | 117 581 714 | 118 477 251 | mCV24201027 | 117 626 679 | CC | TT |
| rs3725409 | 118 309 925 | GG | AA | ||||
| 6 | Clasp1 | 120 285 635 | 120 506 039 | rs3679459 | 120 341 734 | AA | GG |
| rs6216134 | 120 379 270 | CC | TT | ||||
| 7 | Sctr | 121 903 555 | 121 960 115 | rs3723088 | 121 957 599 | AA | GG |
| 8 | Dbi | 122 009 857 | 122 017 656 | rs6302966 | 122 017 145 | GG | AA |
| 9 | En1 | 122 499 258 | 122 504 548 | rs3678634 | 122 503 972 | TT | AA |
| 10 | Ccdc93 | 123 327 644 | 123 398 170 | rs13476083 | 123 358 415 | TT | CC |
| 11 | Dpp10 | 125 228 719 | 125 942 129 | gnf01.123.653 | 125 888 571 | TT | CC |
| 12 | E030049G20Rik | 127 810 213 | 128 727 209 | rs13476098 | 128 464 682 | TT | GG |
| gnf01.126.387 | 128 625 610 | CC | TT | ||||
| 13 | Cxcr4 | 130 484 776 | 130 488 876 | rs8256196 | 130 485 574 | CC | TT |
| 14 | Thsd7b | 131 170 051 | 132 115 855 | rs6281838 | 131 841 777 | TT | CC |
| 15 | Zp3r | 132 473 290 | 132 526 172 | rs6355835 | 132 515 586 | TT | AA |
| 16 | Pigr | 132 723 328 | 132 748 548 | rs13459051 | 132 747 429 | TT | CC |
| 17 | Mapkapk2 | 132 950 281 | 132 994 120 | rs3699561 | 132 988 657 | GG | AA |
| 18 | Srgap2 | 133 181 828 | 133 423 938 | rs6308228 | 133 189 078 | CC | TT |
| rs6329578 | 133 313 859 | TT | CC | ||||
| rs3724826 | 133 401 871 | AA | GG | ||||
| 19 | Ctse | 133 534 891 | 133 572 080 | rs6250833 | 133 558 414 | GG | AA |
| rs4222623 | 133 564 393 | GG | CC | ||||
| 20 | Slc26a9 | 133 640 599 | 133 666 982 | rs13476111 | 133 649 413 | TT | CC |
| rs3718090 | 133 662 206 | TT | CC | ||||
| 21 | Slc45a3 | 133 867 186 | 133 879 541 | rs13459052 | 133 878 180 | CC | TT |
| 22 | Klhdc8a | 134 195 203 | 134 203 934 | rs13476112 | 134 201 085 | CC | TT |
| 23 | Rbbp5 | 134 373 957 | 134 401 467 | rs6350018 | 134 394 446 | CC | GG |
| 24 | Pik3c2b | 134 962 877 | 135 004 025 | rs6241653 | 134 993 149 | CC | TT |
| 25 | LOC100038824 | 135 247 143 | 135 256 900 | rs3022833 | 135 256 658 | GG | CC |
| 26 | Ren1 | 135 247 251 | 135 256 894 | rs3164478 | 135 283 762 | TT | CC |
| 27 | Sox13 | 135 278 877 | 135 320 789 | rs3164478 | 135 283 762 | TT | CC |
| 28 | Adora1 | 136 095 799 | 136 132 008 | rs13476119 | 136 107 190 | AA | GG |
| 29 | Jarid1b | 136 456 772 | 136 529 454 | UT_1_137.539742 | 136 529 119 | GG | CC |
| 30 | Syt2 | 136 543 258 | 136 645 994 | rs3664806 | 136 549 781 | AA | CC |
| UT_1_137.66161 | 136 644 400 | AA | TT | ||||
| 31 | Ppp1r12b | 136 662 520 | 136 852 517 | rs13476122 | 136 766 629 | AA | TT |
| 32 | Lgr6 | 136 882 930 | 137 001 853 | rs6290558 | 136 955 914 | TT | CC |
| 33 | Arl8a | 137 043 411 | 137 052 845 | rs8250053 | 137 052 420 | AA | TT |
| 34 | Rnpep | 137 159 289 | 137 180 606 | rs8270851 | 137 179 674 | AA | TT |
| 35 | Ipo9 | 137 278 892 | 137 327 068 | rs4222666 | 137 279 078 | TT | CC |
