Abstract
Antibody-mediated cell depletion therapy has proven to provide significant clinical benefit in treatment of lymphomas and leukemias, driving the development of improved therapies with novel mechanisms of cell killing. A current clinical target for B-cell lymphoma is CD22, a B-cell–specific member of the sialic acid binding Ig-like lectin (siglec) family that recognizes α2-6–linked sialylated glycans as ligands. Here, we describe a novel approach for targeting B lymphoma cells with doxorubicin-loaded liposomal nanoparticles displaying high-affinity glycan ligands of CD22. The targeted liposomes are actively bound and endocytosed by CD22 on B cells, and significantly extend life in a xenograft model of human B-cell lymphoma. Moreover, they bind and kill malignant B cells from peripheral blood samples obtained from patients with hairy cell leukemia, marginal zone lymphoma, and chronic lymphocytic leukemia. The results demonstrate the potential for using a carbohydrate recognition–based approach for efficiently targeting B cells in vivo that can offer improved treatment options for patients with B-cell malignancies.
Introduction
Each year, approximately 70 000 people are diagnosed with B-cell lymphomas in the United States alone.1 Although therapy with the anti-CD20 antibody rituximab is highly effective, it is not a cure, especially for the indolent lymphoid malignancies, and 22 000 patients die annually.2-5 Thus, novel therapeutics with alternative mechanisms of B-cell killing are needed.2,3 Numerous antibodies for B-cell depletion therapy are in clinical development,3,6-8 and 2 immunotoxins (BL22 and CMC-544) target CD22, a B-lymphocyte–specific receptor. In contrast to CD20, CD22 undergoes constitutive endocytosis and is well suited for efficient delivery of the toxin into the cell.6,8-11 CD22 is also a member of the sialic acid binding Ig-like lectin (siglec) family that recognizes glycan ligands found on glycoproteins and glycolipids, and shows a marked preference for α2-6–linked sialic acid ligands, which interact with the binding site on the extracellular domain of CD22.12,13
As an alternative to antibodies, nanoparticles that are targeted to single cell types have gained attention for their potential to provide selective delivery of therapeutic agents with reduced side effects.14 Liposomal nanoparticles are pharmaceutically proven delivery vehicles that can encapsulate a therapeutic agent and also display ligands that target cell-surface receptors.15 The challenge has been to identify a ligand that provides sufficient selectivity for the targeted cell. Certain high-affinity small-molecule ligands (eg, folate) are efficient at targeting cognate receptors expressed at higher levels on the target cell, but lower expression levels on other cell types reduce selectivity.16,17 Immuno-liposomes use antibodies as targeting agents, but have not to date provided a therapeutic index commensurate with their promise.18,19
Several reports have documented the potential of glycan ligands for targeting glycan-binding proteins that exhibit restricted expression on immune cell subsets or inflamed endothelial cells.20-24 Based on our previous finding that multivalent synthetic high-affinity glycan ligands of CD22 are efficiently bound and internalized by B cells in vitro,25,26 we developed CD22 ligand-decorated liposomal nanoparticles for in vivo targeting B lymphoma cells. In contrast to the approved liposomal doxorubicin (Doxil; Centocor Ortho Biotech Products), which passively delivers doxorubicin (dox) to solid tumors via leaky blood vessels,27,28 dox-loaded liposomes bearing ligands of CD22 are actively targeted to and endocytosed by B cells, and significantly extend life in a murine model of human B-cell lymphoma. Moreover, as described here, the ligand-decorated liposomes recognize and kill malignant B cells in peripheral blood samples from patients with lymphoma in proportion to the amount of CD22 expressed, allowing identification of patient populations that would likely benefit from this targeted chemotherapeutic approach.
Methods
Preparation of liposomes
Distearoyl phosphatidylcholine (DSPC), cholesterol (chol), nitrobenzoxadiazol-phosphoethanolamine (NBD-PE), and polyethyleneglycol-distearoyl phosphoethanolamine (PEG-DSPE) were purchased from Avanti Polar Lipids and NOF Corporation. BPCNeuAc-PEG-DSPE was prepared by coupling 9-N-biphenylcarboxyl-NeuAcα2-6Galβ1-4GlcNAc (BPCNeuAc) sialosides25 with an ethylamine linker to N-hydroxysuccinimide (NHS)–activated pegylated lipids (NOF Corporation; see supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Nontargeted naked liposomes were composed of DSPC:Chol:PEG-DSPE in a 60:35:5 molar ratio. CD22-targeted BPCNeuAc liposomes substituted BPCNeuAc-PEG-DSPE for PEG-DSPE on a mol-for-mol basis. For preparation of liposomes, lipids dissolved in chloroform and dimethyl sulfoxide were mixed and lyophilized for 16 hours. The lipid flakes were hydrated in the cell culture–grade water to achieve a final liposome concentration of 10mM (total phospholipids) before extrusion through polycarbonate membrane filters (Millipore) with controlled pore sizes of 0.4, 0.2, and 0.1 μm. To prepare fluorescently labeled liposomes, 1 mol % of NBD-PE was added into the lipid mixture. Remote loading of dox (Sigma) was obtained using gradients of ammonium sulfate.29 Liposomes were hydrated in 250mM ammonium sulfate followed by extrusion and dialysis against 290mM glucose at 4°C for at least 4 changes of buffer. Dox was added to the liposome suspension in a ratio of dox/phospholipids equaling 1:7.5 (mass/mass) and incubated at 65°C for 40 minutes. The unbound drug was separated from liposomes by passing the suspensions through a Sepharose CL-4B (GE Healthcare) column equilibrated in 5% glucose (approximately 20-mL column for 2-mL sample). Concentration of encapsulated dox was determined using a fluorescence plate reader at excitation (Ex) of 485 nm and emission (Em) of 590 nm after complete lysis of liposomes by 0.5% Triton X-100. The loading efficiency of doxorubicin was greater than 90%. Liposomes prepared using this method had a mean particle size of 100 nm plus or minus 10 nm in diameter and were confirmed by a Malvern instrument.
Cell lines and binding assay
Daudi Burkitt lymphoma cells were maintained in RPMI-1640 (Invitrogen) containing 10% fetal bovine serum (FBS). Wild-type Chinese hamster ovary (CHO) cells or CHO cells expressing human CD22 were cultured in Dulbecco modified Eagle medium (DMEM)/F12 supplemented with 10% FBS and 100 μg/mL phleomycin (Invitrogen) or 500 μg/mL Hygromycin-B (Roche), respectively. CHO cell lines expressing Siglec-F and Siglec-E were prepared essentially as described earlier.30 CHO lines expressing other human or murine siglecs were generously provided by P.R.C. (University of Dundee, Scotland) and Dr Yasuhiro Hashimoto (Riken, Japan). CHO siglec lines were maintained in F10 medium (Invitrogen) supplemented with 10% FBS. TSn cells31 that overexpress human sialoadhesin (Sn) was a gift from Dr Hans Rempel and Dr Lynn Pulliam (University of California, San Francisco). TSn was cultured in RPMI-1640 containing 10% FBS and 1 μg/mL gentamicin (Invitrogen). A20, a murine B-cell line, was maintained in RPMI-1640 with 10% FBS. Unless otherwise stated, the liposome-binding assay was conducted by incubating cells in the mouse or human serum (MP Biomedicals) at 106 cells/100 μL with the presence of fluorescently labeled liposomes at 200μM (total phospholipids). After incubation at 37°C for 1.5 hours, cells were washed followed by fluorescence-activated cell sorter (FACS) analysis.
