Abstract
Recent studies have shown that anemia is commonly observed after exposure to pathogens or pathogen-derived products, which are recognized via Toll-like receptor 9 (TLR9). In the current study, we demonstrate that CpG oligodeoxynucleotide-2006, a TLR9 ligand with phosphodiester (PO; 2006-PO) but not with the phosphorothioate backbone, selectively inhibits the erythroid growth derived from human CD34+ cells. The 2006-PO was internalized by the erythroid progenitors within 30 minutes; however, expression of TLR9 mRNA was not detected in these cells. The 2006-PO directly inhibited burst-forming unit-erythroid growth, resulted in the accumulation of cells in S and G2/M phases, and increased cell size and frequency of apoptotic cells. These features were similar to those observed in erythroid progenitors infected with human parvovirus B19 that causes pure red cell aplasia. The consensus sequence of 2006-PO was defined as 5′-GTTTTGT-3′, which was located in the P6-promoter region of B19 and inhibited erythroid growth in a sequence-specific manner and down-regulated expression of erythropoietin receptor (EPOR) mRNA and EPOR. B19 genome extracted from serum also inhibited erythroid growth and down-regulated expression of EPOR on glycophorin A+ cells. These results provide a possible insight into our understanding of the mechanisms of human parvovirus B19-mediated inhibition of erythropoiesis.
Introduction
Recent studies have shown that anemia is commonly observed after exposure to pathogens or pathogen-derived products,1-4 which are recognized via Toll-like receptor 9 (TLR9). TLR9 has evolved to recognize unmethylated CpG (cytosine linked to a guanine by a phosphate bound) dinucleotides that are relatively common in bacterial and viral genomic DNA, but not in vertebral genomes.5 CpG DNA is generally most active as synthetic single-stranded (ss) oligodeoxynucleotide (ODN) sequences 20 to 30 nucleotides long, containing 2 to 3 CpG motifs with a modified nuclease-resistant backbone, typically a phosphorothioate (PS) backbone in which one of the nonbridging oxygen atoms at each of the natural (wild) phosphodiester (PO) linkages is replaced with a sulfur.5 Of note is that CpG ODN with different backbones and different sequence motifs can induce dramatically different profiles and kinetics of immune activation.6-12
Recently, several lines of evidence have demonstrated a direct link of TLRs and hematopoiesis.13,14 Unlike the ligands for TLRs 2, 4, 7, and 8, the role of CpG-ODN on hematopoiesis has been reported to be indirect via accessory cells. Sparwasser et al15 demonstrated that unmethylated CpG-ODN 2006 triggered extramedullary hematopoiesis by promoting expansion of granulocyte-macrophage colony forming units (CFUs) and early erythroid progenitors, and that enhanced splenic hematopoiesis is the result of CpG-ODN–induced cytokines that mobilize bone marrow progenitor cells to the spleen. Subsequently, Thawani et al16 showed that in vivo administration of unmethylated CpG-ODN exerted anemia in mice. They reported that this CpG-ODN–induced suppression was indirect and required accessory cells, including antigen-presenting cells (APCs), which activated other cells to produce proinflammatory cytokines, and IFN-γ was the major factor mediating erythropoietic suppression. However, the presence of TLR9 on human CD34+ cells, including their progenies and the biologic difference of CpG-ODN backbones, has remained unclear.
Human parvovirus B19 is a small, ssDNA virus that lacks an envelope and is characterized by its target specificity for human erythroid-lineage cells. This specificity is in part the result of the distribution of its receptor known as the P antigen globoside,17,18 which, however, is present not only on erythroblasts but also on megakaryocytes, endothelial cells, synovium, villous trophoblast cells of placental tissues, fetal liver, and heart cells.17,19 Therefore, the precise mechanisms underlying B19-DNA–mediated selective inhibition of erythroid cells remain unclear. It has been reported previously that B19-induced apoptosis and cell-cycle arrest are mediated by nonstructural protein NS1.20-22 However, the G2 arrest has been shown to be induced by UV-inactivated B19.20 These findings suggest that the B19-mediated G2 arrest is induced by B19 DNA in the absence of expression of the viral genome or its proteins.
In the current study, we aimed to precisely characterize the role of the best-characterized ligand for TLR9, CpG-ODN 2006, during hematopoiesis focusing on different backbones. We report that CpG-ODN 2006, containing a PO backbone rather than a PS backbone, shares a consensus sequence with B19 genome and that the consensus sequence selectively inhibits growth and development of human erythroid progenitor cells.
Methods
Reagents
Bovine serum albumin (BSA), Iscove modified Dulbecco medium (IMDM), and propidium iodide were purchased from Sigma-Aldrich. Fetal bovine serum was from HyClone. Penicillin and streptomycin were from Invitrogen. Insulin (porcine sodium, activity 28.9 U/mg) and Triton X-100 were obtained from Wako Pure Chemical Industries. Diaminobenzidine-substrate chromogen and fuchsin-substrate chromogen systems were from Dako Denmark. Interleukin-3 (IL-3), stem cell factor (SCF), and thrombopoietin (TPO) were kind gifts of the Kirin Brewery Co Ltd, and erythropoietin (EPO) and granulocyte colony-stimulating factor (G-CSF) were from Chugai Pharmaceutical Co. Vitamin B12 was from Eisai Co Ltd, and folic acid was from Takeda Pharmaceutical Co Ltd. RNase (type III-A) was from Sigma-Aldrich. LysoTracker and carboxyfluorescein diacetate succinimidyl ester (CFSE) were from Invitrogen. MACS MicroBeads for Indirect Magnetic Labeling was from Miltenyi Biotec. IODO-GEN (1,3,4,6-tetrachloro-3α, 6α-diphenylglycouril) was from Thermo Scientific, and Na 125I was from PerkinElmer Life and Analytical Sciences.
ODN
The specific sequences of ODN used in this study are presented in Table 1. CpG-ODN 2006 (2006-PO), ODN 2137 (2137-PO), and ODN 2243 (2243-PO) all with PO backbone were commercially synthesized by FASMAC and Hokkaido System Science Co Ltd. CpG-ODN 2006 with PS backbone (2006-PS) and ODN 5′ labeled with rhodamine were commercially synthesized by Hokkaido System Science Co Ltd.
CpG ODN tested in this study
| Oligo name . | Sequence . |
|---|---|
| 2006-PO | TCGTCGTTTTGTCGTTTTGTCGTT |
| 2137-PO | TGCTGCTTTTGTGCTTTTGTGCTT |
| 2243-PO | GGGGGAGCATGCTGGGGGGG |
| 2006-PS | TCGTCGTTTTGTCGTTTTGTCGTT |
| Oligo name . | Sequence . |
|---|---|
| 2006-PO | TCGTCGTTTTGTCGTTTTGTCGTT |
| 2137-PO | TGCTGCTTTTGTGCTTTTGTGCTT |
| 2243-PO | GGGGGAGCATGCTGGGGGGG |
| 2006-PS | TCGTCGTTTTGTCGTTTTGTCGTT |
2006-PO indicates CpG ODN 2006 with PO backbone; 2137-PO, CpG ODN 2137 with PO backbone; 2243-PO, CpG ODN 2243 with PO backbone; and 2006-PS, CpG ODN 2006 with PS backbone.