| 36 | Nav1 | 137 335 450 | 137 482 286 | rs3667307 | 137 411 836 | AA | GG |
| 37 | Tnni1 | 137 696 410 | 137 707 553 | rs8260536 | 137 702 249 | CC | AA |
| rs8236489 | 137 706 366 | GG | AA | ||||
| 38 | Tnnt2 | 137 732 982 | 137 748 837 | rs3674857 | 137 733 097 | CC | TT |
| 39 | Cacna1s | 137 949 478 | 138 016 399 | rs13476125 | 138 014 096 | CC | AA |
| 40 | Camsap1l1 | 138 164 700 | 138 242 681 | rs3691222 | 138 214 393 | TT | CC |
| 41 | LOC100042567 | 138 982 190 | 139 267 341 | rs13476129 | 139 032 215 | AA | GG |
| 42 | Crb1 | 141 094 920 | 141 273 618 | rs13476137 | 141 128 063 | CC | TT |
| 43 | F13b | 141 398 304 | 141 420 333 | rs13476138 | 141 417 567 | GG | TT |
| 44 | Kcnt2 | 142 142 834 | 142 506 838 | CEL-1_140236219 | 142 191 594 | TT | CC |
| rs13476140 | 142 246 107 | AA | GG | ||||
| rs13476141 | 142 430 268 | TT | CC | ||||
| 45 | LOC667877 | 146 771 868 | 146 801 392 | rs13476155 | 146 800 236 | TT | GG |
| . | Gene symbol . | Gene start . | Gene stop . | SNP . | SNP position . | B6 genotype . | SJL genotype . |
|---|---|---|---|---|---|---|---|
| 1 | Rnf152 | 107 176 508 | 107 253 247 | rs3707424 | 107 216 898 | CC | TT |
| rs3696200 | 107 241 903 | TT | AA | ||||
| 2 | Pign | 107 417 716 | 107 560 239 | CEL-1_105566001 | 107 541 004 | AA | GG |
| 3 | Serpinb8 | 109 486 648 | 109 505 486 | rs13476039 | 109 503 687 | CC | TT |
| 4 | Cdh19 | 112 785 772 | 112 873 949 | rs3710646 | 112 847 342 | AA | CC |
| 5 | Cntnap5a | 117 581 714 | 118 477 251 | mCV24201027 | 117 626 679 | CC | TT |
| rs3725409 | 118 309 925 | GG | AA | ||||
| 6 | Clasp1 | 120 285 635 | 120 506 039 | rs3679459 | 120 341 734 | AA | GG |
| rs6216134 | 120 379 270 | CC | TT | ||||
| 7 | Sctr | 121 903 555 | 121 960 115 | rs3723088 | 121 957 599 | AA | GG |
| 8 | Dbi | 122 009 857 | 122 017 656 | rs6302966 | 122 017 145 | GG | AA |
| 9 | En1 | 122 499 258 | 122 504 548 | rs3678634 | 122 503 972 | TT | AA |
| 10 | Ccdc93 | 123 327 644 | 123 398 170 | rs13476083 | 123 358 415 | TT | CC |
| 11 | Dpp10 | 125 228 719 | 125 942 129 | gnf01.123.653 | 125 888 571 | TT | CC |
| 12 | E030049G20Rik | 127 810 213 | 128 727 209 | rs13476098 | 128 464 682 | TT | GG |
| gnf01.126.387 | 128 625 610 | CC | TT | ||||
| 13 | Cxcr4 | 130 484 776 | 130 488 876 | rs8256196 | 130 485 574 | CC | TT |
| 14 | Thsd7b | 131 170 051 | 132 115 855 | rs6281838 | 131 841 777 | TT | CC |
| 15 | Zp3r | 132 473 290 | 132 526 172 | rs6355835 | 132 515 586 | TT | AA |
| 16 | Pigr | 132 723 328 | 132 748 548 | rs13459051 | 132 747 429 | TT | CC |
| 17 | Mapkapk2 | 132 950 281 | 132 994 120 | rs3699561 | 132 988 657 | GG | AA |
| 18 | Srgap2 | 133 181 828 | 133 423 938 | rs6308228 | 133 189 078 | CC | TT |
| rs6329578 | 133 313 859 | TT | CC | ||||
| rs3724826 | 133 401 871 | AA | GG | ||||
| 19 | Ctse | 133 534 891 | 133 572 080 | rs6250833 | 133 558 414 | GG | AA |