Fluorescence microscopy
CHO cells expressing CD22 were plated to a coverslip to achieve a 90% confluence followed by incubating with fluorescent liposomes at 37°C for 1 hour. Cells were fixed with 4% paraformaldehyde and stained with PE.Cy5-conjugated anti–human CD22 (BD Pharmingen). For detecting endosomes, fixed cells were permeabilized with 0.05% saponin and stained with anti-EEA1 (BD Pharmingen) followed by AlexaFluor 555–conjugated anti–mouse IgG (Invitrogen). Finally, the specimens were mounted on a slide using mounting solution (Invitrogen) containing DAPI that stains nuclei. Images were taken and processed using a Zeiss fluorescence microscope with a 40×/0.6 oil objective lens (numeric aperature: 0.35; WD: 70 mm), an Axiocam MRm camera (Carl Zeiss), and Axiovision 4 acquisition software.
In vitro cytotoxicity assay
Daudi B lymphoma cells were incubated with dox in free form or entrapped in the naked or BPCNeuAc liposomes at 37°C for 1 hour. Dox concentrations ranging from 1nM to 1mM were examined. Cells were washed and seeded on a 96-well plate at 105 cells/100 μL growth media, which allowed for an additional 48 hours of incubation followed by determining cell viability using CellTiter 96 (Promega) to measure the activity of enzymes that reduce a tetrazole (MTT) to formazan. The maximum cell viability was defined as medium-treated (untreated) cells. The complete killing was determined as Triton X-100 (0.5%) lysed cells. Data were analyzed using Prism nonlinear regression software (GraphPad Software) for the curve-fitting and determination of IC50 values. In the case of the clinical samples, we followed the method described by Kreitman et al8 with modifications. In brief, peripheral blood monocytes were purified using Ficoll-Paque (GE Healthcare) and subjected to liposomal doxorubicin at 10 or 40μM or medium-treated for 1 hour at 37°C. Cells were thoroughly washed and seeded on a 96-well plate at 106 cells/100 μL in RPMI-1640 supplemented with 10% FBS and 2-mercaptoethanol. Cell viability was determined as described on day 5 of incubation.
Mice and pharmacokinetics studies
The Institutional Animal Care and Use Committee of The Scripps Research Institute (TSRI) approved all experimental procedures involving mice. Nonobese diabetic–severe combined immunodeficiency (NOD-SCID) mice were produced by the TSRI breeding colony. Sialoadhesin (Sn) knockout mice were provided by P.R.C. Naive or Daudi tumor-bearing mice (3 mice per group) were intravenously injected with liposomal dox (3 mg/kg). At 0.5, 2, and 25 hours after liposome injection, a sample of blood (100 μL) was drawn from mice by making a tail nick and collected in a tube containing 10 μL of EDTA to prevent plasma from clogging. Plasma (50 μL) was separated from the blood cells by centrifugation and was mixed with 200 μL of 5% Triton X-100, 250 μL of distilled water, and 1500 μL of acidified isopropanol containing 0.75N HCl. Samples were stored at −20°C overnight for protein precipitation. After removal of the precipitated protein, an aliquot (100 μL) of the solution containing plasma samples or dox standards were subjected to the analysis for the concentration of the extracted doxorubicin using a fluorescence plate reader at Ex of 485 nm and Em of 590 nm. The data were analyzed using the Prism software. For macrophage depletion, the SCID mice received 200 μL of liposomal clodronate intravenously32 at 2 days before the injection of the liposomal formulations of dox.
In vivo efficacy studies
Female NOD-SCID mice (6 to 8 weeks old) were injected with 5 × 106 Daudi cells at the lateral tail vein intravenously on day 0 (8-10 per group). At days 1 and 3 after tumor injection, animals received intravenous treatments of PBS or 3 mg/kg dox encapsulated in the naked or BPCNeuAc liposomes (in 150 μL of 5% glucose). Animals were monitored every other day until the end of the study at day 100 and were killed at the onset of hind-leg paralysis. Survival rate was analyzed using a Kaplan-Meier plot. For detecting residual Daudi tumor cells in the bone marrow, paralyzed or long-term survivor animals were killed, followed by the harvest of bone marrow cells from both femurs. Washed cells were stained with anti–human CD19 or isotype antibodies to identify Daudi tumor cells before FACS analysis.
Clinical subjects
The procedures involving human subjects were reviewed and approved by TSRI Institutional Review Board. We obtained normal blood from TSRI's Normal Blood Donor Service and peripheral blood samples from patients seen by physicians of the Scripps Clinic Medical Group. Eligible patients with untreated or progressive lymphomas and leukemias gave informed consent before participation in this study. Whole blood was freshly collected into EDTA-coated tubes and was analyzed within 24 hours. Fluorescent liposomes were directly added into an aliquot of 100 μL of whole blood and incubated at 37°C for 1.5 hours, followed by the lysis of red blood cells. Cells were resuspended in 100 μL of Hanks buffered salt solution (HBSS) buffer containing 5% bovine serum albumin (BSA) and incubated with antibodies for detecting surface markers at room temperature for an additional 30 minutes. The following anti–human antibodies purchased from BD Pharmingen were used in the study: anti-CD5 (L17-F12), anti-CD19 (HIB19), anti-CD20 (L27), anti-CD22 (HIB22), anti-CD103 (Ber-ACT8), and anti-κ (TB28-2) and anti-λ light chains (1-155-2). Cells were washed twice before FACS analysis. Data were analyzed using FlowJo software (TreeStar).
Statistical analysis
We performed analysis of variance (ANOVA) and 2-sample t test for statistical analysis. We determined significant difference in survival studies using Kaplan-Meier plots and the Mantel-Cox rank test.