Antibodies
Fluorescein isothiocyanate (FITC)–labeled monoclonal antibodies (mAbs) specific for CD15 (H198), CD71 (M-A712), and phycoerythrin (PE)–labeled antibodies for CD13 (WM15) and CD71 (M-A712) were purchased from BD Biosciences. PE-CD11c (BU15) was from Immunotech. PE-CDw123 (IL-3Rα; 9F5) and CD61 (VI-PL2) were from BD Biosciences PharMingen. FITC- or PE-labeled glycophorin A (GPA; JC159), PE-CD34 (BIRMA-k3), and FITC-CD61 were from Dako Japan Co. PE-EPO receptor (EPOR; 38409) was from R&D Systems. FITC–annexin V apoptosis detection kit was from Sigma-Aldrich. Rabbit polyclonal antibody to erythroid Krüppel-like factor (EKLF) and mouse mAb to early endosomes (EEA1) were from Abcam. Mouse mAb to α-tubulin (DM 1A) was from Sigma-Aldrich. Alexa Fluor 488 or Alexa Fluor 546 conjugated goat IgG directed against rabbit and mouse IgG were from Invitrogen. Fc-blocking antibody (anti-CD16/32, clone 93) was from eBioscience. Normal mouse and goat sera, rabbit immunoglobulins, and rabbit IgG conjugated with horseradish peroxidase through dextran polymer to IgG were from Cell Signaling Technology.
Cell preparation
G-CSF–mobilized human peripheral blood CD34+ cells were purified from healthy volunteers and stored in liquid nitrogen until use as described previously.23 Informed consent was obtained from each subject before the entry into this study in accordance with the Declaration of Helsinki, and the study was preapproved by the Akita University School of Medicine Committee for the Protection of Human Subjects.
For the generation of erythroid progenitor cells, CD34+ cells were thawed and prepared for the culture, as previously described.23 Cells were cultured in erythroid medium (IMDM containing 20% fetal bovine serum, 10% heat-inactivated pooled human AB serum, 1% BSA, 10 μg/mL of insulin, 0.5 μg/mL vitamin B12, 15 μg/mL folic acid, 50nM β-mercaptoethanol, 50 U/mL penicillin, and 50 μg/mL streptomycin, in the presence of 50 ng/mL IL-3, 50 ng/mL SCF, and 2 IU/mL EPO). Cells were maintained at 37°C in a 5% CO2 incubator as described previously.24 The cells were harvested at indicated days and resuspended in 2 mL of IMDM containing 0.1% BSA. The cells were incubated for 2 hours at 37°C to remove surface EPO before fluorocytometric analysis and radiolabeled EPO-binding measurements.
For the generation of neutrophilic and megakaryocytic progenitor cells, the CD34+ cells were cultured at a density of 2 × 105 to 5 × 105 cells/mL in the erythroid medium, with the exception that SCF, IL-3, and EPO were replaced with either 100 ng/mL G-CSF or 100 ng/mL TPO as described elsewhere.23,25 For the simultaneous generation of erythroid, neutrophilic, and megakaryocytic progenitor cells, CD34+ cells were suspended at a density of 2 × 104 to 3 × 104 cells/mL in the presence of 50 ng/mL IL-3, 50 ng/mL SCF, 2 IU/mL EPO, 100 ng/mL G-CSF, and 100 ng/mL TPO. Plasmacytoid dendritic cells (pDCs, CD11c− CD123+), conventional DCs (cDCs, CD11c+ CD123−), and B cells (CD20+) were isolated from normal human peripheral blood using MoFlo (Dako North America). The yield and viability were measured by dye exclusion using 0.2% trypan blue dye using a hemocytometer.
Semisolid culture of BFU-E
For the burst-forming unit-erythroid (BFU-E) colony assay, purified CD34+ cells were incubated in triplicate at various concentrations, in flat-bottom 24- or 96-well tissue culture plates (Linbro; Flow Laboratories) containing 0.5 mL or 0.05 mL medium with fibrin clots, respectively, as described elsewhere.26 After 14 days in culture, the clots were fixed and stained for hemoglobin.27 Aggregates with greater than 100 hemoglobinized cells were counted as BFU-E colonies.
Flow cytometry
The cells collected from culture were washed twice with IMDM containing 0.3% BSA. The cells were then incubated with FITC- and PE-labeled mAbs, washed twice with staining medium containing 10mM phosphate-buffered saline (PBS, pH7.4), 0.5% BSA, and 2mM EDTA, and analyzed using a FACSCalibur (BD Biosciences), as reported elsewhere.23
Binding of 125I-EPO
Cells in 50 μL of IMDM at 0°C containing 0.1% BSA were incubated in the presence of 2 U/mL of 125I-EPO prepared as reported elsewhere.27,28 After 24 hours of incubation at 0°C, the radioactivity of the cell pellets was counted in a gamma counter (Nuclear Chicago), as reported elsewhere.27,28 Nonspecific binding was determined with a 100-fold excess of unlabeled EPO. Specific binding was calculated by subtracting nonspecific binding from total binding.
Confocal microscopy
Fluorescence staining was imaged using a Confocal Laser Scanning Microscope 510 (LSM510; Carl Zeiss Microscope Systems) equipped with a 100× objective lens and a 10× camera lens (Carl Zeiss Microscope Systems) at zoom 3. Fluorochromes were excited using an argon laser at 488 nm for Alexa 488. Detector slits were configured to minimize cross-talk between channels and processed using software package (LSM510, Version 3.2) and Adobe Photoshop (Adobe Systems).
Cell cycle distribution
Cells were harvested, washed with cold PBS, and fixed in 70% ethanol. The cells were then stored at −20°C until analysis. The fixed cells were centrifuged at 400g, washed with cold PBS twice, and RNase A added at a final concentration of 0.5 mg/mL. The cells were then incubated for 10 minutes at 37°C. Next, 25 μg/mL propidium iodide was added and the cells were incubated for 30 minutes at room temperature in the dark. The cells were analyzed using a FACSCalibur instrument (BD Biosciences) equipped with CellQuest, Version 3.3 software. Multicycle cell-cycle analysis software (Beckman Coulter) was used to determine the percentage of cells in the different cell cycle phases.
Real-time RT-PCR
Total RNA was extracted from 2 × 104 cells per sample using TRizol reagent (Invitrogen). The extracted RNA was then reverse-transcribed using the SuperScript III First-Strand Synthesis System for reverse transcriptase-polymerase chain reaction (RT-PCR; Invitrogen) in a 20-μL reaction volume. cDNA was then subjected to real-time RT-PCR using LightCycler 480 SYBR Green I Master (Roche Applied Science). The relative gene expression levels were normalized with 28S. Primer sequences are presented in Table 2 and were purchased from Nippon Gene Research Laboratories.