| rs4222623 | 133 564 393 | GG | CC | ||||
| 20 | Slc26a9 | 133 640 599 | 133 666 982 | rs13476111 | 133 649 413 | TT | CC |
| rs3718090 | 133 662 206 | TT | CC | ||||
| 21 | Slc45a3 | 133 867 186 | 133 879 541 | rs13459052 | 133 878 180 | CC | TT |
| 22 | Klhdc8a | 134 195 203 | 134 203 934 | rs13476112 | 134 201 085 | CC | TT |
| 23 | Rbbp5 | 134 373 957 | 134 401 467 | rs6350018 | 134 394 446 | CC | GG |
| 24 | Pik3c2b | 134 962 877 | 135 004 025 | rs6241653 | 134 993 149 | CC | TT |
| 25 | LOC100038824 | 135 247 143 | 135 256 900 | rs3022833 | 135 256 658 | GG | CC |
| 26 | Ren1 | 135 247 251 | 135 256 894 | rs3164478 | 135 283 762 | TT | CC |
| 27 | Sox13 | 135 278 877 | 135 320 789 | rs3164478 | 135 283 762 | TT | CC |
| 28 | Adora1 | 136 095 799 | 136 132 008 | rs13476119 | 136 107 190 | AA | GG |
| 29 | Jarid1b | 136 456 772 | 136 529 454 | UT_1_137.539742 | 136 529 119 | GG | CC |
| 30 | Syt2 | 136 543 258 | 136 645 994 | rs3664806 | 136 549 781 | AA | CC |
| UT_1_137.66161 | 136 644 400 | AA | TT | ||||
| 31 | Ppp1r12b | 136 662 520 | 136 852 517 | rs13476122 | 136 766 629 | AA | TT |
| 32 | Lgr6 | 136 882 930 | 137 001 853 | rs6290558 | 136 955 914 | TT | CC |
| 33 | Arl8a | 137 043 411 | 137 052 845 | rs8250053 | 137 052 420 | AA | TT |
| 34 | Rnpep | 137 159 289 | 137 180 606 | rs8270851 | 137 179 674 | AA | TT |
| 35 | Ipo9 | 137 278 892 | 137 327 068 | rs4222666 | 137 279 078 | TT | CC |
| 36 | Nav1 | 137 335 450 | 137 482 286 | rs3667307 | 137 411 836 | AA | GG |
| 37 | Tnni1 | 137 696 410 | 137 707 553 | rs8260536 | 137 702 249 | CC | AA |
| rs8236489 | 137 706 366 | GG | AA | ||||
| 38 | Tnnt2 | 137 732 982 | 137 748 837 | rs3674857 | 137 733 097 | CC | TT |
| 39 | Cacna1s | 137 949 478 | 138 016 399 | rs13476125 | 138 014 096 | CC | AA |
| 40 | Camsap1l1 | 138 164 700 | 138 242 681 | rs3691222 | 138 214 393 | TT | CC |
| 41 | LOC100042567 | 138 982 190 | 139 267 341 | rs13476129 | 139 032 215 | AA | GG |
| 42 | Crb1 | 141 094 920 | 141 273 618 | rs13476137 | 141 128 063 | CC | TT |
| 43 | F13b | 141 398 304 | 141 420 333 | rs13476138 | 141 417 567 | GG | TT |
| 44 | Kcnt2 | 142 142 834 | 142 506 838 | CEL-1_140236219 | 142 191 594 | TT | CC |
| rs13476140 | 142 246 107 | AA | GG | ||||
| rs13476141 | 142 430 268 | TT | CC | ||||
| 45 | LOC667877 | 146 771 868 | 146 801 392 | rs13476155 | 146 800 236 | TT | GG |
A query of the NCBI Build 37 of the mouse genome in January 2008 identified 306 genes within the congenic interval of B6.SJL mice (107-148 mbp). Using the mouse SNP Selector from the Wellcome Trust Centre for Human Genetics, we identified 124 SNPs between B6 and SJL in the same region. A comparison of gene location and SNP location revealed that at least 45 of the genes in this interval contain 1 or more SNPs between the 2 strains.