Results
CD22-targeted liposomes are bound to and internalized by CD22-expressing cells
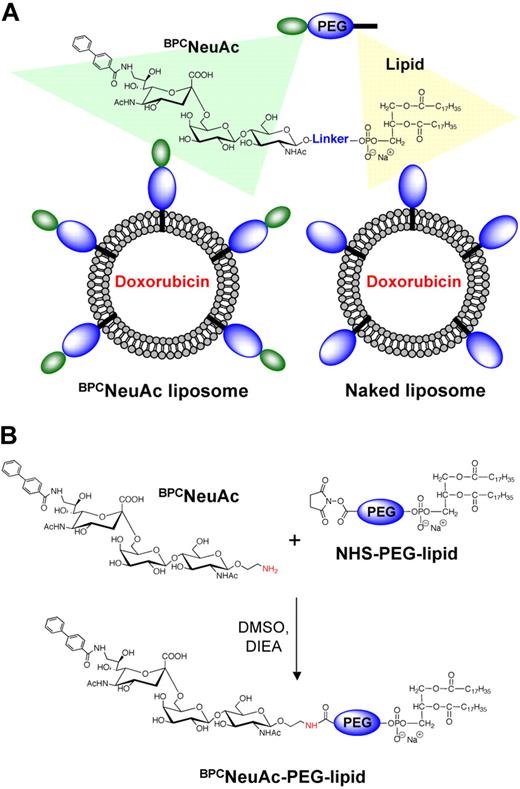
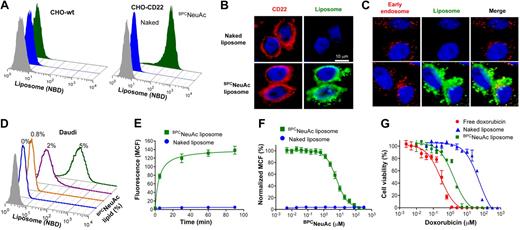
To prepare CD22-targeted liposomes, we coupled a high-affinity glycan ligand of CD22, BPCNeuAc,25 to a commercially available NHS-activated pegylated lipid, and the corresponding BPCNeuAc-pegylated lipid was then incorporated into a liposomal doxorubicin formulation (Figure 1A-B) analogous to that in current clinical use. Fluorescently labeled BPCNeuAc liposomes bound robustly to CD22-expressing but not wild-type CHO cells, whereas the nontargeted “naked” liposomes exhibited no binding to either (Figure 2A). Fluorescence microscopy shows staining of CD22-expressing cells only by BPCNeuAc liposomes (Figure 2B), with diffuse staining of the cell surface, and punctate staining consistent with internalization by endocytosis, as evidenced by costaining with markers of early endosomes (anti-EEA1; Figure 2C) and lysosomes (LysoTracker; supplemental Figure 1).
Liposomal nanoparticles displaying glycan ligands of CD22 for targeting and killing B-cell lymphoma. (A) Schematic illustration of a chemotherapeutic-loaded liposomal formulation comprising BPCNeuAc lipids for active targeting to CD22. (B) Synthesis of BPCNeuAc-pegylated lipids.
Liposomal nanoparticles displaying glycan ligands of CD22 for targeting and killing B-cell lymphoma. (A) Schematic illustration of a chemotherapeutic-loaded liposomal formulation comprising BPCNeuAc lipids for active targeting to CD22. (B) Synthesis of BPCNeuAc-pegylated lipids.
BPCNeuAc liposomes are bound and internalized by CD22-expressing cells. (A) FACS analysis for binding of fluorescently (NBD) labeled liposomes to wild-type CHO cells or CHO cells expressing recombinant human CD22. Cells were incubated with the naked (blue) or BPCNeuAc (green) liposomes or left untreated (gray) at 37°C for 1.5 hours before analysis. (B) CHO cells expressing CD22 were compared for their binding to the fluorescent (green) naked or BPCNeuAc liposomes. CD22 was detected with anti–human CD22 (red), and the nuclei were visualized by staining with DAPI. (C) Fluorescence microscopy analysis of the colocalization of BPCNeuAc liposomes with early endosomes. CHO-CD22 cells were incubated with fluorescent liposomes (green) as described. Early endosomes were visualized by staining with an Alexa Fluor 555–labeled anti-EEA1 (red). (D) Binding of BPCNeuAc liposomes to Daudi human B lymphoma cells. Daudi cells were incubated in mouse serum with liposomes containing 0% to 5% BPCNeuAc lipids or without liposomes (gray) before FACS analysis. (E) BPCNeuAc liposomes rapidly bind to Daudi cells. Fluorescent naked or BPCNeuAc liposomes were added to an aliquot of Daudi cells in mouse serum and incubated at 37°C for the indicated time before FACS analysis. Data are presented as mean channel of fluorescence (MCF) plus or minus SD. (n = 3). (F) Competitive binding of BPCNeuAc liposomes to Daudi cells in the presence of the free BPCNeuAc ligands. Fluorescent naked or BPCNeuAc liposomes were incubated with Daudi cells with the presence of the monovalent BPCNeuAc ligands at indicated concentration. Data were analyzed by FACS and shown as normalized MCF plus or minus SD. (n = 3). (G) Cytotoxicity of dox-loaded liposomes toward Daudi B cells. Cells were subjected to free dox, dox-loaded naked liposomes, or BPCNeuAc liposomes for 1 hour at 37°C. Cells were washed and incubated at 37°C for an additional 48 hours before measuring cell viability. Data shown are means of triplicate plus or minus SD. Representative data from 1 of 3 independent experiments are shown. Data were fitted using the Prism nonlinear regression software.
BPCNeuAc liposomes are bound and internalized by CD22-expressing cells. (A) FACS analysis for binding of fluorescently (NBD) labeled liposomes to wild-type CHO cells or CHO cells expressing recombinant human CD22. Cells were incubated with the naked (blue) or BPCNeuAc (green) liposomes or left untreated (gray) at 37°C for 1.5 hours before analysis. (B) CHO cells expressing CD22 were compared for their binding to the fluorescent (green) naked or BPCNeuAc liposomes. CD22 was detected with anti–human CD22 (red), and the nuclei were visualized by staining with DAPI. (C) Fluorescence microscopy analysis of the colocalization of BPCNeuAc liposomes with early endosomes. CHO-CD22 cells were incubated with fluorescent liposomes (green) as described. Early endosomes were visualized by staining with an Alexa Fluor 555–labeled anti-EEA1 (red). (D) Binding of BPCNeuAc liposomes to Daudi human B lymphoma cells. Daudi cells were incubated in mouse serum with liposomes containing 0% to 5% BPCNeuAc lipids or without liposomes (gray) before FACS analysis. (E) BPCNeuAc liposomes rapidly bind to Daudi cells. Fluorescent naked or BPCNeuAc liposomes were added to an aliquot of Daudi cells in mouse serum and incubated at 37°C for the indicated time before FACS analysis. Data are presented as mean channel of fluorescence (MCF) plus or minus SD. (n = 3). (F) Competitive binding of BPCNeuAc liposomes to Daudi cells in the presence of the free BPCNeuAc ligands. Fluorescent naked or BPCNeuAc liposomes were incubated with Daudi cells with the presence of the monovalent BPCNeuAc ligands at indicated concentration. Data were analyzed by FACS and shown as normalized MCF plus or minus SD. (n = 3). (G) Cytotoxicity of dox-loaded liposomes toward Daudi B cells. Cells were subjected to free dox, dox-loaded naked liposomes, or BPCNeuAc liposomes for 1 hour at 37°C. Cells were washed and incubated at 37°C for an additional 48 hours before measuring cell viability. Data shown are means of triplicate plus or minus SD. Representative data from 1 of 3 independent experiments are shown. Data were fitted using the Prism nonlinear regression software.