Primers used in this study
| Primer . | Direction . | Sequence . |
|---|---|---|
| TLR9 | Forward | 5′-ACC CTG GAA GAG CTA AAC C-3′ |
| Reverse | 5′-CAG TTG CCG TCC ATG AAT-3′ | |
| EPOR | Forward | 5′-TCA TCC TCG TGG TCA TCC T-3′ |
| Reverse | 5′-CCT TCA AAC TCG CTC TCT G-3′ | |
| GATA-1 | Forward | 5′-TGG CCT ACT ACA GGG ACG CT-3′ |
| Reverse | 5′-CTC AGC CGC TCT GTC TTC A-3′ | |
| GATA-2 | Forward | 5′-CCT CCA GCT TCA CCC CTA A-3′ |
| Reverse | 5′-CAC AGG CAT TGC ACA GGT AGT-3′ | |
| EKLF | Forward | 5′-CAG AGG ATC CAG GTG TGA TAG-3′ |
| Reverse | 5′-GCA GGC GTA TGG CTT CTC-3′ | |
| RPS19 | Forward | 5′-GCT CCA TGA CCA AGA TCT AT-3′ |
| Reverse | 5′-GTC CAG ATC TCT TTG TCC CT-3′ | |
| FOG-1 | Forward | 5′-TGC ACA CGG ACA CGC TGA-3′ |
| Reverse | 5′-GTA GAT CTC ACC CTT GGA GCC A-3′ | |
| 28S | Forward | 5′-TGG GTT TTA AGC AGG AGG TG-3′ |
| Reverse | 5′-CCA GCT CAC GTT CCC TAT TA-3′ |
| Primer . | Direction . | Sequence . |
|---|---|---|
| TLR9 | Forward | 5′-ACC CTG GAA GAG CTA AAC C-3′ |
| Reverse | 5′-CAG TTG CCG TCC ATG AAT-3′ | |
| EPOR | Forward | 5′-TCA TCC TCG TGG TCA TCC T-3′ |
| Reverse | 5′-CCT TCA AAC TCG CTC TCT G-3′ | |
| GATA-1 | Forward | 5′-TGG CCT ACT ACA GGG ACG CT-3′ |
| Reverse | 5′-CTC AGC CGC TCT GTC TTC A-3′ | |
| GATA-2 | Forward | 5′-CCT CCA GCT TCA CCC CTA A-3′ |
| Reverse | 5′-CAC AGG CAT TGC ACA GGT AGT-3′ | |
| EKLF | Forward | 5′-CAG AGG ATC CAG GTG TGA TAG-3′ |
| Reverse | 5′-GCA GGC GTA TGG CTT CTC-3′ | |
| RPS19 | Forward | 5′-GCT CCA TGA CCA AGA TCT AT-3′ |
| Reverse | 5′-GTC CAG ATC TCT TTG TCC CT-3′ | |
| FOG-1 | Forward | 5′-TGC ACA CGG ACA CGC TGA-3′ |
| Reverse | 5′-GTA GAT CTC ACC CTT GGA GCC A-3′ | |
| 28S | Forward | 5′-TGG GTT TTA AGC AGG AGG TG-3′ |
| Reverse | 5′-CCA GCT CAC GTT CCC TAT TA-3′ |
TLR9 indicates Toll-like receptor 9; EPOR, erythropoietin receptor; EKLF, erythroid Krüppel-like factor; RPS19, ribosomal protein S19; FOG-1, friend of GATA-1; and 28S, 28S ribosomal gene.
Western blot analysis
CD34+ cells were incubated in erythroid medium for 3 days (day 3 cells) and the dead cells depleted using the Annexin Microbead kit (Milenyi Biotec). Day 3 cells were then treated with or without 2006-PO for 12 hours. Western blot analysis was carried out according to the manufacturer's protocol.
Isolation of the B19 genome
Serum samples containing B19 were collected from donated blood samples provided by the Japanese Red Cross Saitama Blood Center. An aliquot of the serum was used for infection and isolation of viral DNA. This aliquot was layered on 30% sucrose in PBS and centrifuged for 24 hours at 100 000g to precipitate the B19 virion. B19 genome DNA was then extracted from the pellet fraction using the QIAamp DNA Blood Mini Kit (QIAGEN). Cellular DNA was extracted from UT7/Epo-S1 cells20 (a generous gift of Kazuo Sugamura, Tohoku University Graduate School of Medicine, Japan), digested with BamHI, and used as double-stranded cellular DNA (dsDNA). ssDNA was obtained by heat denaturation of dsDNA extracted from UT7/Epo-S1 cells.
B19 infection of erythroid progenitors
CD34+cells were cultured in erythroid medium for 4 days (3 × 105 cells/0.06 mL) and then either mock-infected or mixed with human serum that contained 6 × 109 genome copies of B19. The cell-to-virus ratio was 1:20 000. This mixture was then incubated for 1 hour on ice, diluted in erythroid medium, and further incubated at 37°C in 5% CO2 for 24, 48, or 72 hours.
Statistical analysis
Significant differences between groups were calculated using 1-factor analysis of variance (Scheffé post-hoc test) and the paired t test. Tests were undertaken using Stat View, Version 4.0, and significant differences were defined as P less than .05.
Results
Selective inhibition of erythroid growth by CpG-ODN 2006-PO
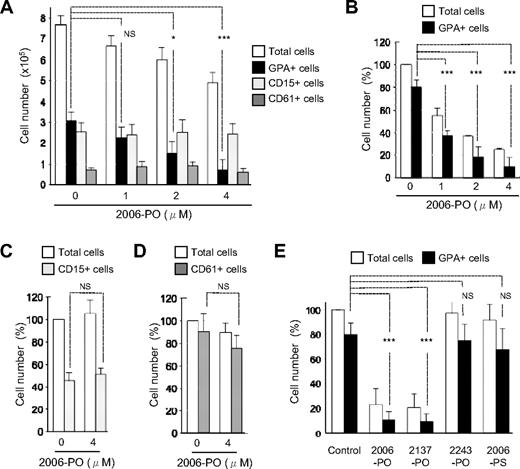
To examine the effects of CpG-ODN 2006-PO (2006-PO) on the growth of hematopoietic progenitors, human CD34+ cells were cultured for 7 days in liquid medium in the presence of multiple cytokines. Under these conditions, simultaneous differentiation of erythroid (GPA+), neutrophilic (CD15+), and megakaryocytic (CD61+) progenitors occurred. We found that 2006-PO inhibited the growth of erythroid progenitors in a dose-dependent manner but did not affect the growth of neutrophilic and megakaryocytic progenitors (Figure 1A). To investigate the specificity of 2006-PO in the inhibition of erythroid growth, we then differentiated CD34+ cells into each of the erythroid (80.1% ± 6.7% pure GPA+ cells, n = 3), neutrophilic (45.9% ± 6.5% pure CD15+ cells, n = 3), and megakaryocytic (90.6% ± 15.9% pure CD61+ cells, n = 3) progenitor lineages. The inhibitory effects of 2006-PO on the growth of erythroid progenitor cells were also evident in a dose-dependent manner and occurred at a half-maximal dose of 1μM (Figure 1B). In contrast, 2006-PO exhibited no inhibitory effects on the growth of neutrophilic (Figure 1C) and megakaryocytic (Figure 1D) progenitors. These data indicate that 2006-PO selectively inhibited erythroid growth. In addition, the absolute number of CD15+ cells generated from 1 × 105 CD34+ cells without 2006-PO was 13.0 × 105 cells in the presence of multiple cytokines (Figure 1A) and 0.6 × 105 cells in the presence of G-CSF alone (Figure 1C), which indicates that multiple cytokines induce a 22-fold of expansion of CD15+ cells compared with that with G-CSF alone. Addition of 2006-PO did not affect the yield of CD15+ cells. These data suggest that 2006-PO does not affect uncommitted CD34+ early progenitors.
Effects of ODN on the growth of hematopoietic progenitors derived from CD34+ cells. (A) CD34+ cells were cultured with IL-3, SCF, EPO, G-CSF, TPO, and various concentrations of 2006-PO ranging from 0 to 4μM. Seven days later, the total cell yield was counted and GPA, CD15, and CD61 expression examined using fluorocytometry. Data are mean ± minus SD of 3 independent experiments. (B-D) CD34+ cells were cultured with IL-3 and SCF in the presence of EPO at a cell density of 2 × 104 cells/mL (B), with G-CSF at a cell density of 3 × 105 cells/mL (C), or with TPO at a cell density of 3 × 105 cells/mL (D) with or without 2006-PO. Seven days later, the cell yields were calculated as a percentage relative to the total number of cells without 2006-PO. Data are mean ± SD of 3 independent experiments. The absolute number of cells generated from CD34+ cells without 2006-PO was 7.6 ± 2.2 × 105 cells (A), 8.6 ± 3.6 × 105 cells (B), 4.0 ± 0.6 × 105 cells (C), and 4.0 ± 0.6 × 105 cells (D). (E) CD34+ cells were cultured in erythroid medium in the presence or absence of various forms of ODN at 4μM. The total cell yield was then calculated as a percentage relative to the total number of cells without ODN (5.3 ± 1.4 × 105 cells/2 × 104 CD34+ cells cultured; n = 4). Data are mean ± SD of 4 independent experiments. *P < .05. ***P < .001. NS indicates no significance.