SNP indicates single nucleotide polymorphism.
Stromal transcriptome variation between B6 and B6.SJL
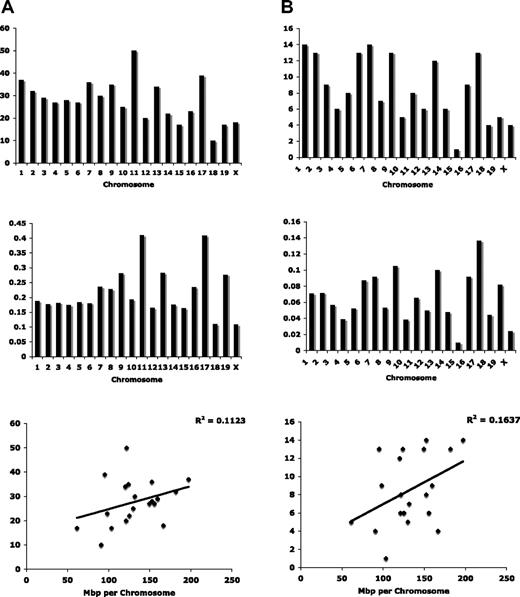
Lastly, we compared gene expression profiles from marrow-derived stromal cells of B6 and B6.SJL to identify potential effects of the SJL-donor congenic interval on the hematopoietic microenvironment. We isolated bone marrow–derived stroma by negative selection for hematopoietic lineage markers and CD45 (“Methods”). Two independently derived stromal cell samples were assayed per strain. The results from the individual samples were pooled by strain and the interstrain results compared. The Illumina Sentrix-Mouse 6 platform, which contains 46 644 probes, allowed us to detect 17 046 expressed genes in stromal cells when using a detection threshold of P = .05 (supplemental Table 2). Among these, we found that 901 were differentially expressed between the strains at a P = .05 stringency (supplemental Table 3), constituting a significant change in 5.3% of expressed stromal genes. Among the 901 differentially expressed genes, 556 are well annotated with known chromosomal locations. Surprisingly, only 37 of the 556 mapped to chromosome 1 (Figure 5A, supplemental Table 3). A more restricted P value cutoff of .01 returned 288 total genes or 1.3% of genes expressed in stroma. Among them, 170 have known chromosomal locations (Figure 5B, supplemental Table 4) and a majority are on chromosome 1 or 7. This relative enrichment for differentially expressed genes on chromosome 1 is expected given the location of the congenic interval. Differentially expressed genes on chromosome 7 and elsewhere are likely the result of polymorphic regulatory elements acting in trans to influence gene expression throughout the genome. Moreover, the genomic distribution of differentially expressed genes was not determined entirely by chromosome size (Figure 5). The correlation between the number of differentially expressed genes and mbp per chromosome was 0.11 and 0.16 for the gene lists created with a .05 and .01 statistical cutoff, respectively. In light of the heterogeneity of the cell sample used for microarray analysis, and the enrichment for differentially expressed transcripts on chromosome 1, we feel that the more restrictive P value of 01 is the most informative in this study, whereas results using the less stringent P value may be of value in pursuing specific candidate regulators of stromal cell gene expression.
Differentially expressed genes between B6 and B6.SJL stroma are distributed throughout the mouse genome. (A) Using a statistical threshold of P < .05, differentially expressed genes are concentrated on mouse chromosomes 11 and 17, and genomic distribution cannot be attributed only to chromosome size. (B) Using a statistical threshold of P < .01, differentially expressed genes are enriched on chromosomes 1 and 7 when displayed relative to chromosome number (top), and on chromosome 17 when displayed relative to chromosome size (middle). Correlation between chromosome size and the number of differentially expressed genes does not account entirely for genomic distribution.
Differentially expressed genes between B6 and B6.SJL stroma are distributed throughout the mouse genome. (A) Using a statistical threshold of P < .05, differentially expressed genes are concentrated on mouse chromosomes 11 and 17, and genomic distribution cannot be attributed only to chromosome size. (B) Using a statistical threshold of P < .01, differentially expressed genes are enriched on chromosomes 1 and 7 when displayed relative to chromosome number (top), and on chromosome 17 when displayed relative to chromosome size (middle). Correlation between chromosome size and the number of differentially expressed genes does not account entirely for genomic distribution.