BPCNeuAc liposomes deliver cytotoxic cargo to Daudi human B lymphoma cells
The human Burkitt lymphoma Daudi B-cell line, commonly used for evaluating drugs for treatment of B-cell lymphomas,33,34 was next used to test the B-cell targeting of the BPCNeuAc liposomes. Binding assays were conducted at 37°C in 100% serum to mimic in vivo conditions. Liposomes without targeting ligands exhibit no detectable binding to Daudi cells, whereas liposomes with BPCNeuAc lipids show increased binding to the cells as the amount of ligand is increased from 0.8 to 5% (Figure 2D). Binding and uptake of BPCNeuAc liposomes to Daudi cells is rapid, saturable, and competitively inhibited by the presence of free glycan ligands of CD22 (Figure 2E-F). When dox (adriamycin), a standard chemotherapeutic drug, was loaded into liposomes, the nontargeted naked liposomes exhibited a 175-fold reduction in cell killing relative to free dox, representing protection of the cells by encapsulation of the drug. In contrast, BPCNeuAc liposomes exhibited a 33-fold higher potency (IC50 = 1.6μM) in cytotoxicity of Daudi cells (Figure 2G) than that of the naked liposomes (IC50 = 53μM), a difference predictive of increased efficacy in vivo.18
Pharmacokinetics and siglec specificity of BPCNeuAc liposomes
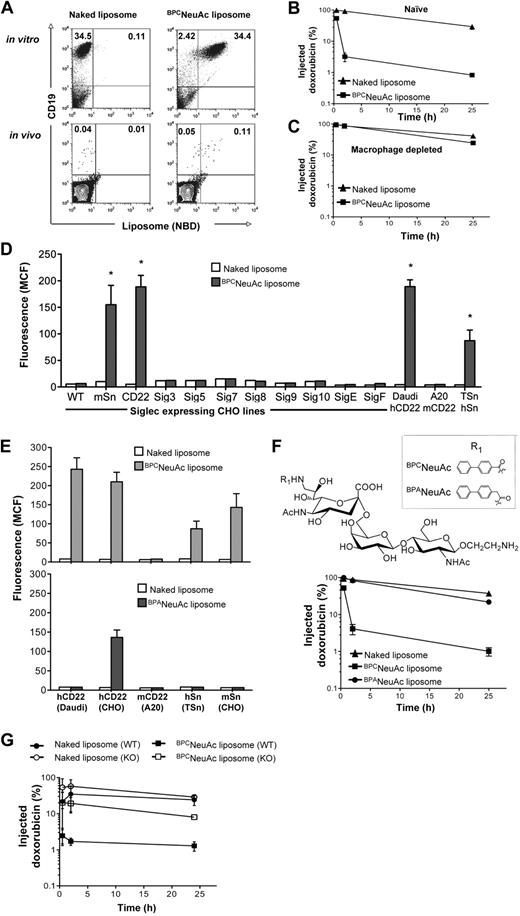
In preparation for in vivo efficacy studies, the specificity of BPCNeuAc liposome binding to Daudi cells was assessed when spiked into an aliquot of mouse whole blood (Figure 3A top panel). Binding is specific for the Daudi cells (hCD19+), with negligible binding to all murine cells (hCD19−), including murine B cells, because murine CD22 does not recognize the BPCNeuAc ligand.25 In addition, BPCNeuAc liposomes injected intravenously 30 minutes after injection of 1 × 106 Daudi cells also efficiently bound the hCD19+ Daudi cells in the blood (Figure 3A bottom panel). These results show BPCNeuAc liposomes efficiently target human B lymphoma cells both in vitro and in vivo.
Pharmacokinetics and siglec specificity of BPCNeuAc liposomes. (A) BPCNeuAc liposomes selectively bind to Daudi cells in mouse blood. In vitro (top panels): Daudi cells were spiked into an aliquot of mouse whole blood followed by addition of fluorescent naked or BPCNeuAc liposomes. Cells were stained with anti–human CD19 to distinguish Daudi cells from other cells in the mouse blood. In vivo (bottom panels): after intravenous injection of Daudi cells, mice were injected with fluorescent naked or BPCNeuAc liposomes. After 2 hours, a blood sample was drawn and the binding of liposomes to Daudi cells was analyzed by FACS. The numbers in the quadrants represent percentages of CD19 Daudi cells that bound or did not bind to liposomes. Shown are data from 1 of 3 independent experiments. (B-C) Dox-loaded liposomes were injected intravenously to the tumor-free SCID mice (3 mice per group) without or with pretreatment of clodronate to deplete tissue macrophages. A sample of blood was withdrawn at 0.5, 2, and 25 hours after liposome injections. The plasma concentration of dox was measured, and data are presented as percentage remaining of the initial injected drug plus or minus SD. (D) FACS analysis for binding of naked or BPCNeuAc liposomes to siglec-expressing CHO lines and Daudi, A20, and TSn cell lines that express hCD22, mCD22 and hSn, respectively. Binding is shown as MCF plus or minus SD (n = 3). Binding degree of BPCNeuAc liposomes to CHO-mSn, CHO-hCD22, Daudi, and TSn cell lines was significant in comparison to the same cell line that was treated with the naked liposomes (*P < .01). (E) Comparison of BPCNeuAc or BPANeuAc liposomes in binding to cell lines expressing hCD22, mCD22, hSn, and mSn. Binding of liposomes is expressed as MCF plus or minus SD (n = 3). (F) Top panel shows that the structures of the trisaccharide ligands designed to be specific for human CD22 are based on the parent compound NeuAcα2-6Galβ1-4GlcNAc, varying the biphenyl substituent at C-9 (R1). Bottom panel shows that BPANeuAc liposomes exhibit a long circulation time in vivo. A sample of blood was withdrawn from mice (n = 3) that received dox-loaded naked, BPCNeuAc, or BPANeuAc liposomes at 0.5, 2, and 25 hours after liposome injections. The plasma concentration of dox was detected using a fluorometer. Data are presented as percentage remaining of the initial dose ± SD. (G) Pharmacokinetics analysis for naked and BPCNeuAc liposomes in wild-type C57BL/6 and Sn knockout mice. Data are presented as percentage remaining of the initial dose ± SD (n = 3).
