Abstract
Spatially and temporally controlled expression of inflammatory mediators is critical for an appropriate immune response. In this study, we define the role for interferon regulatory factor 5 (IRF5) in secretion of tumor necrosis factor (TNF) by human dendritic cells (DCs). We demonstrate that DCs but not macrophages have high levels of IRF5 protein, and that IRF5 is responsible for the late-phase expression of TNF, which is absent in macrophages. Sustained TNF secretion is essential for robust T-cell activation by DCs. Systematic bioinformatic and biochemical analyses of the TNF gene locus map 2 sites of IRF5 recruitment: 5′ upstream and 3′ downstream of the TNF gene. Remarkably, while IRF5 can directly bind to DNA in the upstream region, its recruitment to the downstream region depends on the protein-protein interactions with NF-κB RelA. This study provides new insights into diverse molecular mechanisms employed by IRF5 to regulate gene expression and implicates RelA-IRF5 interactions as a putative target for cell-specific modulation of TNF expression.
Introduction
Tumor necrosis factor (TNF) is one of the major cytokines responsible for effector immune functions. As well as playing a central role in host defense against infection, TNF is a major factor in the pathogenesis of chronic inflammatory disease such as rheumatoid arthritis (RA). Consequently, tightly controlled regulation of its expression is critical for an appropriate immune response. This occurs at the transcriptional and posttranscriptional levels, with transcriptional regulation showing specificity for both stimulus and cell type.1 The NF-κB family of transcription factors (TFs) plays a major role in transcriptional up-regulation of the TNF gene by lipopolysaccharide (LPS) in both mouse and human myeloid cells.2-5
Regulation of transcription for many immune genes in response to Toll-like receptor (TLR) signaling involves a combination of NF-κB and interferon regulatory factor (IRF) factors.6 IRFs appear to provide a mechanism for conferring signal specificity to a variety of target gene subsets, with IRF3 being essential for type I interferon (IFN) response,7 and IRF5 playing a key role in induction of proinflammatory cytokines, including TNF, IL-6, and IL-12.8 Consequently, IRF5−/− mice show resistance to lethal shock induced by CpG-B or LPS.8
Unlike other IRF family members, IRF5 contains 2 nuclear localization signals (NLSs), 1 in the N-terminus and the other in the C-terminus of the protein. This results in low levels of nuclear translocation and therefore weak trans-activation activity of IRF5, even in unstimulated cells.9 The molecular pathways leading to IRF5 activation are not well understood, but it was shown that TLR signaling induces the formation of MyD88-IRF5-TRAF6 complexes,8 and is probably followed by phosphorylation of specific sites within the IRF5 C-terminal autoinhibitory domain.10
Human IRF5 is expressed as multiple spliced variants with distinct cell type–specific expression, cellular localization, differential regulation, and dissimilar functions.11 Moreover, genetic polymorphisms in the IRF5 gene leading to expression of several unique isoforms have been implicated in autoimmune diseases, including systemic lupus erythematosis (SLE), RA and Sjogren syndrome.12-15 IRF5 mRNA expression has been detected in B cells, dendritic cells (DCs), monocytes, and natural killer (NK) cells but not in T cells,11 yet little is known about the IRF5 protein expression in these cells.
Here, we demonstrate that human monocytes acquire high levels of IRF5 protein during differentiation into monocyte-derived DCs (MDDCs) but not monocyte-derived macrophages (MDMs). This leads to a sustained secretion of TNF by MDDCs compared with MDMs and efficient activation of T cells. IRF5 is recruited to both upstream and downstream regions of the gene after LPS induction, and its cooperative action with NF-κB RelA is important for maintaining the TNF gene transcription. Remarkably, IRF5 displays 2 independent modes of transcriptional activity: direct binding to DNA and indirect recruitment via the formation of a protein complex with RelA. Our results provide novel insights into the molecular basis for cell specificity in TNF production by human immune cells and highlight RelA-IRF5 interactions as a novel target for cell-specific modulation of TNF expression.
Methods
Plasmids
Expression constructs encoding full-length human IRF3, IRF5v3/v4, and NF-κB subunits tagged with HA-tag in modified pENTR vector (pBent) were described in Thomson et al.16 IRF5ΔDBD, IRF5A68P mutants were generated. The constructs were recombined into pAD/PL DEST vector (Invitrogen) for adenovirus production and subsequent delivery into human myeloid cells. The IRF5-HA fragment was subsequently transferred into the modified pBent vector containing 1-strep-tag. The 5′wt/3′wt and 5′wt/3′mut TNF luciferase-reporter constructs provided by Mr T. Smallie (Kennedy Institute) were used to generate 5′mut/3′wt and 5′mut/3′mut constructs with mutated sites κB2/2ξ/2a.5 IRF5 DBD (amino acid 1-131) were polymerase chain reaction (PCR) amplified and cloned into bacterial expression vector pET21d (Novagen). All constructs were verified by sequencing. The sequences and restriction maps are available upon request.
Cell culture
All reagents used for cell culture were tested for endotoxin and only used if the endotoxin levels were less than 20 pg/mL (Lonza). All cell cultures were maintained at 37°C in 5% CO2 and 95% humidity in the appropriate media supplemented with 10% fetal calf serum (Gibco) and 1% penicillin/streptomycin (PAA). HEK-293–TLR4-CD14/Md2 cells (Invivogen) were cultured in Dulbecco modified Eagle medium (DMEM; PAA) supplemented with 10 mg/mL blasticidin and 50 mg/mL HygroGold (Invivogen) per the manufacturer's instruction. Enriched populations of human monocytes were obtained from the blood of healthy donors by elutriation as described previously.4 MDMs and MDDCs were obtained after 5 to 7 days of culturing human monocytes in RPMI 1640 (PAA) supplemented with 100 ng/mL macrophage colony-stimulating factor (M-CSF) or 50 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF) and 10 ng/mL IL-4 (Peprotech). MDMs, MDDCs, and cell lines were stimulated with 100 ng/mL LPS (Alexis Biochemicals) unless indicated otherwise.
ELISA
Cytokine secretion was quantified with specific enzyme-linked immunosorbent assays (ELISAs) for human TNF (BD Bioscience), human IFN-γ (BD Bioscience), and human IFN-λ1/IL-29 (R&D Systems) according to manufacturer instructions. Absorbance was read at 450 nm by a spectrophotometric ELISA plate reader (Labsystems Multiscan Biochromic) and analyzed using Ascent Labsystems software. All samples were analyzed in triplicate in a volume of 50 μL.
Mixed lymphocyte reaction
Human MDDCs were plated in 96-well, flat-bottom tissue plates at 2 × 104 cells per well. T lymphocytes were isolated from the blood of healthy donors by elutriation, analyzed by FACS, and used if purity was more than 90%. T lymphocytes were added to MDDCs at 5 × 105 so that the final MDDC/T cell ratio was 1:25. Control cultures contained medium or T lymphocytes, or MDDCs alone. A total of 10 μg/mL anti-TNFR1 antibody (MAB 625; R&D Systems) or IgG control antibody (MAB 002; R&D Systems) was added to the cocultures after 6 hours or 24 hours. Cultures were established in duplicate and incubated at 37°C in 5% CO2 for a total of 72 hours. After culture, supernatants were collected and stored at −20°C for detection of cytokines.
RNA interference
siRNA-mediated knockdown was performed using On-target plus SMART pool reagents (Dharmacon) designed to target human IRF5 and NF-κB RelA. Lipofectamine RNAiMAX (Invitrogen) and DharmaFECT I (Dharmacon) were used as the siRNA transfection reagents for HEK-293–TLR4-Md2/CD14 cells and MDDCs, respectively, according to the manufacturers' instructions. Multiple siRNAs were used to validate the knockdown specificity and exclude off-target effects.
Adenoviral infection
Infections of MDMs were performed in 96-well plates in triplicate at a multiplicity of infection (MOI) of 50:1. Cells were seeded in serum-free, antibiotics-free RPMI containing the desired number of viral particles in a final volume of 50 μL. The plates were incubated overnight at 37°C followed by aspiration of the supernatants and replacement with 100 μL of standard media per well. Cells were allowed to recover for a further 24 hours before experimental assay.
RNA extraction and quantitative real-time RT-PCR
Total RNA was extracted from cells using a QiaAmp RNA Blood mini kit (QIAGEN) according to the manufacturer's instructions. cDNA was synthesized from total RNA using SuperScript III Reverse Transcriptase (Invitrogen) and 18-mer oligo dTs (Eurofins MWG Operon). The gene expression was analyzed by 2-standard curve or ΔΔCt methods where appropriate based on the quantitative real-time PCR with TaqMan primer sets for human TNF and PO (Applied Biosystems) in a Corbett Rotor-gene 6000 machine (Corbett Research Ltd).
Luciferase gene reporter assay
HEK-293–TLR4-CD14/Md2 cells were seeded into polylysine-coated 96-well plates at a density of 30 000 cells per well. Next day, cells were transfected with 10 ng of the indicated expression vector, 50 ng of TNF luciferase reporter, and 50 ng of pEAK8-Renilla using the Lipofectamine 2000 protocol (Invitrogen). Total amount of DNA was kept at 120 ng per well. At 48 hours after transfection, the activity of the reporters were measured using the Dual-Glo Luciferase system (Promega) optimized for 96-well plate format according to the manufacturer's protocol. Each experiment was performed in triplicate.
Nuclear and total protein extracts and Western
Cells were grown on 10-cm2 dishes and exposed to vehicle and agents; reactions were terminated by washing cells twice with ice-cold phosphate-buffered saline (PBS). Cells were then removed by scraping and transferred to Eppendorf tubes. Nuclear or total protein extracts were prepared as previously described.5 Equal amounts of proteins were resolved by Novex Tris-glycine gel (Invitrogen), transferred onto Hybond-N membranes (Amersham Biosciences), and subjected to incubation with antibodies against IRF5 (ab2932; Abcam), followed by detection with horseradish peroxidase (HRP)–conjugated secondary antibodies and the chemiluminescent substrate solution ECL (GE Healthcare).
EMSA
Oligonucleotide probes were radiolabeled with [α-32P]dCTP (Perkin Elmer): κB4 (Forward [F]: agctGGGCATGGGAATTTCCAACTCT; Reverse [R]: agctGAGTTGGAAATTCCCATGCCC); κB4a (F: agctAACTCTGGGAATTCCAATCCTT; R: agctAAGGATTGGAATTCCCAGAGT T); κB4b (F: agctCTTGCTGGGAAAATCCTGCAG; R: agctGCTGCAGGATTTTCCCAGCA AG); ISRE1 (F: agctGAAGCCAAGACTGAAACCAGCATTA; R: agctTAATGCTGGTTT CAGTCTTGGCTTC); ISRE2 (F: agctCCGGGTCAGAATGAAAGAAGAAGG; R: agctC CTTCTTCTTTCATTCTGACC CGGT); ISRE5 (F: agctGGAGAAGAAACCGAGACAGAAGG TG; R: agctCACCTTCTGTC TCGGTTTCTTCTCC). ‘ISRE’16 (F: agctTTTGCTTAGAAAAGAAACATGGTCTC; R: agctGAG ACCATGTTTCTTTTCTAAGCAAA); ‘ISRE’17 (F: agctACATAAACAAAGCCCAACAGAATAT TCC; R: agctGGAATATTCTGTTGGGCTTTGTTTATGT); and PRDI-III(IFN-β) (F: agctGGGAAACTG AAAGGGAAAGTGAAAGTGG; R: agctCCACTTTCACTTTCCCTTTCAGTTTCCC). Binding reactions with 50 ng of bacterially expressed and purified IRF5 DBD and electrophoretic mobility shift assay (EMSA) gel separation were performed as previously described.5
Chromatin immunoprecipitation
A total of 7 × 106 MDDCs or HEK-293-TLR4-CD14/Md2 cells were fixed by adding 1% formaldehyde (final concentration) for 5 minutes at room temperature. Nuclear extracts were subjected to 6 × 12-second pulses of sonication using Vibra-Cell VCX130 (Sonics) at 20% amplitude. For immunoprecipitation reaction, nuclear extracts were precleared with protein G–Sepharose bead slurry (GE Healthcare) for 2 hours, and then incubated with 2 μg of antibodies against IRF5 (ab2932; Abcam), RelA, or Pol II (sc-372 and sc-899; Santa Cruz Biotechnology) overnight at 4°C with rotation. Immunocomplexes were then collected with protein G–Sepharose beads for 30 minutes, rigorously washed, and eluted. For Re–chromatin immunoprecipitation (ChIP), RelA ChIP eluates were subsequently incubated with either IRF5 antibody or no antibody control and processed as described. Cross-linked protein-DNA complexes were reversed by incubating them at 65°C overnight, and DNA fragments were purified using the QIAquick PCR Purification Kit (QIAGEN). The immunoprecipitated DNA fragments were then interrogated by real-time PCR using SYBR Premix Ex Taq II master mix (Takara Bio) and the following primers for TNF locus: control region, (TGTGTGTCTGGGAGTGAGAACT and TCTTCTCAGCTTCTCCTTTGCT); region A, (CCACAGCAATGGGTAGGAGAATGT and GAGGTCCTGGAGGCTCTTTCACT); region B, (GGAAGCCAAGACTGAAACCAGCA and CCGGGAATTCACAGACCCCACT); region C, (TCCCTCCAACCCCGTTTTCT and TAGGACCCTGGAGGCTGAAC); region D, (AACTTTCCAAATCCCCGCCC and GGTGTGCCAACAACTGCCTT); region E, (CAGCAAGGACAGCAGAGGAC and TCCCGGATCATGCTTTCAGT); region F, (GGCAGTCAGTAAGTGTCTCCAA and TACCTACAACATGGGCTACAGG); region G, (ACAGCTTTGATCCCTGACATCT and CTCCGTGTCTCAAGGAAGTCTG); region H, (ATATTCCCCATCCCCCAGGAAACA and CTGCAACAGCCGGAAATCTCACC); region I, (GAGGACCTCACTCAGCCCTT and CGGCAGTTCGGTTCCTTGTT); region J, (ACTGGTCTTTGTGGTGAAGGAG and GAACTAGTGGGCTCAAGTGGTC); region K, (GCTATGATCATGCCACTGTACCC and TACCACATGGTTTTCTCCTGCC); and region L, (GCTGAAAGTCAGCCATGAAGTA and CACTTAGGGTGTCCCATTTAGG). Data were analyzed using Rotogene 6000 software (Corbett Research Ltd). All primer sets were tested for specificity and equal efficiency before use.
Immunoprecipiation
HEK-293–TLR4-CD14/Md2 cells were transfected with ONE-strep–IRF5-HA construct or corresponding empty vector. At 24 hours after transfection cells were fixed with 1% formaldehyde for 10 minutes at room temperature before high salt lysis and affinity purification on Strep-Tactin MacroPrep sepharose (IBA). The eluates were de–cross-linked by incubating at 65°C overnight before separation by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Exogenous IRF5 and endogenous RelA were detected by immunoblotting with anti-HA–HRP (12013819001; Roche) and anti-RelA (sc-372; Santa Cruz Biotechnology). Alternatively, cells were transfected with RelA-FLAG or BAP-FLAG control protein. At 24 hours after transfection, cells were lysed and affinity-purified with anti-FLAG M2 Sepharose beads (Sigma). Exogenous RelA and endogenous IRF5 were detected by immunoblotting with anti-FLAG–HRP (A8592; Sigma) and anti-IRF5 (ab2932; Abcam). Interaction of endogenous RelA and IRF5 was detected by overnight incubation of the cell lysates with goat anti-IRF5 antibody (ab2932; Abcam) or no antibody control before precipitation with protein G beads. IRF5 was detected by immunoblotting with mouse anti-IRF5 antibody (sc-56714; Santa Cruz Biotechnology), while RelA was detected by immunoblotting with anti-RelA. Triton X-100–extracted nuclei and DNase I digestion of chromatin was performed as described previously.17
Bioinformatics and statistical analyses
The nucleotide sequence were inspected with transcription factor binding site searching software JASPAR18 and Genomatrix19 for the presence of putative IFN-sensitive response element (ISRE) sites (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Statistical analysis was performed using 1-way analysis of variance (ANOVA) with Dunnett multiple comparison posttest or Student t test where appropriate (*P < .05; **P < .01; ***P < .001).
Results
Sustained TNF secretion by MDDCs is important for efficient T-cell activation
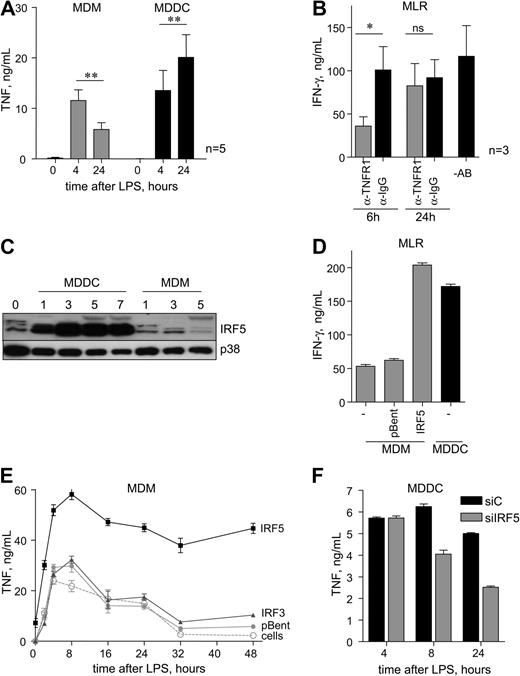
Myeloid cells (eg, macrophages and DCs) are the major producers of the key immune modulator TNF in response to TLR4 stimulation.20 TNF protein is below the limit of detection in the supernatants of resting cells (Figure 1A). After 4 hours of LPS stimulation, TNF is secreted at similar levels in MDMs and MDDCs (early phase). However, a marked difference in TNF production was observed in MDMs and MDDCs stimulated with LPS for 24 hours (late phase). Although the level of TNF significantly decreased in MDMs, there was an increase in TNF levels in MDDCs (Figure 1A) in each individual blood donor (supplemental Figure 1A).
IRF5 protein is highly expressed in MDDCs and control late-phase TNF secretion. (A) Monocyte-derived dendritic cells (MDDCs) and monocyte-derived macrophages (MDMs) were stimulated with lipopolysaccharide (LPS; 10 ng/mL) for 4 hours and 24 hours, and secreted tumor necrosis factor (TNF) was measured by ELISA. Data show means ± standard error of the mean (SEM) of 5 independent experiments each using monocytes derived from a different donor. **P < .01 (Student t test). (B) MDDCs were stimulated with LPS for 2 hours and then were cultured with T lymphocytes. Anti-TNFR1 or anti-IgG control antibodies were added 6 hours or 24 hours after coculture start. IFN-γ secretion was determined by ELISA after 72 hours of coculture. Data show means ± SEM of 3 independent experiments. *P < .05. (C) Cells were collected at day 0 (monocytes); days 1, 3, 5, and 7 (MDDCs) after differentiation with granulocyte-macrophage colony-stimulating factor (GM-CSF; 50 ng/mL) and IL-4 (10 ng/mL); and days 1, 3, and 5 after differentiation with M-CSF (50 ng/mL; MDMs); total protein extracts were then subjected to Western blot analysis. p38 MAPK was used as loading control. Representative blots of 5 independent experiments each using monocytes derived from a different donor. (D) MDMs were left untreated (cells) or infected with adenoviral vectors encoding IRF5 or empty vector (pBent), stimulated with LPS for 2 hours, and cultured with T lymphocytes. IFN-γ secretion was determined by ELISA after 72 hours of coculture. Data show means ± SD and are representative of 3 independent experiments each using MDMs derived from a different donor. (E) MDMs were left untreated (cells) or infected with adenoviral vectors encoding IRF5, IRF3, or empty vector (pBent) and stimulated with LPS for the indicated time. The amount of secreted TNF protein was determined by ELISA. Data show means ± SD and are representative of 3 independent experiments each using MDMs derived from a different donor. (F) MDDCs were transfected with siRNAs targeting IRF5 (siIRF5) and stimulated with LPS (10 ng/mL) for the indicated time. TNF secretion was compared with control cells transfected with nontargeting siRNA (siC). Data shown are the means ± SD and are representative of 2 independent experiments each using MDDCs derived from a different donor.
IRF5 protein is highly expressed in MDDCs and control late-phase TNF secretion. (A) Monocyte-derived dendritic cells (MDDCs) and monocyte-derived macrophages (MDMs) were stimulated with lipopolysaccharide (LPS; 10 ng/mL) for 4 hours and 24 hours, and secreted tumor necrosis factor (TNF) was measured by ELISA. Data show means ± standard error of the mean (SEM) of 5 independent experiments each using monocytes derived from a different donor. **P < .01 (Student t test). (B) MDDCs were stimulated with LPS for 2 hours and then were cultured with T lymphocytes. Anti-TNFR1 or anti-IgG control antibodies were added 6 hours or 24 hours after coculture start. IFN-γ secretion was determined by ELISA after 72 hours of coculture. Data show means ± SEM of 3 independent experiments. *P < .05. (C) Cells were collected at day 0 (monocytes); days 1, 3, 5, and 7 (MDDCs) after differentiation with granulocyte-macrophage colony-stimulating factor (GM-CSF; 50 ng/mL) and IL-4 (10 ng/mL); and days 1, 3, and 5 after differentiation with M-CSF (50 ng/mL; MDMs); total protein extracts were then subjected to Western blot analysis. p38 MAPK was used as loading control. Representative blots of 5 independent experiments each using monocytes derived from a different donor. (D) MDMs were left untreated (cells) or infected with adenoviral vectors encoding IRF5 or empty vector (pBent), stimulated with LPS for 2 hours, and cultured with T lymphocytes. IFN-γ secretion was determined by ELISA after 72 hours of coculture. Data show means ± SD and are representative of 3 independent experiments each using MDMs derived from a different donor. (E) MDMs were left untreated (cells) or infected with adenoviral vectors encoding IRF5, IRF3, or empty vector (pBent) and stimulated with LPS for the indicated time. The amount of secreted TNF protein was determined by ELISA. Data show means ± SD and are representative of 3 independent experiments each using MDMs derived from a different donor. (F) MDDCs were transfected with siRNAs targeting IRF5 (siIRF5) and stimulated with LPS (10 ng/mL) for the indicated time. TNF secretion was compared with control cells transfected with nontargeting siRNA (siC). Data shown are the means ± SD and are representative of 2 independent experiments each using MDDCs derived from a different donor.
Human TNF acting through TNF receptor is involved in DC maturation from bone marrow precursors21,22 and activation of type I helper (Th1), measured by the release of IFN-γ.23 Thus, we examined whether the late-phase secretion of TNF by MDDCs is needed for IFN-γ production by T cells. MDDCs were stimulated with LPS for 2 hours and exposed to human T cells extracted and purified from the peripheral blood of major histocompatibility complex (MHC)–unmatched donors in a mixed lymphocyte reaction (MLR). Antibodies against TNF receptor 1 (TNFR1) or isotype IgG control were added to the reaction. T cells incubated with MDDCs treated with anti-IgG antibodies produced high levels of IFN-γ (Figure 1B), whereas the control reactions (MDDCs or T cells cultured on their own) secreted no detectable IFN-γ. Blocking TNF at 6 hours after setting the MLR reaction resulted in strong reduction of IFN-γ, but no effect was observed when anti-TNFR1 antibodies were added to the reaction after 24 hours, suggesting that most T cells are in an activated state after the prolonged exposure to TNF (Figure 1B).
Thus, the observed sustained expression of TNF by MDDCs might be of benefit to both their maturation and antigen-presenting function and is essential for establishing a robust Th1 phenotype.
IRF5 protein is highly expressed in MDDC and controls late-phase TNF secretion
The observed differential LPS-induced secretion of TNF by human DCs and macrophages (Figure 1A) prompted us to examine the molecular mechanisms of this phenomenon. Myeloid cells from IRF5−/− mice show impaired induction of proinflammatory cytokines, including TNF, upon stimulation by different TLR ligands.8 We hypothesized that the difference in TNF secretion profile in MDDCs and MDMs might be due to the difference in IRF5 expression in these cells. We examined levels of IRF5 protein after human monocyte differentiation into MDMs and MDDCs. No increase in the levels of IRF5 protein was observed in MDMs, even after 5 days of differentiation (Figure 1C). However, expression of IRF5 protein was detected after 1 day of monocyte differentiation into MDDCs and remained at an elevated level until day 7 (Figure 1C). Significantly, whereas at least 3 different IRF5 isoforms were observed in human monocytes, only some of them accounted for high levels of IRF5 in MDDCs: one is likely to be IRF5v3/v4.11
Next, we looked at the effect of ectopic IRF5 expression in MDMs that have low levels of endogenous IRF5 protein (Figure 1C) on T-cell activation. MDMs were infected with adenoviral expression vector encoding HA-tagged IRF5 or the corresponding empty vector pBent. At 48 hours after infection, no significant effect on the resting cells (measured by endogenous IFN-λ1 response) was observed. Exposure of T cells to MDMs with elevated levels of IRF5 protein resulted in increase of IFN-γ secretion to the levels comparable with that of T cells exposed to MDDCs (Figure 1D). Thus, we argued that IRF5 might be responsible for sustained secretion of TNF. To test this hypothesis, MDMs were infected with adenoviral expression vector encoding HA-tagged IRF5 or IRF3 (as a control) or pBent. IRF5-HA and IRF3-HA vectors expressed similar levels of proteins (supplemental Figure 1B), but only IRF5 resulted in a significant increase in TNF secretion (Figure 1D; supplemental Figure 1C), whereas only IRF3 induced IFN-λ1 (supplemental Figure 1D), consistent with the previously published data.16 Strikingly, TNF secretion in MDMs with overexpression of IRF5 remained at a steady sustained level up to 48 hours after LPS stimulation (Figure 1E), similar to that of MDDCs with high levels of endogenous IRF5 (Figure 1C). siRNA-mediated inhibition of IRF5 in MDDCs (supplemental Figure 1E) resulted in reduction of TNF secretion at 8 and 24 hours after LPS stimulation (Figure 1F), supporting the notion that IRF5 may be required for the late-phase TNF expression.
Taken together, these results suggest that sustained TNF secretion by MDDCs leading to robust T-cell activation is likely to be a consequence of a high level of IRF5 protein in these cells.
IRF5 is involved in transcriptional regulation of TNF
We next sought to investigate whether IRF5 is involved in transcriptional regulation of the TNF gene. In human MDDCs, stimulation with LPS resulted in a rapid up-regulation of TNF mRNA expression, which reached the peak between 1 and 2 hours but remained at a steady level until 8 hours after stimulation (Figure 2A). Consistent with the observed differences in protein secretion, TNF mRNA expression in MDMs was characterized by more transient kinetics (supplemental Figure 2A), whereas siRNA-mediated inhibition of IRF5 reduced TNF mRNA expression (Figure 2A). The observed inhibition was statistically significant when analyzed in multiple blood donors (supplemental Figure 2B). In the same cells, siRNA-mediated inhibition of NF-κB RelA, a transcription factor previously shown to be important for an efficient TNF production by human MDDCs,24 resulted in reduction of TNF mRNA expression at the initial phase of gene induction (1-2 hours after LPS stimulation; Figure 2A). Within this time window, depletion of both IRF5 and RelA had the strongest effect on mRNA expression (Figure 2A), indicating that RelA and IRF5 may cooperate in controlling transcription of the TNF gene.
IRF5 is involved in transcriptional regulation of TNF. (A) MDDCs were transfected with siRNAs targeting IRF5 (siIRF5), RelA (siRelA), or both [si(IRF5+RelA)] and stimulated with LPS (10 ng/mL) for the indicated time. TNF mRNA expression was compared with control cells transfected with nontargeting siRNA (siC). Data shown are the means ± SD and are representative of 4 independent experiments each using MDDCs derived from a different donor. (B) HEK-293 cells were cotransfected with the TNF 5′wt/3′wt reporter plasmid and equal amounts of expression plasmids encoding for human IRF5, RelA, IRF3, or empty vector (pBent). At 48 hours after transfection, cells were harvested and luciferase activity was measured as described. Data are presented as a fold over pBent ± SEM from 4 independent experiments. *P < .05; ** P < .01 (1-way ANOVA).
IRF5 is involved in transcriptional regulation of TNF. (A) MDDCs were transfected with siRNAs targeting IRF5 (siIRF5), RelA (siRelA), or both [si(IRF5+RelA)] and stimulated with LPS (10 ng/mL) for the indicated time. TNF mRNA expression was compared with control cells transfected with nontargeting siRNA (siC). Data shown are the means ± SD and are representative of 4 independent experiments each using MDDCs derived from a different donor. (B) HEK-293 cells were cotransfected with the TNF 5′wt/3′wt reporter plasmid and equal amounts of expression plasmids encoding for human IRF5, RelA, IRF3, or empty vector (pBent). At 48 hours after transfection, cells were harvested and luciferase activity was measured as described. Data are presented as a fold over pBent ± SEM from 4 independent experiments. *P < .05; ** P < .01 (1-way ANOVA).
To investigate whether IRF5 can directly modulate transcription of the TNF gene, we used a gene-reporter plasmid in which the luciferase gene was flanked with 1171 nt 5′ upstream and 1252 nt 3′ downstream of the TNF gene. This construct encompassed all evolutionary conserved sequences in the region and contained known κB sites.25,26 It was coexpressed with HA-tagged IRF5, IRF3, and NF-κB subunits in HEK-293 cells, and luciferase activities were compared with empty vector pBent. RelA- and IRF5-transfected cells showed a significant increase in luciferase activity (Figure 2B). Other NF-κB subunits or IRF3 had little or no effect (Figure 2B; supplemental Figure 2C). Of interest, a deletion of the IRF5 DNA-binding domain (IRF5 ΔDBD) or a point alanine to proline mutation in it (IRF5 A68P) previously shown to act as dominant-negative mutants of IRF5,27,28 resulted in a major drop in luciferase activity (supplemental Figure 2D).
We concluded that IRF5 along with RelA is likely to be directly involved in the transcriptional regulation of the human TNF gene. Although the initial phase of TNF induction depends on both factors, only IRF5 appears to be crucial for maintaining prolonged TNF transcription in MDDCs. Moreover, the DBD of IRF5 is required for the optimal level of TNF gene up-regulation.