Discussion
The results of this study emphasize the complex nature of the molecular pathways involved in HSC engraftment and the sensitivity of this process to seemingly subtle genetic modifications in either the donor or the recipient. We found that cotransplanting equal numbers of Ly-5.1 and Ly-5.2 cells produced balanced chimerism in B6.SJL hosts but a 30% to 50% relative deficit in Ly-5.1 cells in the lymphohematopoietic organs of B6 recipients. Detection of a unidirectional engraftment defect in simultaneous comparisons of cotransplanted Ly-5.1 and Ly-5.2 cells in both B6 and B6.SJL recipients suggests that the reason impaired engraftment was observed in Ly-5.1 into Ly-5.212,13 transplants, but not Ly-5.2 into Ly-5.1 transplants,14 is not limited to immunogenicity but reflects a fundamental difference between the 2 transplant models. Simultaneous analysis of the CRU frequency and homing efficiency of B6- and B6.SJL-derived cells transplanted into hosts of the opposite strain revealed that B6.SJL mice possess approximately 4-fold fewer LT-HSCs (15-17 week CRUs) than B6 and a 5-fold reduction in LT-HSCs than can be recovered 24 hours after transplant, constituting a 25% reduction in homing efficiency (Tables 1–2). Poor homing ability of B6.SJL cells is unique to LT-HSCs rather than progenitors that contribute earlier after transplant (Tables 1–2). This explains why the competitive disadvantage of Ly-5.1 cells remains undetectable in hosts repopulated with 2 × 106 cells of each allotype until 15 weeks after transplant, and is greatly amplified by 30 weeks after transplant (Figure 1A-B).
The fact that stem cell numbers between the 2 strains did not differ by in vitro measures suggests that HSC function, rather than absolute stem cell numbers, is what differs between the strains. The CAFC assay requires only that cultured cells be able to migrate under a stromal layer and proliferate to give rise to cobblestone-like clusters of cells, whereas transplantation assays require that cells successfully home to hematopoiesis-supporting sites in the bone marrow and, once there, maintain appropriate interactions with supportive niches. Our data suggest that HSCs from the 2 strains differ not in proliferative capacity or stem cell number but in the capacity for homing and/or the ability to establish optimal interactions with the HSC niche of the opposite strain.
The existence of polymorphic genes within the congenic interval such as cadherin 19 (Cdh19), chemokine receptor 4 (CXCR-4), engrailed 1 (En1), neuron navigator 1 (Nav1), and Slit-Robo GTPase-activating protein 2 (Srgap2) make altered stem cell–microenvironment interactions an attractive hypothesis. Cdh19 could, like N-cadherin, play an important role in communications between the stem cell and the niche.26,27 Numerous studies of HSC homing conducted in the last 10 years confirm that CXCR-4 plays a pivotal role at virtually every stage of homing and engraftment.28-30 En1 is involved in organization of the Drosophilia ovary stem cell niche31 and may have a similar function in regulation of adult stem cell niches. Nav1 is a microtubule-associated protein involved in neuronal migration and may influence hematopoietic cell migration.15 Slit-Robo GTPase-activating proteins, SrGAPs, mediate the signaling between Slit and Robo that orchestrate cytoskeletal rearrangements involved in cell migration.32 Slit-Robo signaling has recently emerged as a key mediator in migration of several cell types including neurons, leukocytes, and various cancer cell populations.33-36 The list of polymorphic genes in this interval provided in Table 3 almost certainly underestimates the number of polymorphisms between the 2 strains. Direct sequence analysis of other genes in this interval would likely reveal polymorphisms between the strains not found in the SNP Selector database. A notable example is the gene encoding Ly-5. Polymorphisms in Ly-5 are the basis for detecting the protein variants on hematopoietic cells, yet it was not identified as polymorphic by the in silico approach.11 Other genes with important implications in HSC biology are also located within the region. For example, the CD34 surface antigen, which is used to negatively select for murine HSCS, is also located within this interval and may too contain strain-specific sequence variations that were undetected in our analysis.