Pharmacokinetics and siglec specificity of BPCNeuAc liposomes. (A) BPCNeuAc liposomes selectively bind to Daudi cells in mouse blood. In vitro (top panels): Daudi cells were spiked into an aliquot of mouse whole blood followed by addition of fluorescent naked or BPCNeuAc liposomes. Cells were stained with anti–human CD19 to distinguish Daudi cells from other cells in the mouse blood. In vivo (bottom panels): after intravenous injection of Daudi cells, mice were injected with fluorescent naked or BPCNeuAc liposomes. After 2 hours, a blood sample was drawn and the binding of liposomes to Daudi cells was analyzed by FACS. The numbers in the quadrants represent percentages of CD19 Daudi cells that bound or did not bind to liposomes. Shown are data from 1 of 3 independent experiments. (B-C) Dox-loaded liposomes were injected intravenously to the tumor-free SCID mice (3 mice per group) without or with pretreatment of clodronate to deplete tissue macrophages. A sample of blood was withdrawn at 0.5, 2, and 25 hours after liposome injections. The plasma concentration of dox was measured, and data are presented as percentage remaining of the initial injected drug plus or minus SD. (D) FACS analysis for binding of naked or BPCNeuAc liposomes to siglec-expressing CHO lines and Daudi, A20, and TSn cell lines that express hCD22, mCD22 and hSn, respectively. Binding is shown as MCF plus or minus SD (n = 3). Binding degree of BPCNeuAc liposomes to CHO-mSn, CHO-hCD22, Daudi, and TSn cell lines was significant in comparison to the same cell line that was treated with the naked liposomes (*P < .01). (E) Comparison of BPCNeuAc or BPANeuAc liposomes in binding to cell lines expressing hCD22, mCD22, hSn, and mSn. Binding of liposomes is expressed as MCF plus or minus SD (n = 3). (F) Top panel shows that the structures of the trisaccharide ligands designed to be specific for human CD22 are based on the parent compound NeuAcα2-6Galβ1-4GlcNAc, varying the biphenyl substituent at C-9 (R1). Bottom panel shows that BPANeuAc liposomes exhibit a long circulation time in vivo. A sample of blood was withdrawn from mice (n = 3) that received dox-loaded naked, BPCNeuAc, or BPANeuAc liposomes at 0.5, 2, and 25 hours after liposome injections. The plasma concentration of dox was detected using a fluorometer. Data are presented as percentage remaining of the initial dose ± SD. (G) Pharmacokinetics analysis for naked and BPCNeuAc liposomes in wild-type C57BL/6 and Sn knockout mice. Data are presented as percentage remaining of the initial dose ± SD (n = 3).
In preliminary pharmacokinetics studies, we observed that the BPCNeuAc liposomes showed faster clearance from the blood relative to the naked liposomes in tumor-free animals (Figure 3B). We deduced that this was likely due to a known cross-reactivity of the BPCNeuAc ligand for another siglec, Sn (Siglec-1).35,36 Because Sn is expressed exclusively on tissue macrophages,37 we repeated the pharmacokinetics in mice depleted of macrophages using liposomal clodronate.32 This macrophage-specific treatment dramatically reduced clearance to that of naked liposomes (Figure 3C), demonstrating that clearance was indeed mediated by macrophages. To investigate this further, we tested binding against a panel of cell lines expressing human and murine siglecs, revealing that the BPCNeuAc liposomes did indeed bind to human (but not murine) CD22 (Siglec-2) and murine/human Sn (Figure 3D). To address the possibility that the sialylated ligand might be abrogating the “stealth” character of the liposomes, leading to nonspecific uptake by macrophages, we prepared liposomes containing a highly related ligand, BPANeuAc, where the BPC substituent was replaced with a 9-N-biphenylacetyl (BPA), which binds poorly to Sn.35,36 As expected, the BPANeuAc liposomes exhibited no binding to Sn while retaining binding to hCD22, albeit with reduced affinity (Figure 3E). These liposomes exhibited reduced clearance equivalent to that of the naked liposomes (Figure 3F), demonstrating that the ligand does not abrogate the stealth character of the liposomes. To confirm that the macrophage clearance of the BPCNeuAc liposomes was indeed mediated by Sn, we conducted pharmacokinetics in Sn-deficient mice,38 which revealed minimal difference in the clearance rate of the BPCNeuAc and naked liposomes (Figure 3G). Thus, the rapid clearance of the BPCNeuAc liposomes is due to a specific and rapid uptake by macrophages mediated predominately by Sn. Although the BPANeuAc liposomes exhibited higher specificity for CD22, their lower avidity reduces their binding to native Daudi cells (Figure 3E), precluding their use for investigation of efficacy in an in vivo model of human lymphoma. Accordingly, we envisioned the use of BPCNeuAc liposomes in a 2-dose protocol, assuming that uptake by macrophages would be blunted after the first dose, in analogy to the use of clodronate that ablated uptake of BPCNeuAc liposomes (Figure 3C).
BPCNeuAc liposomes prolong life in a murine model of human B-cell lymphoma
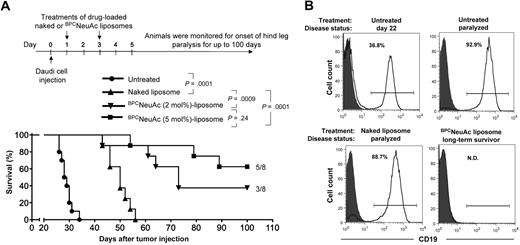
The efficacy of dox-loaded CD22 targeted liposomes was evaluated in a standard Daudi lymphoma model in SCID mice.39 As illustrated by the timeline in Figure 4A, Daudi cells (5 × 106) were injected intravenously and allowed to disseminate for 24 hours, followed by dosing mice on days 1 and 3 with PBS, dox-loaded naked liposomes, or BPCNeuAc liposomes (3 mg dox/kg per dose). As anticipated, pharmacokinetics analysis showed that rapid loss of BPCNeuAc liposomes from blood after the first dose was significantly attenuated after the second dose (supplemental Figure 2). Untreated animals (PBS) had a mean time of survival (MTS) of 28.5 days, whereas the dox-loaded naked liposomes increased the MTS to 50 days. In contrast, liposomes displaying 2% BPCNeuAc ligands exhibited a MTS of 73 days, with 3 of 8 long-term survivors being healthy at the end of the study (day 100), and liposomes with 5% ligands demonstrated a MTS greater than 100 days with 5 of 8 long-term survivors. Both treatments with drug-loaded BPCNeuAc liposomes comprising 2% or 5% ligands proved significant in improving survival rates compared with the treatment of the nontargeted (naked) liposomal regimen. Follow-up analysis revealed that hCD19+ Daudi cells constituted most bone marrow cells in paralyzed untreated or naked liposome–treated animals. In contrast, residual tumor cells were not detectable above background (Ig isotype control) in the bone marrow of the long-term survivors at day 100, further demonstrating the efficacy of the CD22-targeted liposomal regimen (Figure 4B; Table 1).
Efficacy of CD22-targeted liposomes in a xenograft model of the disseminated Daudi human B lymphoma. (A) Top panel shows timeline for the in vivo efficacy study. Bottom panel: tumor-bearing mice that received PBS (untreated, n = 10), dox-loaded naked liposomes (n = 8), or BPCNeuAc liposomes (n = 8) containing 2% or 5% BPCNeuAc ligands were monitored for the onset of hind-leg paralysis for up to 100 days. Survival rate is presented in a Kaplan-Meier plot with indication of numbers of long-term survivor animals. (B) Estimation of residual Daudi lymphoma cells in the bone marrow. Shown are histograms of bone marrow cells isolated from tumor-bearing mice followed by staining with isotype (solid) or anti–human CD19 (line) antibodies to identify infiltrated Daudi cells. Shown are results from 1 of 3 representative mice that received the indicated treatment. Percentages of lymphoid gated CD19+ Daudi cells in the bone marrow are indicated. N.D. (not detected) refers to less than a 0.4% background observed for an IgG isotype control.