Effects of ODN on the growth of hematopoietic progenitors derived from CD34+ cells. (A) CD34+ cells were cultured with IL-3, SCF, EPO, G-CSF, TPO, and various concentrations of 2006-PO ranging from 0 to 4μM. Seven days later, the total cell yield was counted and GPA, CD15, and CD61 expression examined using fluorocytometry. Data are mean ± minus SD of 3 independent experiments. (B-D) CD34+ cells were cultured with IL-3 and SCF in the presence of EPO at a cell density of 2 × 104 cells/mL (B), with G-CSF at a cell density of 3 × 105 cells/mL (C), or with TPO at a cell density of 3 × 105 cells/mL (D) with or without 2006-PO. Seven days later, the cell yields were calculated as a percentage relative to the total number of cells without 2006-PO. Data are mean ± SD of 3 independent experiments. The absolute number of cells generated from CD34+ cells without 2006-PO was 7.6 ± 2.2 × 105 cells (A), 8.6 ± 3.6 × 105 cells (B), 4.0 ± 0.6 × 105 cells (C), and 4.0 ± 0.6 × 105 cells (D). (E) CD34+ cells were cultured in erythroid medium in the presence or absence of various forms of ODN at 4μM. The total cell yield was then calculated as a percentage relative to the total number of cells without ODN (5.3 ± 1.4 × 105 cells/2 × 104 CD34+ cells cultured; n = 4). Data are mean ± SD of 4 independent experiments. *P < .05. ***P < .001. NS indicates no significance.
To examine the effects of different ODNs on erythroid growth, several ODNs were synthesized. The construction of these ODNs focused on the CpG-motif, ODN backbone, and sequence (Table 1). As illustrated in Figure 1E, CpG-ODN (2006-PO) and non–CpG-ODN (2137-PO; similar to 2006 but contained GC instead of CG) exhibited similar inhibitory effects on erythroid growth, and the latter (2137-PO) also specifically inhibited erythroid growth (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). ODN-2243-PO, which contained a different ODN sequence from that of 2006-PO and CpG-ODN with the PS backbone, did not exhibit any inhibitory effects on erythroid growth. These data indicate that the inhibitory effects of 2006-PO on erythroid growth depended on the ODN sequence and backbone but not on the CpG motif.
Immunophenotypic analysis of the cells generated in the presence of 2006-PO revealed that the majority of nonerythroid progenitors (GPA− cells) were CD13+ cells. A relatively small population of CD4+, CD11c+, and CD123+ cells were also observed (supplemental Figure 2).
Internalization of ODN by progenitors derived from CD34+ cells
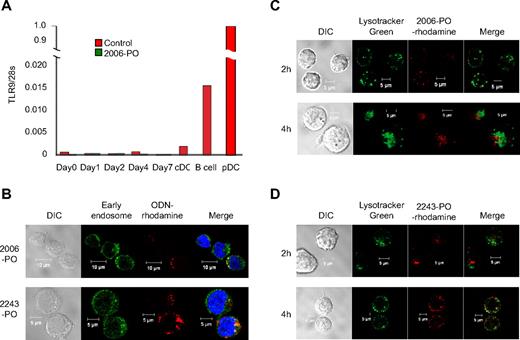
Purified CD34+ cells were cultured in erythroid medium in the presence or absence of 2006-PO, and the TLR9 mRNA expression was monitored at the indicated time points. B cells and pDCs were used as positive controls, whereas cDCs were used as a negative control.29 As illustrated in Figure 2A, pDCs and B cells were found to express TLR9 mRNA. CD34+ cells harvested at days 0 to 4 also appeared to express TLR9; however, the TLR9 expression levels were lower than that of the cDCs. TLR9 expression at day 7 was undetectable, and 2006-PO did not significantly affect the levels of TLR9 mRNA expression. These data suggest that erythroid progenitors do not possess TLR9.
Internalization of ODN by hematopoietic progenitors derived from CD34+ cells. (A) TLR9 expression in pDCs, B cells, cDCs, and hematopoietic progenitors induced erythroid differentiation. CD34+ cells (day 0) were cultured in the liquid phase in erythroid medium with or without 4μM 2006-PO for 1 to 7 days. The relative gene expression levels of TLR9 were normalized with 28S transcripts. The inset represents amplification of the y-axis. The results are representative of 3 independent experiments. (B) ODN-rhodamine and early endosome expression. CD34+ cells were cultured in erythroid medium for 2 days and incubated with rhodamine-conjugated 2006-PO or 2243-PO. Results are representative of 3 independent experiments. (C-D) ODN-rhodamine and lysosome expression. CD34+ cells were cultured in erythroid medium for 2 days, incubated with LysoTracker Green and rhodamine-conjugated 2006-PO (C) or 2243-PO (D) and analyzed using confocal microscopy. Results are representative of 3 independent experiments.
Internalization of ODN by hematopoietic progenitors derived from CD34+ cells. (A) TLR9 expression in pDCs, B cells, cDCs, and hematopoietic progenitors induced erythroid differentiation. CD34+ cells (day 0) were cultured in the liquid phase in erythroid medium with or without 4μM 2006-PO for 1 to 7 days. The relative gene expression levels of TLR9 were normalized with 28S transcripts. The inset represents amplification of the y-axis. The results are representative of 3 independent experiments. (B) ODN-rhodamine and early endosome expression. CD34+ cells were cultured in erythroid medium for 2 days and incubated with rhodamine-conjugated 2006-PO or 2243-PO. Results are representative of 3 independent experiments. (C-D) ODN-rhodamine and lysosome expression. CD34+ cells were cultured in erythroid medium for 2 days, incubated with LysoTracker Green and rhodamine-conjugated 2006-PO (C) or 2243-PO (D) and analyzed using confocal microscopy. Results are representative of 3 independent experiments.
To examine the intracellular localization of ODNs, purified CD34+ cells were cultured in erythroid medium for 2 days and were incubated with rhodamine-labeled 2006-PO or 2243-PO (ODN-PO with a different ODN sequence to that of 2006-PO) for 30 minutes. The cells were then incubated with anti–early endosome antibody (EEA1). The 2006- and 2243-ODN-rhodamine were found to occasionally colocalize with the green fluorescence of the early endosomes, indicating that ODN was internalized within 30 minutes and was partially targeted to the early endosome regardless of the differences in sequence and biologic activity (Figure 2B). When LysoTracker Green was used as a lysosome tracker in day 2 cells, 2006-PO (Figure 2C) and 2243-PO (Figure 2D) were partially colocalized with the lysosomes after a 2-hour incubation. At 4 hours after incubation with ODN-rhodamine, 2243-PO and 2006-PO were partially colocalized with the vesicular LysoTracker Green, suggesting that a portion of the ODN had escaped degradation in the lysosomes. In addition, the half-life of 2006-PO in serum was estimated at 4 hours, whereas that of 2006-PS exceeded more than 24 hours (supplemental Figure 3). The PS backbone appeared to increase the nonspecific ODN binding to a wide variety of serum proteins, as reported previously.30
2006-PO inhibits the early stages of erythroid growth
To further understand the kinetics of erythroid inhibition by 2006-PO, purified CD34+ cells were cultured in erythroid medium in the presence or absence of 2006-PO and the surface phenotypes observed at different time points (Figure 3A). The total number of cells recovered in both the absence and presence of 2006-PO was comparable after 3 days in culture. The cell yield substantially increased in the cultures that did not contain 2006-PO and was decreased in the cultures that did contain 2006-PO after 4 days in culture. We also found that the main cell population that was decreased after treatment with 2006-PO was the GPA+ cell population. These findings indicated that 2006-PO affected CD34+ cell development by inhibiting erythroid growth in the early stage of development. When purified CD34+ cells were cultured for 7 days in erythroid medium and 2006-PO was added at the indicated time points (Figure 3B), addition of 2006-PO to the medium resulted in significant inhibitory effects on the generation of erythroid progenitors as late as 4 days after treatment. In contrast, 2006-PO did not affect the growth of more mature erythroid progenitor cells in the stage of CFU-erythroid (CFU-E)27 (Figure 3C). These findings suggest that 2006-PO inhibits the erythroid progenitor cells in the stage of BFU-E.