IRF5 is recruited to the 5′ upstream and 3′ downstream regions of the TNF gene in response to LPS stimulation
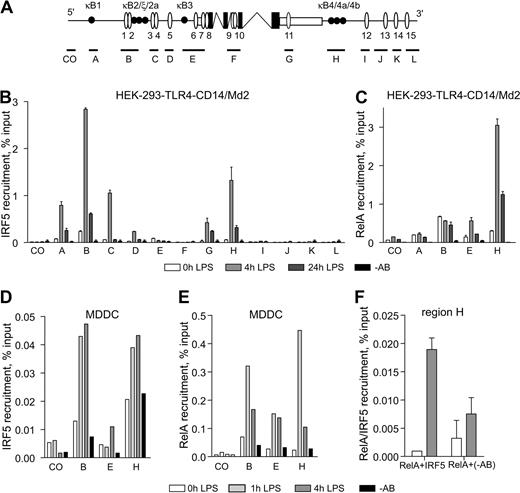
To further address the involvement of IRF5 in the TNF gene regulation, we systematically analyzed the recruitment of IRF5 to the TNF locus. A well-conserved N-terminal DBD of IRF factors recognizes a class of DNA sequences known as ISRE, 15 of which were computationally mapped to this locus together with known κB sites: κB1, κB2/ζ/2a, κB3, and κB4/4a/4b (Figure 3A; supplemental Table 1). A series of primers spanning the locus and encompassing ISRE sites was designed and used in the quantitative ChIP assay (Figure 3A).
IRF5 is recruited to the 5′ upstream and 3′ downstream region of TNF. (A) Schematic of the TNF locus. Protein coding and noncoding exons are shown in black and white. Putative ISREs are allocated as white ovals; κB sites are shown as black circles. The approximate amplicon size of primer sets spanning the TNF locus (A to L) are indicated by black lines. CO indicates a control primer set containing neither an ISRE nor a κB site. (B-C) HEK-293–TLR4-Md2/CD14 cells were left unstimulated or stimulated with LPS (1 μg/mL) for 4 hours and 24 hours and analyzed by ChIP with antibodies specific to IRF5 (B) or RelA (C). (D-E) MDDCs were left unstimulated or stimulated with LPS for 1 hour and 4 hours, followed by ChIP with antibodies specific to IRF5 (D) or RelA (E). (F) Corecruitment of RelA and IRF5 to region H was assessed by re-ChIP analysis with antibodies against RelA followed by IRF5-specific antibodies. (B-F) Data show mean percentage input relative to genomic DNA (gDNA) plus or minus SD of a representative experiment. −AB indicates a no-antibody control.
IRF5 is recruited to the 5′ upstream and 3′ downstream region of TNF. (A) Schematic of the TNF locus. Protein coding and noncoding exons are shown in black and white. Putative ISREs are allocated as white ovals; κB sites are shown as black circles. The approximate amplicon size of primer sets spanning the TNF locus (A to L) are indicated by black lines. CO indicates a control primer set containing neither an ISRE nor a κB site. (B-C) HEK-293–TLR4-Md2/CD14 cells were left unstimulated or stimulated with LPS (1 μg/mL) for 4 hours and 24 hours and analyzed by ChIP with antibodies specific to IRF5 (B) or RelA (C). (D-E) MDDCs were left unstimulated or stimulated with LPS for 1 hour and 4 hours, followed by ChIP with antibodies specific to IRF5 (D) or RelA (E). (F) Corecruitment of RelA and IRF5 to region H was assessed by re-ChIP analysis with antibodies against RelA followed by IRF5-specific antibodies. (B-F) Data show mean percentage input relative to genomic DNA (gDNA) plus or minus SD of a representative experiment. −AB indicates a no-antibody control.
HEK-293–TLR4-CD14/Md2 cells responsive to LPS were used to investigate the effect of LPS stimulation on recruitment of IRF5 to the TNF locus. Increased occupancy of IRF5 was observed at regions A, B, C, G, and H 4 hours after LPS stimulation, followed by a decrease after 24 hours (Figure 3B). Taking into consideration the average ChIP fragment size of around 500 bp and the proximity of the sequences amplified, some degree of overlap in regions A through C was inevitable and might have accounted for the observed symmetrical distribution of enrichment at regions A, B, and C. Whereas the enrichment of IRF5 signal at region B was expected due to the presence of putative ISRE 1/2 that can interact with IRF5 in vitro (supplemental Figure 3A), it was surprising to observe the recruitment of IRF5 at region H because this region contains no putative ISREs (Figure 3A). Moreover, we observed no IRF5-DNA binding at κB4/4a/4b binding sites in region H or at additional nonconsensus ‘ISRE’16 and ‘ISRE’17 sites in the vicinity of region H (supplemental Table 1; supplemental Figure 3A). We also investigated the recruitment of NF-κB RelA to the TNF locus and observed LPS-induced binding of RelA to regions B, E, and H (Figure 3C), which correlated with the distribution of multiple NF-κB–binding regions.
Next, we validated the pattern of IRF5 and RelA binding to the TNF locus in MDDCs stimulated with LPS for 0, 1, and 4 hours. Strong enrichment in both IRF5 and RelA recruitment was observed at regions B and H (Figure 3D-E), reproducible in 5 independent blood donors (supplemental Figure 3B). Importantly, LPS stimulation resulted in rapid transcription of the full-length nascent TNF transcript (estimated by recruitment of Pol II to the TNF 3′ downstream region H), which was robustly maintained at least up to 4 hours after stimulation (supplemental Figure 3D).
In summary, in response to LPS stimulation IRF5 along with RelA is efficiently recruited to the 5′ upstream and 3′ downstream regions of the human TNF gene. Significantly, the lack of putative ISRE binding sites in the 3′ downstream region of the gene strongly suggested that recruitment of IRF5 to this region might be mediated via its interactions with other TFs or accessory proteins.
IRF5 forms specific physical interactions with RelA
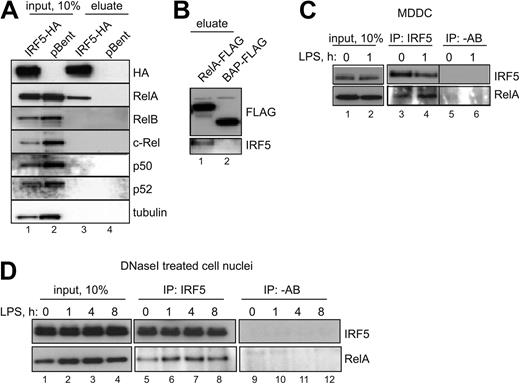
To tease out whether IRF5 recruitment to region H may be mediated via its interactions with RelA, we performed sequential ChIP analysis of the region and found that IRF5 recruitment was codependent on RelA after LPS stimulation (Figure 3F). This finding prompted us to investigate whether IRF5 and RelA interact physically.
IRF5 with an N-terminal ONE-strep–tag and a C-terminal HA-tag was expressed in HEK-293 cells. Figure 4 shows that in conditions similar to ChIP analysis (ie, in vivo cross-linking with formaldehyde), ectopically expressed IRF5, purified over a Strep-Tactin column, efficiently pulls down endogenous RelA (Figure 4A, compare lanes 3 and 4). To determine whether this interaction was specific, we immunoblotted for other NF-κB family members: Rel-B, c-Rel, p50 and p52, or a control protein tubulin. None of these resulted in a positive interaction (Figure 4A). Furthermore, we conducted a complementary experiment in which human RelA containing C-terminal FLAG tag was expressed in HEK-293 cells and immunoprecipitated in the absence of cross-linking agents on anti-FLAG Sepharose. Specific interactions between ectopically expressed RelA and endogenous IRF5 were observed. No interaction was detected between a control FLAG-tagged bacterial alkaline phosphatase (BAP) and IRF5 (Figure 4B, compare lanes 1 and 2).