Efficacy of CD22-targeted liposomes in a xenograft model of the disseminated Daudi human B lymphoma. (A) Top panel shows timeline for the in vivo efficacy study. Bottom panel: tumor-bearing mice that received PBS (untreated, n = 10), dox-loaded naked liposomes (n = 8), or BPCNeuAc liposomes (n = 8) containing 2% or 5% BPCNeuAc ligands were monitored for the onset of hind-leg paralysis for up to 100 days. Survival rate is presented in a Kaplan-Meier plot with indication of numbers of long-term survivor animals. (B) Estimation of residual Daudi lymphoma cells in the bone marrow. Shown are histograms of bone marrow cells isolated from tumor-bearing mice followed by staining with isotype (solid) or anti–human CD19 (line) antibodies to identify infiltrated Daudi cells. Shown are results from 1 of 3 representative mice that received the indicated treatment. Percentages of lymphoid gated CD19+ Daudi cells in the bone marrow are indicated. N.D. (not detected) refers to less than a 0.4% background observed for an IgG isotype control.
Detection of residual Daudi tumor cells in the bone marrow of the paralyzed or the long-term survivor mice
| Treatment group . | Disease status . | N . | Murine cells (hCD19−), % . | Daudi B cells (hCD19+), % . |
|---|---|---|---|---|
| Untreated | 22 d | 3 | 52.4 | 47.6 ± 9.1 |
| Untreated | Paralyzed | 3 | 5.0 | 95.1 ± 3.1 |
| Dox naked liposomes | Paralyzed | 3 | 9.5 | 90.5 ± 3.3 |
| Dox-BPCNeuAc (2%) liposomes | Survivor | 3 | 99.7 | ND |
| Dox-BPCNeuAc (5%) liposomes | Survivor | 5 | 99.8 | ND |
| Treatment group . | Disease status . | N . | Murine cells (hCD19−), % . | Daudi B cells (hCD19+), % . |
|---|---|---|---|---|
| Untreated | 22 d | 3 | 52.4 | 47.6 ± 9.1 |
| Untreated | Paralyzed | 3 | 5.0 | 95.1 ± 3.1 |
| Dox naked liposomes | Paralyzed | 3 | 9.5 | 90.5 ± 3.3 |
| Dox-BPCNeuAc (2%) liposomes | Survivor | 3 | 99.7 | ND |
| Dox-BPCNeuAc (5%) liposomes | Survivor | 5 | 99.8 | ND |
ND indicates not detectable compared with isotype control (< 0.4%).
BPCNeuAc liposomes target B cells from blood of patients with lymphoma
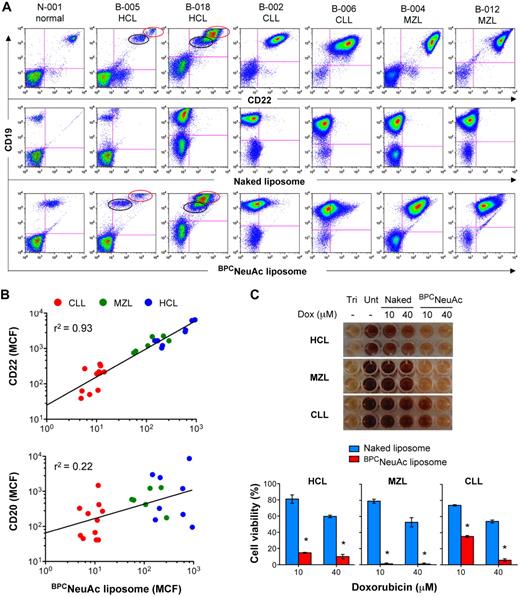
We next analyzed the ability of the BPCNeuAc liposomes to bind to neoplastic B cells from patients with leukemia/lymphoma. As summarized in Table 2, blood samples were obtained from 4 healthy donors and 30 patients with hairy cell leukemia (HCL), chronic lymphocytic leukemia (CLL), or splenic marginal zone lymphoma (MZL). These are 3 indolent lymphoproliferative disorders, known to express varying levels of CD22 (HCL, high; MZL, moderate; and CLL, low).40,41 Although most B cells in CLL and MZL samples are neoplastic cells, in the case of HCL, it is not uncommon to observe the normal and neoplastic B cells segregating into 2 distinct populations upon costaining with anti-CD22 and another B-cell marker (eg, CD19; Figure 5A). When this was observed, neoplastic B cells were distinguished from the normal B cells by staining with anti-CD103 (expressed on most HCL cells and usually present on MZL cells), anti-CD5 (expressed on CLL cells), and anti-κ and anti-λ light chain probes42 (data not shown).
Patient characteristics
| Participant no. . | Disease type . | Disease status . | Lymphocyte counts, K/μL . | CD20 level on B cells, MCF . | CD22 level on B cells, MCF . | Binding of BPCNeuAc liposomes* . |
|---|---|---|---|---|---|---|
| N-001 | Normal | NA | 2.5 | 1185 | 1739 | ++ |
| N-002 | Normal | NA | 1.8 | 958 | 1708 | ++ |
| N-003 | Normal | NA | 2.1 | 1226 | 1126 | ++ |
| N-004 | Normal | NA | 1.6 | 1086 | 1137 | ++ |
| B-001† | HCL | Remission | 3.1 | 2471 | 1208 | +++ |
| B-003 | HCL | Stable | 0.9 | 3017 | 1665 | +++ |
| B-005 | HCL | Untreated | 1.1 | 8613 | 6128 | ++++ |
| B-009 | HCL | Untreated | 1.1 | 1202 | 2971 | ++++ |
| B-017 | HCL | Progression | 0.9 | 154 | 1651 | +++ |
| B-018 | HCL | Relapsed | 14.1 | 327 | 1035 | +++ |
| B-021† | HCL | Remission | 1.8 | 95 | 6445 | ++++ |
| B-022 | HCL | Relapsed | 1.8 | 219 | 3322 | ++++ |
| B-002 | CLL | Stable | 23 | 418 | 324 | + |
| B-006 | CLL | Progression | 97.7 | 271 | 217 | + |
| B-007 | CLL | Progression | 36.4 | 1476 | 359 | + |
| B-008† | CLL | Stable | 54.3 | 232 | 50 | +/− |
| B-013 | CLL | Untreated | 50.1 | 150 | 219 | + |
| B-015† | CLL | Stable | 171 | 329 | 39 | +/− |
| B-016 | CLL | Progression | 70.5 | 67 | 192 | + |
| B-020 | CLL | Progression | 47.3 | 41 | 66 | + |
| B-023 | CLL | Untreated | 27.1 | 55 | 63 | +/− |
| B-024 | CLL | Untreated | 15.3 | 41 | 202 | + |
| B-025 | CLL | Untreated | 81.4 | 45 | 268 | +/− |
| B-027 | CLL | Untreated | 58.3 | 56 | 235 | + |
| B-029† | CLL | Progression | 78.8 | 42 | 56 | +/− |
| B-030† | CLL | Progression | 130 | 54 | 351 | + |
| B-004 | MZL | Progression | 80 | 1274 | 2246 | +++ |
| B-010 | MZL | Progression | 34.3 | 1219 | 2157 | +++ |
| B-011 | MZL | Stable | 22.8 | 564 | 838 | ++ |
| B-012† | MZL | Progression | 97.8 | 421 | 1190 | +++ |
| B-014† | MZL | Progression | 74.9 | 587 | 750 | ++ |
| B-019 | MZL | Untreated | 54 | 176 | 1648 | +++ |
| B-026 | MZL | Progression | 10.1 | 987 | 1126 | +++ |
| B-028 | MZL | Untreated | 1.1 | 536 | 1098 | +++ |
| Participant no. . | Disease type . | Disease status . | Lymphocyte counts, K/μL . | CD20 level on B cells, MCF . | CD22 level on B cells, MCF . | Binding of BPCNeuAc liposomes* . |