2006-PO inhibits erythroid growth in the early stages of development. (A) Kinetics of erythroid growth. CD34+ cells were cultured in erythroid medium with (●) or without (○) 2006-PO. At the indicated days, the cells were collected, washed, and counted. The total cell yields are represented as a percentage relative to the total number of cells without 2006-PO on the seventh day (4.9 ± 0.2 × 105 cells, n = 3). Insets: Amplification of the y-axis. (B) CD34+ cells were cultured for 7 days in erythroid medium. At the indicated days, 2006-PO was added to the medium. The total cell yields are represented as a percentage relative to the total number of cells without 2006-PO (7.4 ± 3.8 × 105 cells, n = 3). (C) Effects of 2006-PO on CFU-E. CD34+ cells were cultured in erythroid medium. After 7 days in culture, the cells were collected and washed, and then cultured for a further 5 days (until day 12) in the presence of EPO, with or without 2006-PO. (A-C) Data are mean ± SD of 3 independent experiments. (D) Purified CD34+ cells were cultured in semisolid medium with IL-3, SCF, and EPO, with or without ODN. After 14 days in culture, the clots were observed directly (top panel) and then fixed and stained for hemoglobin (bottom panel). (E) BFU-E and small erythroid colonies were then differentially counted under light microscopy. The number of colonies is a representative of 2 independent experiments and the mean ± SD of triplicate culture. *P < .05. ***P < .001. NS indicates no significance.
2006-PO inhibits erythroid growth in the early stages of development. (A) Kinetics of erythroid growth. CD34+ cells were cultured in erythroid medium with (●) or without (○) 2006-PO. At the indicated days, the cells were collected, washed, and counted. The total cell yields are represented as a percentage relative to the total number of cells without 2006-PO on the seventh day (4.9 ± 0.2 × 105 cells, n = 3). Insets: Amplification of the y-axis. (B) CD34+ cells were cultured for 7 days in erythroid medium. At the indicated days, 2006-PO was added to the medium. The total cell yields are represented as a percentage relative to the total number of cells without 2006-PO (7.4 ± 3.8 × 105 cells, n = 3). (C) Effects of 2006-PO on CFU-E. CD34+ cells were cultured in erythroid medium. After 7 days in culture, the cells were collected and washed, and then cultured for a further 5 days (until day 12) in the presence of EPO, with or without 2006-PO. (A-C) Data are mean ± SD of 3 independent experiments. (D) Purified CD34+ cells were cultured in semisolid medium with IL-3, SCF, and EPO, with or without ODN. After 14 days in culture, the clots were observed directly (top panel) and then fixed and stained for hemoglobin (bottom panel). (E) BFU-E and small erythroid colonies were then differentially counted under light microscopy. The number of colonies is a representative of 2 independent experiments and the mean ± SD of triplicate culture. *P < .05. ***P < .001. NS indicates no significance.
To investigate whether the inhibitory effects of 2006-PO mediated inhibition of the clonal development of BFU-E, purified CD34+ cells were cultured in semisolid erythroid medium for 14 days, with or without different concentrations of 2006-PO or 2243-PO. 2006-PO administered at a concentration of 4μM completely abolished the formation of visible BFU-E colonies, whereas 2243-PO promoted it (Figure 3D top panel). Microscopic observation of the clots stained for hemoglobin (Figure 3D bottom panel) showed that the number of BFU-E colonies was greatly reduced compared with that formed in the control treated cells (Figure 3E).
2006-PO directly inhibits erythroid growth, produces large erythroblasts, retards cell division, and induces G2/M accumulation and apoptosis of erythroblasts
To exclude the possibility that 2006-PO may inhibit BFU-E growth by nonerythroid cells, we investigated the clonal basis of BFU-E growth, in both the presence and absence of 0.5μM 2006-PO, using limiting dilution analysis.31 Purified CD34+ cells were plated at a variety of concentrations in fibrin clots as indicated in Figure 4A, and the percentage of nonresponder wells plotted against the number of cells per well. Straight lines through the origin for BFU-E colonies could be constructed, indicating that 2006-PO directly inhibited BFU-E during clonal development.
2006-PO directly inhibits BFU-E growth. (A) Limiting dilution analysis of BFU-E growth from purified CD34+ cells in semisolid medium. CD34+ cells were cultured in fibrin clots in 96-well flat-bottom plates in the presence of IL-3, SCF, and EPO, with (●) or without (○) 2006-PO. After 14 days, the clots were fixed, stained for hemoglobin, and the number of clots in which BFU-E colonies did not form counted as BFU-E colony-negative wells. These data were plotted against the number of CD34+ cells plated into the wells originally. Each point represents the values obtained from 60 wells. The results are representative of 3 independent experiments. (B) Morphology and size distribution of the generated cells. Purified CD34+ cells were cultured in erythroid medium, with or without 2006-PO for 7 days and subjected to May-Grünwald-Giemsa staining (×1000). Results are representative of 5 independent experiments. Scale bars represent 10 μm. (C) Top panel: Purified CD34+ cells were cultured in erythroid medium. At the indicated days, the cells were harvested and analyzed by fluorocytometry. Results are representative of 3 independent experiments. Bottom panel: CFSE dilution assay. Purified CD34+ cells were incubated with CFSE and cultured in erythroid medium. At the indicated days, the cells were harvested and analyzed by fluorocytometry. Results are representative of 2 independent experiments. (D-E) Purified CD34+ cells were cultured in erythroid medium with or without 2006-PO for 7 days. Results are representative of 2 independent experiments. (D) Cell-cycle analysis of the generated cells. (E) The cells were labeled with propidium iodide and annexin V and analyzed using fluorocytometry.
2006-PO directly inhibits BFU-E growth. (A) Limiting dilution analysis of BFU-E growth from purified CD34+ cells in semisolid medium. CD34+ cells were cultured in fibrin clots in 96-well flat-bottom plates in the presence of IL-3, SCF, and EPO, with (●) or without (○) 2006-PO. After 14 days, the clots were fixed, stained for hemoglobin, and the number of clots in which BFU-E colonies did not form counted as BFU-E colony-negative wells. These data were plotted against the number of CD34+ cells plated into the wells originally. Each point represents the values obtained from 60 wells. The results are representative of 3 independent experiments. (B) Morphology and size distribution of the generated cells. Purified CD34+ cells were cultured in erythroid medium, with or without 2006-PO for 7 days and subjected to May-Grünwald-Giemsa staining (×1000). Results are representative of 5 independent experiments. Scale bars represent 10 μm. (C) Top panel: Purified CD34+ cells were cultured in erythroid medium. At the indicated days, the cells were harvested and analyzed by fluorocytometry. Results are representative of 3 independent experiments. Bottom panel: CFSE dilution assay. Purified CD34+ cells were incubated with CFSE and cultured in erythroid medium. At the indicated days, the cells were harvested and analyzed by fluorocytometry. Results are representative of 2 independent experiments. (D-E) Purified CD34+ cells were cultured in erythroid medium with or without 2006-PO for 7 days. Results are representative of 2 independent experiments. (D) Cell-cycle analysis of the generated cells. (E) The cells were labeled with propidium iodide and annexin V and analyzed using fluorocytometry.