IRF5 specifically interacts with RelA. (A) HEK-293–TLR4-Md2/CD14 cells were transfected with human IRF5 tagged with ONE-strep–tag (N-terminus) and HA-tag (C-terminus; lanes 1 and 3) or an empty vector pBent (lanes 2 and 4) and fixed with formaldehyde. Cross-links were reversed by heating and immunoblotted for bait IRF5 (anti-HA antibodies), or NF-κB subunits and tubulin. (B) HEK-293–TLR4-Md2/CD14 cells were transfected with RelA-FLAG (lane 1) or BAP-FLAG (lane 2). Cell lysates were immunoprecipated with M2 anti-FLAG Sepharose and immunobloted for bait RelA (anti-FLAG antibodies) or IRF5. (C) MDDCs were stimulated with LPS for 1 hour or left untreated. The endogenous interaction between RelA and IRF5 was examined by immunoprecipitation (IP) with anti-IRF5 antibody and immunoblotting with anti-RelA antibody. −AB indicates a mock IP. (D) Nuclear pellet from Triton-extracted HEK-293–TLR4-Md2/CD14 cells was solubilized with DNaseI, and endogenous interaction between RelA and IRF5 was examined after IP as in panel C.
IRF5 specifically interacts with RelA. (A) HEK-293–TLR4-Md2/CD14 cells were transfected with human IRF5 tagged with ONE-strep–tag (N-terminus) and HA-tag (C-terminus; lanes 1 and 3) or an empty vector pBent (lanes 2 and 4) and fixed with formaldehyde. Cross-links were reversed by heating and immunoblotted for bait IRF5 (anti-HA antibodies), or NF-κB subunits and tubulin. (B) HEK-293–TLR4-Md2/CD14 cells were transfected with RelA-FLAG (lane 1) or BAP-FLAG (lane 2). Cell lysates were immunoprecipated with M2 anti-FLAG Sepharose and immunobloted for bait RelA (anti-FLAG antibodies) or IRF5. (C) MDDCs were stimulated with LPS for 1 hour or left untreated. The endogenous interaction between RelA and IRF5 was examined by immunoprecipitation (IP) with anti-IRF5 antibody and immunoblotting with anti-RelA antibody. −AB indicates a mock IP. (D) Nuclear pellet from Triton-extracted HEK-293–TLR4-Md2/CD14 cells was solubilized with DNaseI, and endogenous interaction between RelA and IRF5 was examined after IP as in panel C.
Next, we examined whether an interaction between the endogenous RelA and IRF5 could be detected in MDDCs, and if this interaction may be induced by LPS stimulation. IRF5 was immunoprecipitated from the cells stimulated with LPS for 0 or 1 hour using anti-IRF5 antibodies. The Western blot for RelA revealed a specific interaction with IRF5 (Figure 4C). A densitometry analysis of quantities of the bait and target proteins indicated that the quantity of RelA bound to IRF5 was somewhat higher in LPS-stimulated cells (Figure 4C lane 4).
Finally, we asked the question whether the observed RelA-IRF5 interactions are dependent on the simultaneous binding of both TFs to DNA (ie, RelA and IRF5 interact only when bound to corresponding κB and ISRE binding sites in proximity to each other). To address this, we extracted nuclei from HEK-293–TLR4-CD14/Md2 cells stimulated with LPS for 0, 1, 4, and 8 hours and subjected the chromatin to DNaseI digestion. Subsequent precipitation of endogenous immune complexes with anti-IRF5 antibodies revealed that RelA interacted with IRF5 even in the absence of DNA bridging (Figure 4D lanes 5-8). Once again, the number of RelA-IRF5 complexes increased with LPS stimulation, corresponding to the rise in nuclear RelA (Figure 4D lanes 1-4).
In summary, IRF5 can specifically interact with RelA but not other 4 NF-κB subunits. This interaction is not dependent on IRF5 binding to DNA, and the quantity of RelA-IRF5 complexes is increased in response to LPS stimulation. Thus, we hypothesized that IRF5 recruitment to the 3′ downstream region of the TNF gene lacking putative ISRE sites is a consequence of direct physical interactions between DNA-bound RelA and IRF5.
RelA is required for IRF5-dependent trans-activation of the TNF gene
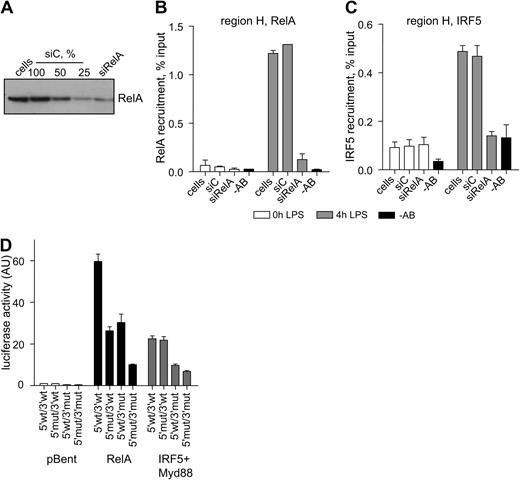
To test this hypothesis, we first analyzed IRF5 recruitment to the TNF locus in the cells in which the levels of RelA were significantly reduced. In HEK-293–TLR4-CD14/Md2 cells, siRNA-mediated knockdown of RelA resulted in approximately 75% reduction in RelA protein (Figure 5A) and about a 10-fold decline in its recruitment to region H after 4 hours of LPS stimulation (Figure 5B). As predicted, the IRF5 recruitment to the same region was prevented (Figure 5C). Of interest, when we analyzed RelA and IRF5 recruitment to region B in RelA-depleted cells, we observed only partial reduction in IRF5 recruitment (supplemental Figure 4), consistent with the view that IRF5 can bind directly to DNA at this region (supplemental Figure 3A).
RelA is required for IRF5-mediated activation of TNF. (A-C) HEK-293–TLR4-Md2/CD14 cells were transfected with siRNA against RelA (siRelA) or with nontargeting siRNA (siC) and used in ChIP analysis of RelA and IRF5 recruitment. Data indicate mean percentage input relative to gDNA ± SD of a representative experiment. −AB indicates a no-antibody control. (A) A total of 75% of RelA protein was degraded estimated by serial dilutions of the siC control sample analyzed by Western blotting. (B) Reduction in LPS-induced RelA recruitment to region H in siRelA-treated cells. (C) Reduction in LPS-induced IRF5 recruitment to region H in siRelA-treated cells. (D) HEK-293–TLR4-Md2/CD14 cells were transfected with the RelA, IRF5, and MyD88 expression constructs together with the TNF 5′ upstream/luciferase/TNF 3′ downstream reporter plasmids: 5′wt/3′wt indicates wild-type construct; 5′mut/3′wt indicates mutated κB2 (GTGAATTCCC → tTGAATTCCC), κBξ (GTGATTTCAC → aTccTTTCAC), and κB2a (GGGCTGTCCC → taGCTGTGCCC) sites in the TNF 5′ upstream; 5′wt/3′mut indicates mutated κB4 (GGGAATTTCC → cGcAATgTgC) and κB4a (GGGAATTCCA → cGcAAgTgCA) sites in the TNF 3′ downstream; and 5′mut/3′mut indicates all κB sites mutated. Data show means ± SD and are a representative of 3 independent experiments, each performed in triplicate.
RelA is required for IRF5-mediated activation of TNF. (A-C) HEK-293–TLR4-Md2/CD14 cells were transfected with siRNA against RelA (siRelA) or with nontargeting siRNA (siC) and used in ChIP analysis of RelA and IRF5 recruitment. Data indicate mean percentage input relative to gDNA ± SD of a representative experiment. −AB indicates a no-antibody control. (A) A total of 75% of RelA protein was degraded estimated by serial dilutions of the siC control sample analyzed by Western blotting. (B) Reduction in LPS-induced RelA recruitment to region H in siRelA-treated cells. (C) Reduction in LPS-induced IRF5 recruitment to region H in siRelA-treated cells. (D) HEK-293–TLR4-Md2/CD14 cells were transfected with the RelA, IRF5, and MyD88 expression constructs together with the TNF 5′ upstream/luciferase/TNF 3′ downstream reporter plasmids: 5′wt/3′wt indicates wild-type construct; 5′mut/3′wt indicates mutated κB2 (GTGAATTCCC → tTGAATTCCC), κBξ (GTGATTTCAC → aTccTTTCAC), and κB2a (GGGCTGTCCC → taGCTGTGCCC) sites in the TNF 5′ upstream; 5′wt/3′mut indicates mutated κB4 (GGGAATTTCC → cGcAATgTgC) and κB4a (GGGAATTCCA → cGcAAgTgCA) sites in the TNF 3′ downstream; and 5′mut/3′mut indicates all κB sites mutated. Data show means ± SD and are a representative of 3 independent experiments, each performed in triplicate.
Next, we examined the effect of site-specific mutations in the κB sites on the ability of IRF5 to activate the TNF gene. A panel of 4 gene-reporter constructs was used in this analysis: (1) 5′wt/3′wt (as in Figure 2B); (2) 5′mut/3′wt (mutated κB2/2ξ/2a sites in the TNF 5′ upstream); (3) 5′wt/3′mut (mutated κB4/4a sites in the 3′ TNF downstream); and (4) 5′mut/3′mut (mutated all κB sites just described). The reporter constructs were coexpressed with HA-tagged IRF5 and RelA in HEK-293 cells, and luciferase activities were compared with empty vector pBent. As expected, removal of either 5′ upstream or 3′ downstream κB sites diminished the ability of RelA to drive the gene-reporter activity (Figure 5D). However, the trans-activation of the reporter constructs by IRF5 (supported by ectopically expressed Myd88) appeared to be largely unaffected by mutations in the 5′ upstream κB sites, suggesting that IRF5 does not use κB2/2ξ/2a sites for its binding to the TNF 5′ upstream and is likely to involve the identified ISRE 1 and 2 sites. However, the trans-activation of the reporter construct with mutations in κB4/4a sites by IRF5 was significantly reduced, indicating that IRF5 activity depends on NF-κB binding to this region (Figure 5B). Low amounts of endogenous RelA detected in the nuclei of resting HEK-293–TLR4-CD14/Md2 cells (data not shown) appeared to provide a necessary DNA anchor for IRF5.
Thus, IRF5 recruitment to the TNF 3′ downstream region is mediated by way of a complex assembly with RelA and does not involve a direct contact to DNA. Importantly, another mode of function of IRF5 in TNF regulation is a direct recruitment to the TNF gene 5′ upstream. The 2 functional modes also imply the possibility of a higher order enhancer structure at the TNF locus, possibly involving IRF5-RelA–mediated intrachromosomal looping.
Discussion
Production of the key immune modulator TNF is both cell and stimulus specific. Myeloid cells are the major producers of TNF in response to TLR4 stimulation.20 Consequently, a tight control of the amount and duration of TNF expression by these cells is critical for a self-limited immune response. Here, we aimed to understand the molecular bases of differential TNF expression in human DCs and macrophages. We demonstrate that IRF5 appears to be a defining factor in maintaining the TNF gene transcription in MDDCs. Remarkably, we unravel a complex molecular mechanism used by IRF5 to control the human TNF gene expression: 2 spatially separated regulatory regions and 2 independent modes of actions are involved.
IRF5 is highly expressed in MDDCs but not other myeloid cells (Figure 1). During differentiation MDDCs acquire a particular phenotype, characterized among other markers by higher levels of RelB and c-Rel.29 Important for understanding the mechanisms of sustained TNF expression, RelB was previously shown to replace RelA at the promoters of macrophage-derived chemokine and Epstein-Barr virus (EBV)–induced molecule 1 ligand chemokine genes and to prolong their transcription in MDDCs.30 We also observed an increase in the RelB and c-Rel levels during monocyte differentiation into MDDCs, but not into MDMs (supplemental Figure 5A). However, neither RelB nor c-Rel was able to drive transcription of TNF (supplemental Figure 2), whereas RelA, whose level was similar in all human myeloid cell types (supplemental Figure 5A), had a strong trans-activating effect (Figure 2). This led us to conclude that RelA was likely to participate in the initial phase of TNF activation, which is indistinguishable between MDDCs and MDMs, but other MDDC-specific factors, such as IRF5, may contribute to the observed extended expression of TNF in MDDCs. Indeed, forced expression of IRF5 in MDMs led to prolonged TNF secretion (Figure 1), whereas depletion of IRF5 in MDDCs resulted in reduction of TNF expression, particularly at a later time (4 hours) after LPS stimulation (Figure 2; supplemental Figure 2). Although we cannot formally rule out other factors that might feed into the TNF expression system at a later time, the ability of IRF5 to activate the TNF gene-reporter construct (Figure 2) and its efficient recruitment to the TNF locus (Figure 3) strongly suggest a direct role for IRF5 in TNF gene induction in response to LPS.
TNF is an early primary response gene whose mRNA expression in MDDCs is induced approximately 100-fold within 30 minutes after LPS treatment (supplemental Figure 2). The genomic locus encompassing the TNF gene is open to regulatory proteins and in murine bone marrow–derived macrophages (BMDMs) does not require nucleosome remodeling complexes for its activation.31 Consistent with this notion, we find a significant accumulation of Pol II molecules at the transcription start site (TSS) of the gene even in resting MDDCs (supplemental Figure 6), akin to the results obtained in mouse BMDMs.32,33 LPS stimulation, however, results in a robust recruitment of RelA and IRF5 to both 5′ upstream region B and 3′ downstream region H (Figure 3) and in a significant induction of Pol II recruitment to the 3′ downstream region of the gene (supplemental Figure 6). This suggests an increase in production of full-length nascent TNF transcripts upon LPS stimulation of MDDCs, in addition to induction of splicing of already generated nascent transcripts reported by Hargreavas et al.32
The recruitment of IRF5 to the 5′ upstream region is likely to involve direct binding to DNA via the identified ISRE sites (supplemental Figure 3), whereas the recruitment of IRF5 to the 3′ downstream region is mediated via protein-protein interactions with RelA (Figure 4). These interactions are induced after stimulation of MDDCs with LPS, while no other NF-κB subunits appear to complex with IRF5 (Figure 4). Previous studies demonstrated that IRF3, another member of the IRF family, forms in vitro interactions with RelA via its Rel homology domain (RHD).34 Considering that the RHD is a highly conserved domain present in all NF-κB proteins, the exclusiveness of IRF5 interactions with RelA is somewhat surprising. Further work is needed to map the interface of RelA-IRF5 interactions.
Regions B and H are characterized by high level of sequence conservation,25,35 and contain cell type–specific DNaseI hypersensitivity sites.35,36 Moreover, the TNF 5′ upstream and 3′ downstream regions have been shown to physically interact by forming an intrachromosomal loop, the topology that could promote the reinitiation of transcription.35 This model may be of a particular relevance to TNF expression by MDDCs, in which a cooperative action of RelA and IRF5 at both the 5′ upstream and downstream regions appears to be essential for maintaining TNF gene transcription over a prolonged period of time. Here, the locus circularization may be directed via newly unraveled protein-protein interactions between RelA and IRF5 (Figure 4). The observed DNA-binding–independent corecruitment of IRF5 to the 3′ downstream region (Figure 5) further supports the possibility of high-order enhancer structure at the locus (Figure 6).
A model for IRF5-RelA–mediated induction of TNF in myeloid cells. LPS-induced recruitment of NF-κB RelA-containing complexes (gray ovals) to the 5′ upstream and 3′ downstream regions leads to transient TNF expression in MDMs. The mechanisms of transmitting the activating signal from 3′ downstream to Pol II requires further investigation. IRF5 binding to DNA at the 5′ upstream and to RelA at the 3′ downstream establishes the possibility for region circularization and recycling of Pol II molecules, leading to sustained TNF expression in MDDCs.