|---|---|---|---|---|---|---|
| N-001 | Normal | NA | 2.5 | 1185 | 1739 | ++ |
| N-002 | Normal | NA | 1.8 | 958 | 1708 | ++ |
| N-003 | Normal | NA | 2.1 | 1226 | 1126 | ++ |
| N-004 | Normal | NA | 1.6 | 1086 | 1137 | ++ |
| B-001† | HCL | Remission | 3.1 | 2471 | 1208 | +++ |
| B-003 | HCL | Stable | 0.9 | 3017 | 1665 | +++ |
| B-005 | HCL | Untreated | 1.1 | 8613 | 6128 | ++++ |
| B-009 | HCL | Untreated | 1.1 | 1202 | 2971 | ++++ |
| B-017 | HCL | Progression | 0.9 | 154 | 1651 | +++ |
| B-018 | HCL | Relapsed | 14.1 | 327 | 1035 | +++ |
| B-021† | HCL | Remission | 1.8 | 95 | 6445 | ++++ |
| B-022 | HCL | Relapsed | 1.8 | 219 | 3322 | ++++ |
| B-002 | CLL | Stable | 23 | 418 | 324 | + |
| B-006 | CLL | Progression | 97.7 | 271 | 217 | + |
| B-007 | CLL | Progression | 36.4 | 1476 | 359 | + |
| B-008† | CLL | Stable | 54.3 | 232 | 50 | +/− |
| B-013 | CLL | Untreated | 50.1 | 150 | 219 | + |
| B-015† | CLL | Stable | 171 | 329 | 39 | +/− |
| B-016 | CLL | Progression | 70.5 | 67 | 192 | + |
| B-020 | CLL | Progression | 47.3 | 41 | 66 | + |
| B-023 | CLL | Untreated | 27.1 | 55 | 63 | +/− |
| B-024 | CLL | Untreated | 15.3 | 41 | 202 | + |
| B-025 | CLL | Untreated | 81.4 | 45 | 268 | +/− |
| B-027 | CLL | Untreated | 58.3 | 56 | 235 | + |
| B-029† | CLL | Progression | 78.8 | 42 | 56 | +/− |
| B-030† | CLL | Progression | 130 | 54 | 351 | + |
| B-004 | MZL | Progression | 80 | 1274 | 2246 | +++ |
| B-010 | MZL | Progression | 34.3 | 1219 | 2157 | +++ |
| B-011 | MZL | Stable | 22.8 | 564 | 838 | ++ |
| B-012† | MZL | Progression | 97.8 | 421 | 1190 | +++ |
| B-014† | MZL | Progression | 74.9 | 587 | 750 | ++ |
| B-019 | MZL | Untreated | 54 | 176 | 1648 | +++ |
| B-026 | MZL | Progression | 10.1 | 987 | 1126 | +++ |
| B-028 | MZL | Untreated | 1.1 | 536 | 1098 | +++ |
MCF indicates mean channel fluorescence of malignant B lymphocytes; HCL, hairy cell leukemia; CLL, chronic lymphocytic leukemia; MZL, marginal zone lymphoma; and NA, not applicable.
Binding of BPCNeuAc liposomes to B cells was determined as MCF intensity: ++++ indicates MCF > 500; +++, 500 > MCF > 100; ++, 100 > MCF > 50; +, 50 > MCF > 10; and +/−, MCF < 10.
Patients previously treated with rituximab.
CD22-targeted liposomes bind to and kill malignant cells from patients with B-cell lymphomas or leukemias. (A) Binding of BPCNeuAc liposomes to patient B cells. Shown are representative samples from 1 healthy donor and 6 patients with HCL, CLL, or splenic MZL. Lymphocytes were gated based on the forward- and side-scatter characteristics. B-cell populations detected with anti–human CD19 were analyzed for binding of anti–human CD22 (top row), naked liposomes (middle row), and CD22-targeted BPCNeuAc liposomes (bottom row). CD22-bright HCL (red circle) was distinguished from normal B cells (black circle). (B) Correlation of binding of BPCNeuAc liposomes with CD22 or CD20 expression on the B-cell lymphomas. The diagonal lines represent linear regression that was analyzed using Prism software and the values of goodness of fit (r2) are indicated (n = 25). (C) Cytotoxicity of the dox-loaded BPCNeuAc liposomes toward malignant B cells. Top panel shows results of the viability of blood lymphocytes evaluated by the standard MTT assay after treatments of dox-loaded naked or BPCNeuAc liposomes with dox concentrations at 10 or 40μM. Cells left untreated (Unt) were defined as the maximal cell viability. Complete cell killing was determined from the Triton X-100 lysed cells (Tri). Bottom panel shows percentages (means of triplicate ± SD) of the viable blood lymphocytes after treatment with dox-loaded naked or BPCNeuAc liposomes. *P < .05 compared with control treatments of naked liposomes. Representative data from 1 of 4 samples are shown.
CD22-targeted liposomes bind to and kill malignant cells from patients with B-cell lymphomas or leukemias. (A) Binding of BPCNeuAc liposomes to patient B cells. Shown are representative samples from 1 healthy donor and 6 patients with HCL, CLL, or splenic MZL. Lymphocytes were gated based on the forward- and side-scatter characteristics. B-cell populations detected with anti–human CD19 were analyzed for binding of anti–human CD22 (top row), naked liposomes (middle row), and CD22-targeted BPCNeuAc liposomes (bottom row). CD22-bright HCL (red circle) was distinguished from normal B cells (black circle). (B) Correlation of binding of BPCNeuAc liposomes with CD22 or CD20 expression on the B-cell lymphomas. The diagonal lines represent linear regression that was analyzed using Prism software and the values of goodness of fit (r2) are indicated (n = 25). (C) Cytotoxicity of the dox-loaded BPCNeuAc liposomes toward malignant B cells. Top panel shows results of the viability of blood lymphocytes evaluated by the standard MTT assay after treatments of dox-loaded naked or BPCNeuAc liposomes with dox concentrations at 10 or 40μM. Cells left untreated (Unt) were defined as the maximal cell viability. Complete cell killing was determined from the Triton X-100 lysed cells (Tri). Bottom panel shows percentages (means of triplicate ± SD) of the viable blood lymphocytes after treatment with dox-loaded naked or BPCNeuAc liposomes. *P < .05 compared with control treatments of naked liposomes. Representative data from 1 of 4 samples are shown.
All blood samples were assessed for their levels of B-cell expression of CD22 and the binding to the BPCNeuAc liposomes. In all cases, control naked liposomes bound no better to the CD19+ B cells than the non–B cells (Figure 5A; supplemental Figure 3). In contrast, B cells from all patients bound to CD22-targeted liposomes in proportion to their CD22 expression, regardless of their prior treatment history. The CD22-targeted liposomes were confirmed not to cross-react with neutrophils, known to express high levels of CD33 (Siglec-3; supplemental Figure 4). B cells from the patients with HCL and MZL bound strongly to the BPCNeuAc liposomes, whereas B cells from patients with CLL bound at lower levels. Using mean channel fluorescence (MCF) as a quantitative measure, binding of BPCNeuAc liposomes strongly correlated with the expression of CD22, whereas there was no correlation with binding to CD20, which is the target of the anti-CD20 antibody rituximab used in standard treatment of B-cell lymphomas (Figure 5B).
Despite variation of CD22 expression, malignant B cells from patients with HCL, MZL, and CLL were sensitive to cell killing by the dox-loaded BPCNeuAc liposomes in an in vitro assay8 (Figure 5C). The assay involves exposure of the peripheral blood leukocytes to the liposomal preparations for 1 hour, followed by incubation of the cells for 5 days in media. Cell viability was assessed using a standard MTT assay. Whereas HCL and MZL were susceptible to both low (10μM) and high (40μM) concentrations of the targeted liposomal dox, CLL cells were efficiently killed only by the higher dox concentration. Taken together, the results indicate that while binding of the CD22-targeted liposome to malignant B cells is proportional to CD22 expression on the cell surface, even low levels of expression on CLL cells are sufficient to effect cell killing. Because the targeted liposomes breach the cells by an endocytic mechanism, it is anticipated that the therapeutic benefit will be synergistic with anti-CD20 rituximab and other immune-mediated therapies.
Discussion
Two families of glycan-binding proteins, the siglecs and C-type lectins, are recognized as attractive targets for cell-directed therapies because they exhibit restricted expression to 1 or a few cell types, and are predominately expressed on leukocytes or endothelial cells that can be exploited for modulation of the immune system and treatment of inflammatory disease.24,43 A disadvantage is that glycan-binding proteins typically have low affinity for their glycan ligands, which pose constraints for the design of ligand-based targeting agents.44,45 In this report, we demonstrate that dox-loaded liposomal nanoparticles displaying high-affinity glycan ligands of human CD22 (Siglec-2) efficiently target human B cells in an in vivo murine model of human lymphoma.
We have found that multivalent presentation of high-affinity CD22 ligands on liposomes is the only multivalent platform to date that generates sufficient avidity to target B cells in a complex biologic milieu. It is well documented that endogenous cis ligands on B cells mask binding of synthetic ligands to CD22 in trans unless cells are first treated with sialidase.44,46 However, we have previously demonstrated that multivalent display of the high-affinity ligand (BPCNeuAc) on a polyacrylamide polymer could complete with cis ligands, and to bind to CD22 on native B cells.25 Similarly, a heterobifunctional ligand comprising an antigen, nitrophenol (NP), coupled to the BPCNeuAc ligand could use an anti-NP antibody as a scaffold to assemble immune complexes with CD22 on B cells.26 However, subsequent analysis showed that neither of these multivalent complexes could bind to B cells in 100% serum, abrogating their use for targeting B cells in vivo (data not shown). In contrast, the ligand-decorated liposomes not only bound to native cells, but also bound well to B cells in serum in vitro and in vivo. Thus, the liposomes provide a robust scaffold that is unique for targeting CD22 on B cells in a physiologically relevant setting.
Liposomal formulations of doxorubicin are currently approved for treatment of solid tumors, acting to prolong circulatory half-life, and concentrate in tumor tissue by escaping the blood through leaky blood vessels. In contrast, BPCNeuAc liposomes loaded with dox effectively target B cells by binding to and being endocytosed by CD22, carrying their toxic cargo into the cell (Figure 2). The effect of active targeting is clearly seen when comparing in vivo efficacy of the “naked” and ligand-decorated liposomes in a disseminated model of human B-cell lymphoma on SCID mice (Figure 4A). The ligand-targeted liposomes significantly prolonged the lives of tumor-bearing animals compared with the nontargeted liposomal dox. The results support the strategy for developing a new use for an approved drug by converting the delivery mechanism to active targeting and uptake by a targeted cell. This concept is of direct therapeutic interest for treatment of non-Hodgkin lymphoma or other B-cell lymphomas, with more than a dozen clinical trials completed or in progress evaluating liposomal dox in place of dox for standard cyclophosphamide, doxorubicin, prednisone, and vincristine (CHOP) chemotherapy.47-49
Although the BPCNeuAc liposomes proved strong efficacy in the B-cell lymphoma model, there is room for further improving the selectivity and effectiveness of the targeting ligand. These liposomes have a known cross-reactivity with Sn (Siglec-1) expressed on tissue macrophages, which effectively compete for liposomes in the in vivo experiments, limiting the amount available for targeting to the human B lymphoma. No other siglec appears to play a significant role in the clearance of liposomes in wild-type mice, because rapid clearance is not seen in macrophage-depleted mice and in Sn knockout mice (Figure 3). Thus, liposomes bearing ligands that do not bind to Sn and retain affinity for CD22 would be expected to exhibit significantly improved targeting of human B cells.
Several reports combined with the results presented here suggest that glycan ligand-decorated liposomes may provide a robust platform with unique properties for targeted delivery of therapeutic agents to cells that express the cognate glycan-binding protein.20-24,43 We suggest that design of glycan ligands that will target a single glycan-binding protein is within reach, and that additional examples of successful targeting using the pharmaceutically proven liposomal nanoparticle platform will stimulate their consideration as an option to antibodies for targeted therapeutics.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Kelly Bethel for her assistance in analyzing patient B lymphoma cells using FACS and Laurie Sampson for patient sample collections. We thank Norihito Kawasaki for providing clonal CD22-expressing CHO cells and Emma McKenzie for assistance in handling Sn knockout mice. We also thank Anna Tran-Crie and Mary O'Reilly for assistance in manuscript preparation and review.
This work was supported by National Institutes of Health grants R01-AI050143 and R01-GM060938.
National Institutes of Health
Authorship
Contribution: W.C.C. and J.C.P. designed the reported experiments; W.C.C. prepared liposomes and performed all in vitro and in vivo experiments; G.C.C. synthesized the CD22 ligand coupled to phospholipids; D.S.S. and A.S. provided patient blood samples and clinical insights; P.R.C. provided Sn knockout mice and scientific inputs; W.C.C. wrote the initial draft of the manuscript; and all authors participated in the analysis of the data and contributed to manuscript revisions.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: James C. Paulson, The Scripps Research Institute, 10550 North Torrey Pines Rd, Mailcode MEM-L71, La Jolla, CA 92037; e-mail: jpaulson@scripps.edu.