After 7 days in culture, the size of the erythroid cells generated in the presence of 2006-PO appeared to be larger than those generated without 2006-PO when observed using light microscopy (Figure 4B left and middle panels). Size distribution analysis of these GPA+ cells revealed that 2006-PO treatment promoted larger erythroid progenitors compared with control cells (Figure 4B right panel). Phenotypic analysis of CD34+ cells cultured in the erythroid medium showed that 2006-PO treatment decreased CD71+ GPA+ cells after 4 days in culture (Figure 4C top panel). When CD34+ cells were stained with CFSE and cultured in erythroid medium with or without 2006-PO, the GPA+ cells exhibiting a high CFSE intensity were observed in the presence of 2006-PO after 4 days in culture (Figure 4C bottom panel). These findings suggested that 2006-PO retarded erythroid progenitor cell proliferation. When purified CD34+ cells were cultured with 2006-PO for 7 days, 2006-PO resulted in the accumulation of cells in the S and G2/M phase. This result was accompanied by an increase in both the apoptotic fraction (Figure 4D) and the number of apoptotic cells that were annexin V–positive (Figure 4E).
Taken together, 2006-PO both directly and selectively inhibits erythroid cell development by promoting the growth of large erythroid progenitors that demonstrate reduced cell division and accumulation in the S and G2/M phase. These results were accompanied by an increased frequency of apoptosis. In combination, the majority of these features were similar to those observed in erythroid progenitor cells infected with B19,20-22 a virus that causes pure red cell aplasia (PRCA) with giant erythroblasts. Thus, we subsequently searched for the 2006-PO consensus sequence within the B19 genome.
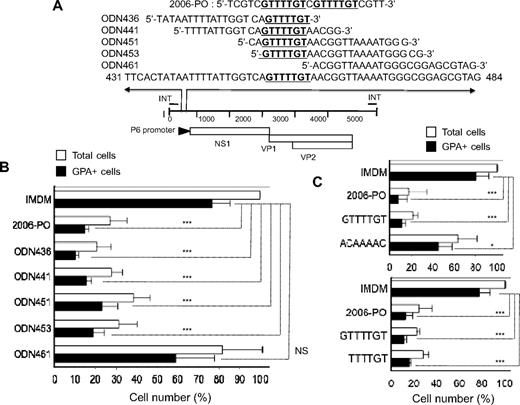
B19 (453 5′-GTTTTGT-3′ 459) consensus sequence is shared with 2006-PO and inhibits erythroid growth
We found that the 2006-PO consensus sequence 5′-GTTTTGT-3′ was located within the P6-promoter region of B19 (Figure 5A). To examine the effects of synthetic ODN that shared the same nucleotide length as 2006-PO (24 nucleotides) and the identical sequence to B19, purified CD34+ cells were cultured in erythroid medium for 7 days with or without 4μM synthetic ODN. The synthetic ODN-436, -441, -451, and -453 that contained the consensus sequence inhibited erythroid growth. These ODNs were found to inhibit growth by 86% plus or minus 2% (P < .001), 79% plus or minus 3% (P < .001), 69% plus or minus 10% (P < .001), and 75% plus or minus 7% (P < .001) compared with control growth, respectively (Figure 5B). ODN-461 that lacked the consensus sequence did not show any significant inhibitory effects on erythroid growth (23% ± 24% compared with control growth, P > .05). These results suggested that the 24 nucleotide sequences containing the 5′-GTTTTGT-3′ consensus sequence of the B19 DNA inhibited erythroid progenitor growth in a sequence-dependent manner. To confirm the minimum consensus sequence length required for erythroid growth, purified CD34+ cells were cultured in erythroid medium for 7 days with or without 4μM 2006-PO, 5′-GTTTTGT-3′ or 5′-ACAAAAC-3′, the latter being the complimentary sequence of 5′-GTTTTGT-3′ (Figure 5C top panel). ODN containing the GTTTTGT inhibited erythroid growth in a similar fashion to that observed by 2006-PO. ODNs containing ACAAAAC also inhibited erythroid growth, but only to a minimal degree. A similar effect was also observed with 5′-TTTTGT-3′ (Figure 5C bottom panel). These results indicated that the consensus DNA sequence of the human parvovirus B19 was shared with 2006-PO and inhibited erythroid growth.
Consensus sequence with 2006-PO in human parvovirus B19 genome (453 5′-GTTTTGT-3′ 459) inhibits erythroid growth. (A) Map of the B19 genome, highlighting the region that includes the consensus sequence (5′-GTTTTGT-3′) and the sequences of synthetic ODN. (B) Effects of synthetic B19 ODN with PO backbone on erythroid growth. Purified CD34+ cells were cultured in erythroid medium, with or without ODN. After 7 days in culture, the generated cells were collected, washed, and counted. The cell yields are presented as a percentage relative to the total number of cells without ODN. Data are mean ± SD of 3 independent experiments. The total cells generated from 2 × 104 CD34+ cells were 4.3 ± 0.8 × 105 cells. (C) Effects of ODN 5′-GTTTTGT-3′ and its complementary ODN 5′-ACAAAAC-3′ (top panel), and 5′-TTTTGT-3′ (bottom panel) with PO backbone on erythroid growth. Purified CD34+ cells were cultured in erythroid medium, with or without ODN. After 7 days in culture, the generated cells were collected, washed, and counted. The total cell yields are presented as a percentage relative to the total number of cells without ODN. Data are mean ± SD of 3 independent experiments. The absolute number of cells generated from 2 × 104 CD34+ cells was 5.4 ± 2.1 × 105 cells (top panel) and 6.8 ± 0.8 × 105 cells (bottom panel). *P < .05. ***P < .001. NS indicates no significance.
Consensus sequence with 2006-PO in human parvovirus B19 genome (453 5′-GTTTTGT-3′ 459) inhibits erythroid growth. (A) Map of the B19 genome, highlighting the region that includes the consensus sequence (5′-GTTTTGT-3′) and the sequences of synthetic ODN. (B) Effects of synthetic B19 ODN with PO backbone on erythroid growth. Purified CD34+ cells were cultured in erythroid medium, with or without ODN. After 7 days in culture, the generated cells were collected, washed, and counted. The cell yields are presented as a percentage relative to the total number of cells without ODN. Data are mean ± SD of 3 independent experiments. The total cells generated from 2 × 104 CD34+ cells were 4.3 ± 0.8 × 105 cells. (C) Effects of ODN 5′-GTTTTGT-3′ and its complementary ODN 5′-ACAAAAC-3′ (top panel), and 5′-TTTTGT-3′ (bottom panel) with PO backbone on erythroid growth. Purified CD34+ cells were cultured in erythroid medium, with or without ODN. After 7 days in culture, the generated cells were collected, washed, and counted. The total cell yields are presented as a percentage relative to the total number of cells without ODN. Data are mean ± SD of 3 independent experiments. The absolute number of cells generated from 2 × 104 CD34+ cells was 5.4 ± 2.1 × 105 cells (top panel) and 6.8 ± 0.8 × 105 cells (bottom panel). *P < .05. ***P < .001. NS indicates no significance.