A model for IRF5-RelA–mediated induction of TNF in myeloid cells. LPS-induced recruitment of NF-κB RelA-containing complexes (gray ovals) to the 5′ upstream and 3′ downstream regions leads to transient TNF expression in MDMs. The mechanisms of transmitting the activating signal from 3′ downstream to Pol II requires further investigation. IRF5 binding to DNA at the 5′ upstream and to RelA at the 3′ downstream establishes the possibility for region circularization and recycling of Pol II molecules, leading to sustained TNF expression in MDDCs.
Why is TNF secretion maintained for longer in MDDCs than in MDMs (Figure 1)? DCs are professional antigen-presenting cells (APCs) that are crucial for both innate and adaptive responses to infection. They sense invading pathogens and respond by secreting various cytokines as well as by up-regulating the expression of MHC II and costimulatory molecules, essential for efficient antigen presentation to T cells.37 The mature DCs migrate to the draining lymph nodes, where they initiate Th1 differentiation. TNF acting through the TNF receptor is involved in DC maturation from bone marrow precursors.21,22 A recent study demonstrated that TNF blockade impaired DC survival and function in RA.38 Our data showing that TNF produced by DCs is a key factor in human Th1 activation support this study. Moreover, it is the late-phase TNF secretion that is needed to achieve the full activation potential (Figure 1). Macrophages, on the other hand, do not migrate to the draining lymph nodes but accumulate in large numbers at a site of inflammation, secrete inflammatory cytokines, and attract other immune cells via chemotaxis.39 Thus, a mechanism which would restrain the degree and duration of TNF secretion by macrophages would be important for ensuring the resolution of acute inflammatory response, thereby limiting tissue damage.
Another question is how IRF5 is activated in MDDCs by TLR4 signaling. Takaoka et al demonstrated that ectopically expressed IRF5 translocates to the cell nuclei in response to LPS, and that this translocation is dependent on the presence of Myd88.8 We observed endogenous nuclear IRF5 even in resting MDDCs or HEK-293–TLR4-CD14/Md2 cells, and its level was not increased after LPS stimulation (supplemental Figure 5B), although we could not exclude the possibility of active nuclear export-import of IRF5 induced by phosphorylation at the previously described serine residues40 due to the lack of phosphospecific antibodies. In the same cells, endogenous IRF3 showed a clear pattern of induced nuclear translocation (supplemental Figure 5B), which corresponded to its phosphorylated form (data not shown). It is worth noting, however, that an ectopically expressed mutant of IRF5 in which the described critical serines at positions 427 and 4309,40 were substituted with alanines was still transcriptionally active in the TNF reporter assay (G.R., unpublished data, 2009), suggesting that IRF5 may not need to be phosphorylated at these residues to activate transcription. We also did not observe any loss in IRF5 trans-activating potential when lysines 401 and 402, implicated in another Myd88-induced posttranslational modification of IRF5, K63-linked polyubiquitination,41 were substituted with arginines (G.R., unpublished data, 2009). Because the overexpression data generated in the HEK-293 cell line may be misleading, we plan to test these and other mutants of IRF5 in complementation experiments in the cells from IRF5-deficient mice to elucidate the impact of these mutations on IRF5 activation and function in vivo.
Regulation of IRF5 activity is an important issue because the excessive activation of this protein may lead to pathology. Interestingly, another member of the IRF family, IRF4, was shown to act as a negative regulator of TLR signaling by inhibiting the production of selected IRF5-dependent genes, including TNF, via direct competition with IRF5 for interactions with Myd88.42 In mice, IRF4 was observed to be differentially expressed in DCs and regulate the development of a specific DC subset, conventional DCs.43 In humans, IRF4 was also found to be expressed in MDDCs but not MDMs,44 suggesting that a self-controlled IRF5-IRF4 regulatory system might have developed to finely modulate TLR signaling pathways and production of IRF5-dependent inflammatory cytokines.
In summary, sustained TNF secretion in human MDDCs is mediated by cooperative action of IRF5 and RelA at the 5′ upstream and 3′ downstream regions of the TNF gene. TLR4 stimulation induces protein-protein interactions between RelA and IRF5 and allows for DNA-independent recruitment of IRF5 to the TNF 3′ downstream region. IRF5 may assist in formation of a high-order enhancer structure linking together the regulatory regions in the TNF 5′ upstream and 3′ downstream and allowing for maintaining of transcription over a longer time (Figure 6). Based on the resistance of IRF5−/− mice to lethal endotoxic shock, impaired production of proinflammatory cytokines, and deficiency in Th1 immune response, IRF5 was proposed as a target for therapeutic interventions.8 Here, we define RelA-IRF5 interactions as a putative target for cell-specific modulation of TNF expression and possible other selected inflammatory mediators.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to Dr Daniel Wong (Wellcome Trust Centre for Human Genetics, Oxford, United Kingdom) for help with IRF5 DBD expression and purification, Mr Tim Smallie (Kennedy Institute of Rheumatology [KIR], London, United Kingdom) for providing 5′wt/3′wt and 5′wt/3′mut TNF gene-reporter constructs, and Dr Fui G. Goh and Mr Scott Thomson (KIR) for advice and useful suggestions on RNAi and ChIP. We also thank Drs Jonathan Dean, Matt Pierce, and Lynn Williams (KIR) for critical reading of the manuscript and helpful comments.
The research leading to these results was supported by MRC collaborative grant 82189 to I.A.U. and has received funding from the European Community's Seventh Framework Program FP7/2007-2013 under grant agreement no. 222008.
Authorship
Contribution: T.K., D.S., G.R., K.B., and A.L. performed research; T.K., D.S., and I.A.U. designed research and analyzed data; and T.K., D.S., and I.A.U. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Irina A. Udalova, Kennedy Institute of Rheumatology Division, Faculty of Medicine, Imperial College of Science, Technology and Medicine, 65 Aspenlea Rd, London, W6 8LH, United Kingdom; e-mail: i.udalova@imperial.ac.uk.
References
Author notes
T.K. and D.S. contributed equally to this work.

![Figure 2. IRF5 is involved in transcriptional regulation of TNF. (A) MDDCs were transfected with siRNAs targeting IRF5 (siIRF5), RelA (siRelA), or both [si(IRF5+RelA)] and stimulated with LPS (10 ng/mL) for the indicated time. TNF mRNA expression was compared with control cells transfected with nontargeting siRNA (siC). Data shown are the means ± SD and are representative of 4 independent experiments each using MDDCs derived from a different donor. (B) HEK-293 cells were cotransfected with the TNF 5′wt/3′wt reporter plasmid and equal amounts of expression plasmids encoding for human IRF5, RelA, IRF3, or empty vector (pBent). At 48 hours after transfection, cells were harvested and luciferase activity was measured as described. Data are presented as a fold over pBent ± SEM from 4 independent experiments. *P < .05; ** P < .01 (1-way ANOVA).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/115/22/10.1182_blood-2010-01-263020/4/m_zh89991052850002.jpeg?Expires=1767706684&Signature=acns4OZVQVDmz~b1aj1n~TAtWhczSrTKbbkcoTZk0QbmdFf27-9YGLsSrW9kD2djs2Qmhwdlr57~59dhptkQ33UJZLM~QTFpkVEzIV0mIS5pn5E~an2kxJZz4oBW~KMridNsihOweFL8AgBBmr6KpWNWCM26b9L0nGKbdWRLwp3rimfGyyLNXith5I4ZSaGGgDPJgQZ14o1~QNgiZIKUvr7KxAKxIznbVu5MVaBGWzAL774IM5ABaAu69a6MiG0lZe~PYJkOIVsSPweAnQL-yFuwxNAdDFYxnQb43VKdrgZZJ9rPFKBKhGsiAA9GemY0nOlCWJBU3EdZzOv5ysT2Og__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)