Given the subtlety of the phenotype and the numerous polymorphic genes within the B6.SJL congenic interval, it is likely that variations in expression or function of the product of these and/or other genes underlie engraftment differences between B6.SJL- and B6-derived HSCs. It is also possible that polymorphisms in Ly-5 influence HSC function, although this has never been demonstrated. Although Ly-5 is typically used as only a marker, it has important functions in the hematopoietic system, particularly in T-cell development37-39 and T-cell chemotaxis via CXCR-4.40
One might have expected substantial overlap between the list of genes containing SNPs (supplemental Table 3) and those identified as differentially expressed in stromal cells (supplemental Tables 3-4), although this was not the case. In fact, the only gene common to supplemental Tables 3 and 4 is Rnpep, or aminopeptidase B, which has no known role in engraftment or stem cell–microenvironment interactions. It is likely that polymorphisms between the 2 strains influence the bone marrow microenvironment at the level of protein structure or function, or by acting in trans to regulate gene expression throughout the genome. Our finding that differentially expressed genes are distributed throughout the genome supports this hypothesis (Figure 5). Identification of polymorphic transcription factors in the congenic interval and corresponding differentially expressed target genes may provide insight into regulatory pathways governing regulation of stromal gene expression. Congenic mice offer the advantage of reducing the possible genetic influences on a particular phenotype to a restricted portion of the genome. Although the B6.SJL congenic strain was not generated for such studies, it has allowed us to detect a stromal cell gene expression profile that is distinct from the parental B6 strain, although they differ genetically by only approximately 1% of the genome.
In aggregate, this work demonstrates that B6-derived Ly-5.2 cells and B6.SJL-derived Ly-5.1 cells differ in their capacity for engraftment of lethally irradiated B6 but not B6.SJL hosts, owing in part, if not entirely, to the 33% relative increase in B6 homing efficiency. The strains exhibit distinct patterns of gene expression in bone marrow stroma, and show that the B6.SJL congenic interval harbors at least 45 polymorphic SJL passenger genes. As the body of knowledge regarding interactions between stem cells and the niche expands, it will likely reveal several avenues by which hematopoietic stem cell engraftment is being influenced in the Ly-5 model. The Ly-5 marker system, however, will continue to serve as an excellent resource in these and other transplantation studies when appropriate experimental design is used. Many studies are designed such that Ly-5 markers distinguish only between donor and recipient cells, thus cells bearing alternate Ly-5 allotypes are not directly competed. Such a design eliminates the possibility that Ly-5 allotypic variation or polymorphic passenger loci confound the results. Because HSCs from B6 and B6.SJL do not contribute equally to engraftment of B6.SJL recipients, the selection of donor-recipient combinations must be considered in all Ly-5 transplant studies. During a time when reciprocal transplantation schemes are often used to distinguish between the biologic effects of HSCs versus cells of the niche, special precautions must be taken when using the Ly-5 marking system. Specifically, if transplantation studies are used to measure the effects of genetic modifications, age, or treatment in B6 or B6.SJL mice, they must be interpreted relative to unmanipulated controls studies performed according to the same transplantation scheme. In addition to the inherent differences between B6- and B6.SJL-derived HSCs and stroma, it is possible that experimental interventions influence the stem cells and/or microenvironment of B6 mice differently than B6.SJL congenic mice in significant ways.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We acknowledge the helpful discussions of genetic variation and bioinformatics resources provided by Robert W. Williams, assistance with statistical analysis provided by Tiantian Sun, and the technical assistance of Hung-Chi Liang and Shawn Smith in the preparation of this paper.
This work was supported by grants R36AG029669 (A.W.), R01HD060915, and R01AG024950 (G.V.Z.) from the National Institutes of Health, and funding from the Ellison Medical Foundation (G.V.Z.).
National Institutes of Health
Authorship
Contribution: A.W. contributed to the design of experiments, conducted experiments, analyzed results, and wrote the paper; Y.L. designed and conducted experiments and analyzed results; C.F.S. assisted in conducting experiments; B.J.S. performed statistical analyses; and G.V.Z. designed experiments, analyzed results, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gary Van Zant, Markey Cancer Center, Division of Hematology/Oncology, University of Kentucky, Rm cc414, 800 Rose St, Lexington, KY 40536-0093; e-mail: gvzant1@uky.edu.