2006-PO inhibits the expression of EPOR mRNA, EPOR, and EKLF protein
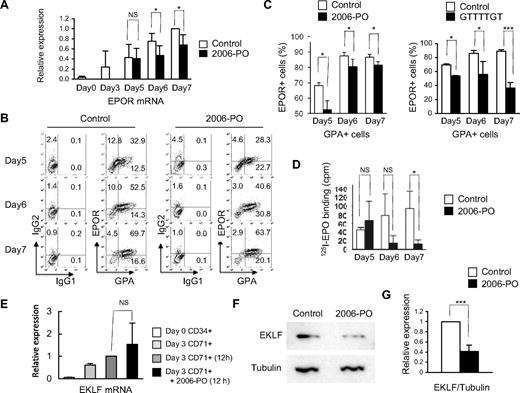
It is well established that several transcription factors and genes are required for commitment to the erythroid lineage. We therefore examined the effects of 2006-PO on the expression of GATA-2,32 GATA-1,33,34 friend of GATA-1 (FOG-1),35 EKLF,36 ribosomal protein S19 (RPS19),37 and EPOR. For this purpose, CD34+ cells were incubated for 3 days in erythroid medium (day 3 cells) and CD71+ cells purified using MiniMACS. The CD71+ cells were then incubated with or without 2006-PO in erythroid medium for 12 hours, with the exception of the EPOR gene that required several days in culture to be activated. We found that 2006-PO inhibits EPOR mRNA expression but did not affect the expression of any of other genes examined (Figure 6A,E; supplemental Figure 4). Fluorocytometric analysis showed that 2006-PO significantly decreased the expression of EPOR in the GPA+ cells (Figure 6B-C left panel). A similar effect was also observed with ODN-GTTTTGT (Figure 6C right panel). 125I-EPO binding assay showed a decrease of the specific binding of 125I-EPO to the cells treated with 2006-PO (Figure 6D). Western blot analysis revealed that 2006-PO resulted in a significant decrease in EKLF protein levels (Figure 6F-G).
2006-PO down-regulates the expression of EPOR and EKLF. (A) Quantitative analysis of EPOR using real time RT-PCR. Purified CD34+ cells were cultured for 3 days in erythroid medium and the cells enriched as CD71+ cells. The CD71+ cells were then cultured in erythroid medium, with or without 2006-PO. At the indicated days, the cells were harvested and mRNA extracted from the cells and subjected to real-time RT-PCR. (B-C) Fluorocytometric analysis of EPOR expression. At the indicated days, the cells were harvested and EPOR expression on GPA+ cells analyzed using fluorocytometry. Data are representative of 3 independent experiments (B) and are mean ± SD (C left panel). (C right panel) Parallel experiments in which 2006-PO was replaced with ODN 5′-GTTTTGT-3′. (D) Specific binding of 125I-EPO. Purified CD34+ cells were cultured for 3 days in erythroid medium and the cells were then cultured in erythroid medium, with or without 2006-PO. At the indicated days, the cells were harvested and subjected to 125I-EPO binding. The cells were collected in IMDM at 0°C containing 0.1% BSA, and 4 × 105 cells in 50 μL were incubated in the presence of 2 U/mL 125I-EPO. Data are mean ± SD of triplicate measurement; representative data of 2 experiments are shown. (E) Quantitative analysis of EKLF transcripts by real-time RT-PCR. CD71+ cells were incubated in erythroid medium with or without 2006-PO, the cells harvested, and mRNA extracted and quantified by real-time RT-PCR. (F) Western blot analysis of EKLF. The cells were harvested from the parallel experiments shown in panel E, and the lysates separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to nylon membranes. Western blot analyses were performed using an anti-EKLF antibody. (G) Relative expression of EKLF. Data are the mean ± SD of 3 independent experiments. *P < .05. ***P < .001. NS indicates no significance.
2006-PO down-regulates the expression of EPOR and EKLF. (A) Quantitative analysis of EPOR using real time RT-PCR. Purified CD34+ cells were cultured for 3 days in erythroid medium and the cells enriched as CD71+ cells. The CD71+ cells were then cultured in erythroid medium, with or without 2006-PO. At the indicated days, the cells were harvested and mRNA extracted from the cells and subjected to real-time RT-PCR. (B-C) Fluorocytometric analysis of EPOR expression. At the indicated days, the cells were harvested and EPOR expression on GPA+ cells analyzed using fluorocytometry. Data are representative of 3 independent experiments (B) and are mean ± SD (C left panel). (C right panel) Parallel experiments in which 2006-PO was replaced with ODN 5′-GTTTTGT-3′. (D) Specific binding of 125I-EPO. Purified CD34+ cells were cultured for 3 days in erythroid medium and the cells were then cultured in erythroid medium, with or without 2006-PO. At the indicated days, the cells were harvested and subjected to 125I-EPO binding. The cells were collected in IMDM at 0°C containing 0.1% BSA, and 4 × 105 cells in 50 μL were incubated in the presence of 2 U/mL 125I-EPO. Data are mean ± SD of triplicate measurement; representative data of 2 experiments are shown. (E) Quantitative analysis of EKLF transcripts by real-time RT-PCR. CD71+ cells were incubated in erythroid medium with or without 2006-PO, the cells harvested, and mRNA extracted and quantified by real-time RT-PCR. (F) Western blot analysis of EKLF. The cells were harvested from the parallel experiments shown in panel E, and the lysates separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to nylon membranes. Western blot analyses were performed using an anti-EKLF antibody. (G) Relative expression of EKLF. Data are the mean ± SD of 3 independent experiments. *P < .05. ***P < .001. NS indicates no significance.
B19 down-regulates the expression of EPOR
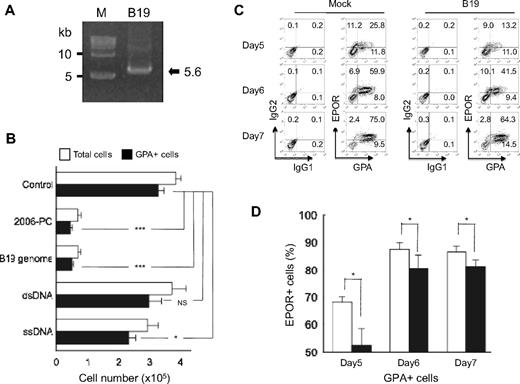
To examine the effects of B19 genome DNA on erythroid growth, B19 genome was extracted from the sera of patients with acute B19 infection (Figure 7A). When CD34+ cells were differentiated to the erythroid lineage for 7 days in either the presence or absence of native single-stranded B19 genome, B19 genome significantly inhibited erythroid growth to a level similar to that observed with 2006-PO (Figure 7B). dsDNA derived from UT7/Epo-S1 cells exhibited no effects on erythroid growth, whereas ssDNA demonstrated a mild inhibition. These data indicated that nonsynthetic B19 genome inhibited erythroid growth.
Native single-stranded B19 genome inhibits erythroid growth and down-regulates EPOR expression. (A) Purification of the B19 genome. B19 genome DNA was extracted from the virions. An equal molar ratio of plus and minus strands were then annealed and analyzed by agarose gel electrophoresis (arrow) and compared with molecular size markers (M). (B) Effects of B19 genome on erythroid growth. Purified CD34+ cells were cultured in erythroid medium with or without native single-stranded B19 genome DNA, dsDNA, and ssDNA derived from UT7/Epo-S1 cells. After 7 days in culture, the generated cells were collected, washed, and counted. Data are mean ± SD of 3 triplicates. (C-D) B19 down-regulated EPOR expression. Purified CD34+ cells were cultured for 4 days in erythroid medium, the cells harvested, and then incubated with serum containing B19. The cells were then cultured in erythroid medium. At the indicated days, the cells were harvested and EPOR expression on GPA+ cells analyzed using fluorocytometry. Data are representative of 3 independent experiments (C) and are mean ± SD (D). *P < .05. ***P < .001. NS indicates no significance.
Native single-stranded B19 genome inhibits erythroid growth and down-regulates EPOR expression. (A) Purification of the B19 genome. B19 genome DNA was extracted from the virions. An equal molar ratio of plus and minus strands were then annealed and analyzed by agarose gel electrophoresis (arrow) and compared with molecular size markers (M). (B) Effects of B19 genome on erythroid growth. Purified CD34+ cells were cultured in erythroid medium with or without native single-stranded B19 genome DNA, dsDNA, and ssDNA derived from UT7/Epo-S1 cells. After 7 days in culture, the generated cells were collected, washed, and counted. Data are mean ± SD of 3 triplicates. (C-D) B19 down-regulated EPOR expression. Purified CD34+ cells were cultured for 4 days in erythroid medium, the cells harvested, and then incubated with serum containing B19. The cells were then cultured in erythroid medium. At the indicated days, the cells were harvested and EPOR expression on GPA+ cells analyzed using fluorocytometry. Data are representative of 3 independent experiments (C) and are mean ± SD (D). *P < .05. ***P < .001. NS indicates no significance.
Several lines of evidence have demonstrated that the most permissive target cells for B19 are erythroid progenitors, BFU-E and CFU-E, and erythroblasts.38-40 We therefore used cells derived from CD34+ cells that had been cultured in erythroid medium for 4 days as the target cells for B19. After incubating cells in serum containing B19 for 1 hour, the cells were further cultured in erythroid medium for various periods. The expression of EPOR in GPA+ cells was found to be down-regulated in cells cultured with B19 compared with that observed in mock-infected cells (Figure 7C-D). These data suggested that B19 infection in erythroid progenitor cells down-regulated EPOR expression, a result that is consistent with that indentified in the erythroid cells exposed to 2006-PO.
Discussion
It is well established that unmethylated CpG-ODN induces APCs to produce cytokines, such as IL-12, IL-6, IL-1, and TNF-α.41 It has been previously reported that the biologic effect of unmethylated CpG-ODN 2006 on hematopoietic progenitor cells is indirect and is exerted by accessory cells, including APCs that produce proinflammatory cytokines.15,16 Unfortunately, the precise backbone expressed by unmethylated CpG-ODN was not described in these reports. In our study, the limiting dilution analysis of BFU-E growth indicated that 2006-PO directly inhibited clonal development and proliferation of BFU-E. This finding appears to be inconsistent with that reported by other investigators. These differing biologic effects were probably the result of the differences in both the sequence and ODN backbone used in this study.
Depending on the precise DNA sequence, ODNs may function as a decoy or as siDNA and result in the reduction of target gene mRNA levels.5 The EPOR DNA sequence has been shown to possess 5′-ACAAAAC-3′, a sequence complimentary to 5′-GTTTTGT-3′. This sequence is located upstream of the first TATA-box in EPOR (4713-4716). The 5′-GTTTTGT-3′ consensus sequence is not identified in the EPOR DNA. Therefore, the inhibition of EPOR mRNA expression suggests that the sequence, 5′-GTTTTGT-3′, acts as a siDNA for EPOR gene, and results in the down-regulation of EPOR expression and the decrease of EPO binding. EPO represents the main and the specific cytokine involved in the control of erythropoiesis. It is most probable that 2006-PO selectively inhibits erythroid growth through the inhibition of EPOR mRNA expression. A modest inhibition of the expression of EPOR mRNA suggests that other unknown mechanisms may also exist.
Among the erythroid lineage-related genes examined in this study, only FOG-1 possesses 5′-ACAAAAC-3′, but GATA-2, GATA-1, FOG-1, EKLF, and RPS19 do not. The reason why 2006-PO did not inhibit FOG-1 mRNA transcription remains unclear; however, it might be explained by the possibility that the antigene effect of ODN depends on multiple factors, including optimal length, hybridization capacity, and secondary conformations.42 In addition, 2137-PO containing the GpC-motif, rather than the CpG-motif, also inhibited erythroid growth, which led us to confirm that the 5′-TTTTGT-3′ motif also inhibits erythroid growth. A sequence complimentary to 5′-TTTTGT-3′ (5′-ACAAAA-3′) is included in the same site as that of 5′-ACAAAAC-3′ in the EPOR gene. ODNs also act as an antisense, forming a duplex with the mRNA and inhibiting its translation or processing, consequently inhibiting protein biosynthesis.42 However, mRNA of the erythroid lineage-related genes examined in this study do not possess 5′-ACAAAAC-3′, a sequence complimentary to 5′-GTTTTGT-3′. Two sites of 5′-ACAAAA-3′, a sequence complimentary to 5′-TTTTGT-3′, are present in GATA-2 mRNA, but 2006-PO treatment did not affect the transcriptional level of GATA-2 mRNA. The mechanism of reduced expression of EKLF protein by 2006-PO remains unclear. Down-regulation of EPOR may cause instability of EKLF protein through the differentiation arrest of the erythroid lineage.43
Additional human DNA viruses also express the consensus sequence 5′-GTTTTGT-3′ and include the Epstein-Barr virus (EBV, 10 sites), cytomegalovirus (27 sites), and adenovirus (19 sites). Among these viruses, EBV has been reported to be associated with PRCA, although the frequency of PRCA is extremely low. Socinski et al reported a patient who developed PRCA after EBV infection.44 In their case, depletion of bone marrow T cells by E-rosetting resulted in an increase in CFU-E in the patient's bone marrow and addition of bone marrow T cells significantly suppressed autologous CFU-E from T cell–depleted bone marrow. These results suggested that the PRCA associated with chronic EBV infection may be mediated by cytotoxic T lymphocytes.44 The role of the 5′-GTTTTGT-3′ sequence present in these dsDNA viruses during hematopoiesis is unclear. In addition to the target specificity of B19 for human erythroid cells, the major difference between these viruses and B19 is that the former represents a dsDNA virus, whereas the latter (B19) is an example of a ssDNA virus. These differences may result in the pathophysiologic functions. dsDNA but not ssDNA derived from either pathogens or the host activates both immune cells (macrophages and DCs) and nonimmune cells when introduced into the cytoplasm by transfection.45,46 In addition, dsDNA activates a set of genes, including those encoding major histocompatibility complex, costimulatory molecules, and immunoproteasome subunits, as well as the transcription factors STAT1.45,46 Evidence suggests that the induction of interferon-inducible genes and up-regulation of costimulatory molecules by dsDNA are mediated in part by a TLR9-independent pathway47 and that double-stranded B-form DNA but not Z-form DNA stimulated mouse and human stromal and DCs, resulting in the production of type I interferon and chemokines by a TLR9-independent pathway.48 These findings suggest that these dsDNA viruses may be processed by a different pathway than that of ssDNA viruses.
In conclusion, the data presented in the current study describe, for the first time, the presence of a ssDNA consensus sequence in both synthetic ODN-2006 and the B19 genome and that this site selectively inhibits erythroid growth. The inhibition of erythroid growth by the consensus sequence was also accompanied by the inhibition of EPOR mRNA expression, suggesting a potential mechanism for lineage-specific inhibition of erythroid progenitor cells by B19 DNA. Further work is necessary to understand the complete mechanism of ODN with PO backbone in association with the pathology of B19.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Sanford B. Krantz for helpful discussions and comments and Keiko Iwamoto, Hiromi Kataho, and Etsuko Kobayashi (Department of Hematology, Nephrology, and Rheumatology, Akita University Graduate School of Medicine) for their valuable technical assistance.
This study was supported in part by the Global Center of Excellence Program of the Ministry of Education, Science, Technology, Sports, and Culture of Japan (Grants-in-Aid 20591144) and the Idiopathic Disorders of Hematopoietic Organs Research Committee of the Ministry of Health, Labor and Welfare of Japan (research grant).
Authorship
Contribution: Y.-M.G., K.I., and K.S. designed and performed the experiments, analyzed data, and prepared the manuscript; M.H., H.T., H.O., Y.M., J.Y., and K.U. performed experiments and helped write the manuscript; and T.O., N.O., W.X., and K.K. analyzed and interpreted data and helped write manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kenichi Sawada, Department of Hematology, Nephrology, and Rheumatology, Akita University Graduate School of Medicine, Hondo 1-1-1, Akita 010-8543, Japan; e-mail: ksawada@doc.med.akita-u.ac.jp.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal