Abstract
Gene-targeting experiments report that the homeodomain-interacting protein kinases 1 and 2, Hipk1 and Hipk2, are essential but redundant in hematopoietic development because Hipk1/Hipk2 double-deficient animals exhibit severe defects in hematopoiesis and vasculogenesis, whereas the single knockouts do not. These serine-threonine kinases phosphorylate and consequently modify the functions of several important hematopoietic transcription factors and cofactors. Here we show that Hipk2 knockdown alone plays a significant role in terminal fetal liver erythroid differentiation. Hipk1 and Hipk2 are highly induced during primary mouse fetal liver erythropoiesis. Specific knockdown of Hipk2 inhibits terminal erythroid cell proliferation (explained in part by impaired cell-cycle progression as well as increased apoptosis) and terminal enucleation as well as the accumulation of hemoglobin. Hipk2 knockdown also reduces the transcription of many genes involved in proliferation and apoptosis as well as important, erythroid-specific genes involved in hemoglobin biosynthesis, such as α-globin and mitoferrin 1, demonstrating that Hipk2 plays an important role in some but not all aspects of normal terminal erythroid differentiation.
Introduction
Terminal erythropoiesis is a complex but well-coordinated process encompassing many significant cellular changes, including: 4 or 5 stereotypical cell divisions of colony-forming unit-erythroid progenitors that result in decrease in cell size; increase in chromatin condensation and final enucleation; increase in hemoglobin and other specialized proteins; and alterations in cell-surface receptor expression.1 Such involved tissue-specific expression programs require intricate transcriptional regulatory networks that include the dynamic modulation of gene expression through posttranslational modification of transcription factors, cofactors, and chromatin-modifying enzymes in response to stimuli and environmental conditions. Phosphorylation is one of the most prevalent of these reversible modifications and can regulate transcription by altering transcription factor and cofactor stability, subcellular localization, ability to bind to chromatin remodeling complexes, ability to complex with and recruit core transcriptional machinery, and even (for some regulators) the ability to bind DNA.2
The homeodomain-interacting protein kinases (Hipks) are a family of 4 highly conserved serine-threonine kinases that were originally identified through their interaction with homeobox factors as enzymes that regulate transcription.3 The kinase domains of HIPK1, HIPK2, and HIPK3 show greater than 90% homology, the homeobox-interacting domains more than 70%; HIPK4 is the least conserved. Hipk1 and Hipk2 transcripts are expressed in many tissues, with higher expression in such tissues as neurons (the amygdala and prefrontal cortex), skeletal and smooth muscle, thyroid, and the hematopoietic system (Novartis Gene Expression Atlas, http://biogps.gnf.org). Thus far, most of the information on the functions of these kinases in hematopoiesis comes from studies performed on HIPK2 using genetically modified mice and in vitro hematopoietic cell lines. Hipk1/Hipk2 double-knockout embryos are embryonic-lethal between embryonic day (E) 9.5 and E12.5 and exhibit defects in neuronal development, blood vessel formation, and definitive hematopoiesis,4,5 measured as fewer colony-forming cells isolated from the yolk sac and para-aortic-splanchnopleural region of double-knockout embryos than from control animals. Blood counts or fetal liver hematopoiesis have not yet been evaluated in the double knockouts, so the effects on terminal hematopoiesis are unknown. Mice deficient in either Hipk1 or Hipk2 alone were initially reported to be grossly normal and fertile,5,6 but a more recent study has shown that Hipk2−/− mice are significantly smaller than their wild-type littermates through adulthood but show no difference in hematology, blood chemistry, or pathology.7
Studies on the Hipk2−/− mouse have also implicated HIPK2 in the regulation of cell proliferation and apoptosis. Survival and proliferation defects were observed in single neurons in the Hipk2 knockout,5 and mouse embryonic fibroblasts from these mice also showed reduced proliferation with accumulation in the G0/G1 phase of the cell cycle.7 HIPK2 also binds several regulators involved in cell survival: the tumor suppressor p53,8-10 antiapoptotic transcription factors Brn3a5 and CtBP,11 as well as the oncogenes c-Myb12 and c-Ski13 ; Hipk2 is also required for TGFβ-dependent survival of murine dopaminergic neurons14 and helps to regulate the G1/S transition in epidermal cells through interactions with β-catenin and CtBP.15
Many other binding partners of HIPK2 play significant roles in the regulation of hematopoietic differentiation, including Smad1-4, p300, AML1, MOZ, Groucho, HDAC1, c-Myb, c-Jun, c-Fos, and Pax62,4,12,16,17 ; knockdown of HIPK2 in hematopoietic cell lines recently uncovered a role for Hipk2 in cell-cycle arrest during non-DNA damage conditions, such as terminal differentiation and growth factor deprivation.18 Given the hematopoietic defects in the Hipk1−/−Hipk2−/− double-knockout as well as these recent findings, we examined the role that these kinases play specifically in terminal erythropoiesis. We found that Hipk1 and Hipk2 (but not Hipk3) are highly induced in murine fetal liver erythropoiesis during the same transition that the most important erythroid-specific genes, such as the globins and heme biosynthetic enzymes, are also induced. Using retroviral shRNA knockdown experiments, we show that Hipk2 is crucial for normal terminal erythroid proliferation and regulates some aspects of terminal erythroid differentiation, specifically hemoglobin accumulation and enucleation, but not surface expression of erythroid markers TER119 or CD71. We also found that Hipk2 is required for expression of some proliferation-, apoptosis-, and cell cycle–related transcripts as well as many genes involved in heme synthesis and hemoglobin production. It is clear that Hipk2 plays a significant role in terminal fetal liver erythropoiesis, but we cannot yet draw conclusions about the role of Hipk1 in terminal erythroid development.
Methods
Cells
The retrovirus-packaging cell line 293T was maintained in Dulbecco modified Eagle medium containing 10% fetal bovine serum (FBS; Invitrogen). Fetal liver cells were isolated from C57BL/6 embryos (The Jackson Laboratory) and resuspended by pipetting in erythroid-differentiation medium (Iscove modified Dulbecco medium containing 20% FBS [Invitrogen], 2mM l-glutamine [Invitrogen], and 10−4M β-mercaptoethanol [Sigma-Aldrich]). Single-cell suspensions were prepared by passing the dissociated cells through 70-μm cell strainers. All studies were approved by the Whitehead Institute for Biomedical Research Animal Care and Use Committee.
Erythroid precursor isolation and culture
Total fetal liver cells were labeled with biotin-conjugated anti-TER119 antibody (1:100; BD Biosciences PharMingen) and biotin-conjugated anti-CD11b (Mac-1, 1:100; eBioscience), and TER119/Mac1-negative cells were purified through a StemSep column per the manufacturer's instructions (StemCell Technologies). Purified cells were seeded in fibronectin-coated wells (BD Discovery Labware) at a cell density of 1 to 2 × 105/mL. On the first day, the purified cells were cultured in Iscove modified Dulbecco medium containing 15% FBS (StemCell Technologies), 1% detoxified bovine serum albumin (StemCell Technologies), 250 μg/mL holo-transferrin (Sigma-Aldrich), 10 μg/mL recombinant human insulin (Sigma-Aldrich), 2mM l-glutamine (Invitrogen), 10−4M β-mercaptoethanol (Sigma-Aldrich), and 10 U/mL Epo (Amgen) as previously described.19 On the second day, this medium was replaced with erythroid-differentiation medium (described in “Cells”). Cells were counted at 24 and 48 hours using a hemacytometer, and aliquots were also harvested for various studies, described below in “Immunostaining, flow cytometric analysis of erythroid differentiation and enucleation, and FACS.”
Retroviral constructs
MSCV-pgkGFP-U3-U6P-Bbs vector (murine stem cell retroviral vector-pgk promoter-GFP-U6 promoter shRNA) was a gift from Dr Biao Luo (Broad Institute, Cambridge, MA). The human U6 promoter had been inserted at an NheI site into U3 of the 3′-LTR. The U6 promoter is in the opposite orientation to the retroviral LTR and the GFP gene. Two back-to-back BbsI sites were used as cloning sites for each shRNA. An shRNA against the firefly luciferase gene was also cloned into an identical vector as the control. Candidate shRNA sequences for each gene were obtained from the online Broad Institute RNAi consortium shRNA library (http://www.broadinstitute.org/rnai/trc/lib, Cambridge, MA).
Generation of retroviral supernatants and infection of primary cells
The 293T cells were seeded on 100-mm dishes 1 day before transfection. The 293T cells were then cotransfected with 10 μg retroviral plasmid and 5 μg pCL-Eco vector using the Fugene-6 reagent (Roche Diagnostics) according to the manufacturer's protocol. Retroviral supernatants were collected 48 hours after transfection and stored in aliquots at −80°C. To titer the packaged viruses, a series of dilutions of the supernatants were used to infect NIH-3T3 cells; titers of 107 to 108 infectious units per milliliter were routinely obtained. For infection of purified TER119-negative fetal liver cells, 1 × 105 to 5 × 105 cells were resuspended in 1 mL thawed viral supernatant containing 4 μg/mL Polybrene (Sigma-Aldrich) and centrifuged at 241.6g for 1 hour at 25°C.
Western blotting
For most experiments, 3 × 106 cells GFP+ cells were sorted from each sample after 36 hours of retroviral infection and used to prepare protein extracts using modified radio immunoprecipitation assay buffer supplemented with Complete protease inhibitors (Roche Diagnostics). Protein was run on 4% to 12% Bis-Tris NuPAGE precast gels with MOPS running buffer (Invitrogen) and transferred to nitrocellulose membrane. Primary antibodies used were: rabbit polyclonal to glyceraldehyde-3-phosphate dehydrogenase at 1:1000 (Santa Cruz Biotechnology), mouse monoclonal to Hipk2 at 1:500 (Abcam), and rabbit polyclonal to Hipk1 at 1:500 (Santa Cruz Biotechnology). Secondary antibodies used were: TrueBlot horseradish peroxidase anti–mouse IgG at 1:2000 (eBioscience) and TrueBlot horseradish peroxidase anti–rabbit IgG at 1:2000 (eBioscience). Blots were scanned in as .pic images and quantified using ImageJ software (http://rsbweb.nih.gov/ij; National Institutes of Health, Bethesda, MD).
Immunostaining, flow cytometric analysis of erythroid differentiation and enucleation, and FACS
Either freshly isolated fetal liver cells or retrovirally transduced fetal liver cells that had been cultured in vitro were immunostained with phycoerythrin (PE)–conjugated anti-CD71 at 1:200 (BD Biosciences PharMingen), and allophycocyanin-conjugated anti-TER119 at 1:200 (BD Biosciences PharMingen) antibodies. Cells were also stained with 10 μg/mL Hoechst 33342 for 15 minutes at room temperature, and propidium iodide was added to exclude dead cells from analysis. Flow cytometry was carried out on a FACS LSRII (BD Biosciences) and analyzed using FlowJo software (TreeStar). Retrovirally transduced cells were sorted for GFP+ expression using a FACSAria machine (BD Biosciences) with reduced pressure. Sorted GFP+ cells were harvested for various studies described below in the following 4 sections.
Cytospin preparation, histology staining, and hemoglobin quantification
After sorting, 5 × 103 to 2 × 104 GFP+ cells from each sample were spun onto slides for 3 minutes at 71.7g (Cytospin 3, Thermo Shandon) and air-dried. Cells were fixed in ice-cold methanol for 2 minutes and stained with dilute 3,3-diaminobenzidine Giemsa stains according to the manufacturer's recommendations (Sigma-Aldrich). Color microscopy images were obtained using Axiovision LE software, Version 4.7.2 (Carl Zeiss), using an Axiophot (Carl Zeiss) microscope, equipped with an Axiocam MR color digital camera (Carl Zeiss) and a Plan NEOFLUAR 40×/1.30 oil lens (Carl Zeiss). Images were taken using brightfield microscopy at room temperature. Positively stained cells were quantified using 5 independent black-and-white fields captured at 40× magnification using phase-contrast: benzidine-positive (nonwhite) cells were counted; color shots were taken using light microscopy with the same microscope, camera, lens, and software. Hemoglobin was quantified by lysis of 1 × 106 GFP+ cells in 200 μL of the Drabkin reagent and analysis by spectrophotometric reading at 540 nm.
Cell-cycle and apoptosis assays
For cell-cycle analysis, GFP+ cells were isolated, washed with phosphate-buffered saline containing 2% FBS, and then permeabilized in 100% ethanol. Cells were resuspended in staining solution containing 50 μg/mL propidium iodide and incubated at 37°C for 30 minutes in the dark. The percentages of cells in G1/S/G2 were then calculated by flow cytometry using the Dean-Jett-Fox model within the FloJo software package (TreeStar). For apoptosis, GFP+ cells were labeled with PE-conjugated annexin V and 7-amino-actinomycin D (7-AAD) using the apoptosis detection kit (BD Biosciences PharMingen) according to the manufacturer's protocol. Apoptotic cells were identified as annexin V-PE-positive and 7-AAD-negative by flow cytometry. For apoptosis analysis, gating is on GFP+ cells, but cell-cycle analysis is not (in the cell-cycle assay, permeabilizing with ethanol causes GFP to leak out).
RNA isolation and quantitative PCR
GFP+ cells from each knockdown sample, or cells from each differentiation stage R1 to R5, were harvested for mRNA isolation 36 hours after retroviral infection or at day E14.5, respectively. Total RNA was isolated with the RNeasy micro kit (QIAGEN) and then reverse-transcribed using the Affinity Script QPCR cDNA synthesis kit with random primers (Stratagene). Quantitative polymerase chain reaction (PCR) was performed using SYBR Green real-time PCR on the ABI Prism 7900 sequence detection system (Applied Biosciences). Primers used are listed in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Normalization was performed against RPS3, a ubiquitous ribosomal gene, and each sample was compared with the luciferase control. Samples used were from at least 3 separate RNA isolations.
Microarray sample preparation, hybridization, and analysis
Total RNA isolated from each sample (described in “RNA isolation and quantitative PCR”) was analyzed for quality using a bioanalyzer (2100 Bioanalyzer; Agilent Technologies), according to the manufacturer's instructions. For each hybridization, 300 ng total RNA was reverse-transcribed using oligo(deoxythymidine)12-18 (oligo(dT)12-18) primer (Invitrogen) using the Superscript II enzyme kit (Invitrogen) and labeled using the Micromax ASAP labeling kit (PerkinElmer Life and Analytical Sciences). For Hipk knockdown microarray analysis, control samples were labeled with cyanin-3-uridine 5′-triphosphate and retroviral samples with cyanin-5-uridine 5′-triphosphate from the labeling kit. Control cDNA and each retroviral sample cDNA were hybridized to a dual-color mouse whole genome oligo expression microarray (Agilent Technologies), according to the manufacturer's instructions. Slides were scanned using the Agilent scanner and processed using Agilent's Genepix software. Resulting intensities were normalized using global mean normalization, and log ratios compared with the control sample were determined using linear-fit modeling with limma, both using R and Bioconductor.20 For erythropoiesis expression profiling, cDNA from each stage R1 to R5 was labeled and hybridized to the Affymetrix GeneChip Mouse Genome 430 2.0 array (3′ mouse whole genome expression array). Affymetrix slides were then scanned using the GeneChip Scanner 3000 7G and processed using Affymetrix's GeneChip Operating Software, Version 1.1.1 (Affymetrix). Resulting intensities were normalized using global mean normalization, and log ratios compared with the R2 stage were determined using linear-fit modeling with limma, both using R and Bioconductor.20 Significantly changed genes were defined as at least a 2-fold increase or decrease from control and with P values less than .05.
Results
Hipk1 and Hipk2 mRNA are highly induced during the same stage of erythropoiesis as other important, erythroid-specific transcripts
Fetal liver erythroid differentiation can be studied quantitatively and the precursors sorted into successive erythroid differentiation stages by fluorescence-activated cell sorter (FACS); each of the distinct populations R1 to R5 (Figure 1A) corresponds morphologically to consecutive stages of in vivo erythroid differentiation, progressing from predominately primitive progenitor cells (including blast-forming units-erythroid and colony-forming units-erythroid) in R1, to proerythroblasts and early basophilic erythroblasts in R2, to early and late basophilic erythroblasts in R3, to polychromatophilic and orthochromatophilic erythroblasts in R4, to late orthochromatophilic erythroblasts and reticulocytes in R5.19
Induction of Hipk1 and Hipk2 during in vivo erythropoiesis. (A) Freshly isolated E14.5 murine fetal liver cells analyzed by FACS with antibodies staining for CD71 and Ter119. Regions R1 through R5 are defined by their staining patterns: CD71medTer119low, CD71highTer119low, CD71highTer119high, CD71medTer119high, and CD71lowTer119high, respectively. (B) Quantitative RT-PCR on mRNA isolated from each corresponding stage in panel A using primers against Hipk1 and Hipk2. (C) Quantitative RT-PCR using primers against other erythroid-specific genes: GATA1, FoxO3, Klf1, and α (Hba-a1)– and β (Hbb-b1)–globins. The relative amounts shown for each transcript are compared with the level of the corresponding transcript in R2 cells; n = 3 (mean ± 2 SEM).
Induction of Hipk1 and Hipk2 during in vivo erythropoiesis. (A) Freshly isolated E14.5 murine fetal liver cells analyzed by FACS with antibodies staining for CD71 and Ter119. Regions R1 through R5 are defined by their staining patterns: CD71medTer119low, CD71highTer119low, CD71highTer119high, CD71medTer119high, and CD71lowTer119high, respectively. (B) Quantitative RT-PCR on mRNA isolated from each corresponding stage in panel A using primers against Hipk1 and Hipk2. (C) Quantitative RT-PCR using primers against other erythroid-specific genes: GATA1, FoxO3, Klf1, and α (Hba-a1)– and β (Hbb-b1)–globins. The relative amounts shown for each transcript are compared with the level of the corresponding transcript in R2 cells; n = 3 (mean ± 2 SEM).
To determine which networks are active in the transcriptional regulation of terminal erythropoiesis, comprehensive expression profiling was performed on these primary fetal liver erythroid populations R1 to R5 (Figure 1A) and analyzed for changes in transcripts encoding DNA-binding factors and their modulators (data not shown). Microarray results have been submitted to the Gene Expression Omnibus under accession number GSE2039. Based on these microarray results, mRNA transcripts of 2 regulators, Hipk1 and Hipk2, were found to be highly induced during the R2/R3 transition (Figure 1B). The greatest induction of erythroid-specific transcripts (such as α- and β-globins, Gata1, Fog1, FoxO3, and Klf1 [EKLF], ankyrin and β-spectrin) also occurs during this same transition, just as the glycoprotein TER119 is being induced, which, interestingly, coincides with a decrease in erythropoietin receptor mRNA and subsequent erythroid precursors' decreasing requirement for erythropoietin.19 Quantitative reverse transcription real-time (RT)-PCR on mRNA isolated from these sorted populations confirms this induction for Gata1, Foxo3, Klf1, and α- and β-globins (Figure 1C).
shRNA-mediated knockdown of Hipk2 inhibits terminal proliferation of erythroid precursors
To examine the roles of Hipk1 and Hipk2 in erythroid differentiation, retroviral vectors containing shRNA hairpins against Hipk1 and Hipk2 were constructed. Quantitative RT-PCR results (Figure 2A-B) show the extent and specificity of each of the shRNA hairpins: sample shHK2a shows specific Hipk2 knockdown (∼ 85%) and does not affect Hipk1, whereas sample shHK1a shows specificity for Hipk1 (∼ 80% knockdown) and not Hipk2. The 2 constructs (a and b) showed similar effects on proliferation (data not shown), so shHK1a and shHK2a were used throughout the remainder of the experiments and are referred to as shHK1 and shHK2. Western blot and quantification of the bands (Figure 2C) show the specificity and extent of shRNA knockdown of HIPK1 and HIPK2 at the protein level after 36 hours of infection. There was more significant knockdown of HIPK2 at the protein level. Because the retroviral vectors also contain GFP, viral infection rate was measured via GFP expression by flow cytometry: these 2 constructs consistently showed at least a 75% to 80% rate of infection (data not shown).
Specific knockdown of Hipk2 inhibits terminal erythroid proliferation. RNAi short hairpins designed to specifically target Hipk1 (shHK1a and b) or Hipk2 (shHK2a and b) were cloned into retroviruses and infected into Ter119-negative fetal liver cells before culture in Epo-containing media. The control sample refers to a nonspecific shRNA hairpin directed against the firefly luciferase gene. (A-B) Quantitative RT-PCR on RNA isolated from infected cells after 48 hours was performed using primers against Hipk1 (A) or Hipk2 (B); n = 3 (mean ± 2 SEM). Expression levels were normalized to RPS3 and then compared with control-infected cells. (C) Western blot showing reduced HIPK1 and HIPK2 protein by 36 hours as a result of each specific shRNA knockdown; quantification of the bands is included below the blot. (D) Cell counts measured at 24 and 48 hours after retroviral infection; fold increase compared with equal starting cell numbers (2 × 105) at day 0, set to 1; n = 5 (mean ± 2 SEM). *P < .05.
Specific knockdown of Hipk2 inhibits terminal erythroid proliferation. RNAi short hairpins designed to specifically target Hipk1 (shHK1a and b) or Hipk2 (shHK2a and b) were cloned into retroviruses and infected into Ter119-negative fetal liver cells before culture in Epo-containing media. The control sample refers to a nonspecific shRNA hairpin directed against the firefly luciferase gene. (A-B) Quantitative RT-PCR on RNA isolated from infected cells after 48 hours was performed using primers against Hipk1 (A) or Hipk2 (B); n = 3 (mean ± 2 SEM). Expression levels were normalized to RPS3 and then compared with control-infected cells. (C) Western blot showing reduced HIPK1 and HIPK2 protein by 36 hours as a result of each specific shRNA knockdown; quantification of the bands is included below the blot. (D) Cell counts measured at 24 and 48 hours after retroviral infection; fold increase compared with equal starting cell numbers (2 × 105) at day 0, set to 1; n = 5 (mean ± 2 SEM). *P < .05.
To measure the effects of Hipk knockdown on cell growth, cell counts were determined at 24 and 48 hours after retroviral shRNA infection (Figure 2D) during in vitro culture in Epo-containing media. By 48 hours, the Hipk2 knockdown sample showed a statistically significant inhibition in growth (∼ a 2- to 3-fold reduction in cell number), whereas Hipk1 knockdown had very little effect.
Accumulation in S phase and increased apoptosis of erythroblasts after shRNA knockdown of Hipk1 or Hipk2
To help determine the mechanisms responsible for the significant defect in cell growth after specific knockdown of Hipk2, both cell-cycle status and levels of apoptosis after Hipk1 or Hipk2 depletion were analyzed by flow cytometry. At 24 hours, there was not much difference in cell-cycle status between cells infected with knockdown or control constructs (Figure 3A-B), and approximately 50% of the cells were cycling in S phase. However, by 36 hours, most control cells started to arrest in G1, as would be expected for cells about to extrude their nuclei,21 but more of the Hipk1 and Hipk2 knockdown cells had accumulated in S phase. At this point, the difference between shHK2 and control cells was statistically significant (Figure 3A-B; 36 and 48 hours). By 48 hours, most cells with Hipk1 knockdown also arrested in G1, but a larger percentage of Hipk2-deficient cells still remained in S phase.
Hipk knockdown causes cell accumulation in S phase and increased apoptosis. Ter119-negative fetal liver cells infected with shRNA-containing retroviruses were cultured in Epo-containing media. Aliquots of cells were removed at 24, 36, and 48 hours and stained with propidium iodide for FACS analysis of cell cycle, or annexin V and 7-AAD for FACS analysis of apoptosis. (A-B) Cell cycle: percentage of cells from each infection in S phase (A) or G1 (B); n = 3 (mean ± 2 SEM). *P < .05. **P < .01. (C-D) Apoptosis: percentage of cells that are actively undergoing apoptosis but not dead (C; 7-AAD-negative, annexin V-positive) or dead cells (D; 7-AAD, annexin V–double-positive); n = 3 (mean ± 2 SEM). *P < .05.
Hipk knockdown causes cell accumulation in S phase and increased apoptosis. Ter119-negative fetal liver cells infected with shRNA-containing retroviruses were cultured in Epo-containing media. Aliquots of cells were removed at 24, 36, and 48 hours and stained with propidium iodide for FACS analysis of cell cycle, or annexin V and 7-AAD for FACS analysis of apoptosis. (A-B) Cell cycle: percentage of cells from each infection in S phase (A) or G1 (B); n = 3 (mean ± 2 SEM). *P < .05. **P < .01. (C-D) Apoptosis: percentage of cells that are actively undergoing apoptosis but not dead (C; 7-AAD-negative, annexin V-positive) or dead cells (D; 7-AAD, annexin V–double-positive); n = 3 (mean ± 2 SEM). *P < .05.
Similarly, there was a significant increase in the percentage of apoptotic and dead cells after knockdown of Hipk2, but only after 36 hours (Figure 3C-D). There was also an increase in apoptotic and dead cells in Hipk1-deficient cells at 24 and 36 hours, but this difference was not significant, which correlates with the absence of significant effect of Hipk1 depletion on cell growth (Figure 2D).
shRNA knockdown of Hipk2 does not affect induction of Ter119 or down-regulation of CD71 but does inhibit enucleation
We next evaluated the extent of differentiation of shRNA-infected erythroid precursors by flow cytometry. Fetal liver erythroblasts develop a specific pattern of forward and side scatter as they differentiate (they tend to get smaller and increase their nuclear-cytoplasmic ratio with each cell division) and first gain Ter119 expression and then lose CD71 expression over 48 hours of in vitro culture.19 The forward scatter/side scatter (FSC/SSC) pattern (Figure 4A) and CD71/Ter119 expression (Figure 4B) of the major cell population were not significantly affected by knockdown of either Hipk2 or Hipk1. In contrast (Figure 4C), the extent of enucleation was significantly inhibited by Hipk2 knockdown alone.
Specific knockdown of Hipk2 inhibits enucleation but not expression of erythroid-specific surface markers. Ter119-negative fetal liver cells infected with shRNA-containing retroviruses were cultured in Epo-containing media. Aliquots of cells from each sample were removed at 48 hours and stained with anti-Ter119 and anti-CD71 antibodies (to assay differentiation) or anti-Ter119 antibody and DAPI-Hoechst (to assay enucleation). (A) Forward scatter (FSC) versus side scatter (SSC) plots for control and Hipk2 knockdown cultures show no significant differences in the major cell populations. (B) Terminal erythroid differentiation as assayed by FACS; n = 4 (mean ± 2 SEM). (C) Extent of enucleation as assayed by FACS: enucleated cells (reticulocytes, circled), are defined by their lack of DNA but positive expression of Ter119, and are distinguishable from erythroblasts (solid arrow). Extruded nuclei (dashed arrow) have the lowest Ter119 signal but highest Hoechst signal because of their high DNA content and loss of membrane proteins. Quantification of enucleated cells is from 4 replicate experiments (mean ± 2 SEM). **P < .01.
Specific knockdown of Hipk2 inhibits enucleation but not expression of erythroid-specific surface markers. Ter119-negative fetal liver cells infected with shRNA-containing retroviruses were cultured in Epo-containing media. Aliquots of cells from each sample were removed at 48 hours and stained with anti-Ter119 and anti-CD71 antibodies (to assay differentiation) or anti-Ter119 antibody and DAPI-Hoechst (to assay enucleation). (A) Forward scatter (FSC) versus side scatter (SSC) plots for control and Hipk2 knockdown cultures show no significant differences in the major cell populations. (B) Terminal erythroid differentiation as assayed by FACS; n = 4 (mean ± 2 SEM). (C) Extent of enucleation as assayed by FACS: enucleated cells (reticulocytes, circled), are defined by their lack of DNA but positive expression of Ter119, and are distinguishable from erythroblasts (solid arrow). Extruded nuclei (dashed arrow) have the lowest Ter119 signal but highest Hoechst signal because of their high DNA content and loss of membrane proteins. Quantification of enucleated cells is from 4 replicate experiments (mean ± 2 SEM). **P < .01.
shRNA knockdown of Hipk2 reduces hemoglobin accumulation
One possible explanation why enucleation may be inhibited by Hipk2 depletion is that enucleation may be coupled with other processes of terminal erythroid differentiation and might only commence once differentiation is complete. To evaluate this possibility, an important measure of terminal erythroid differentiation other than expression of TER119 and CD71 was evaluated: accumulation of hemoglobin.
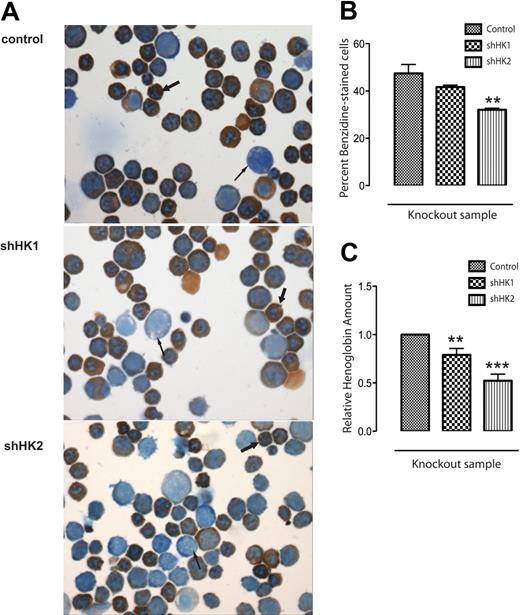
To examine hemoglobin accumulation in the knockdown samples, retrovirally infected (GFP+) erythroid progenitors were isolated at 36 hours, fixed on slides, and stained with benzidine-Giemsa (Figure 5A), which demarcates hemoglobin-containing (benzidine-positive) cells. There were more unstained erythroblasts (thin arrow) and less fully differentiated, highly stained cells (thick arrow) in the shHK2 knockdown sample than in the control. Quantitation of stained cells (Figure 5B) reflects that Hipk1 and Hipk2 knockdown both decrease the percentage of benzidine-positive cells, although Hipk2 knockdown does so to a greater extent. This was confirmed by lysing the cells and using the Drabkin reagent, which revealed that the hemoglobin quantity per cell is also inhibited significantly by Hipk2 knockdown (Figure 5C).
Specific knockdown of Hipk2 reduces hemoglobin accumulation. GFP+ cells were sorted from fetal liver erythroid cultures grown in Epo-containing media 36 hours after shRNA retroviral infection. (A) Slides made from cytospins of infected cells were fixed and stained with benzidine-Giemsa (original magnification 40×). Positively stained cytoplasm reflects the presence of hemoglobin: the more differentiated orthochromatophilic erythroblasts (thick arrow) show more hemoglobin staining than the remaining early erythroblasts (thin arrow). (B) Quantification of positively stained, hemoglobinized cells, expressed as the percentage of stained cells in 5 unique 40× fields for each sample; n = 5 (mean ± SEM). **P < .01. (C) Equal numbers (1 × 106) of GFP+ cells were lysed with 200 μL Drabkin reagent and spectrophotometric readings made at 540 nm; n = 5 (mean ± SEM). **P < .01; ***P < .001.
Specific knockdown of Hipk2 reduces hemoglobin accumulation. GFP+ cells were sorted from fetal liver erythroid cultures grown in Epo-containing media 36 hours after shRNA retroviral infection. (A) Slides made from cytospins of infected cells were fixed and stained with benzidine-Giemsa (original magnification 40×). Positively stained cytoplasm reflects the presence of hemoglobin: the more differentiated orthochromatophilic erythroblasts (thick arrow) show more hemoglobin staining than the remaining early erythroblasts (thin arrow). (B) Quantification of positively stained, hemoglobinized cells, expressed as the percentage of stained cells in 5 unique 40× fields for each sample; n = 5 (mean ± SEM). **P < .01. (C) Equal numbers (1 × 106) of GFP+ cells were lysed with 200 μL Drabkin reagent and spectrophotometric readings made at 540 nm; n = 5 (mean ± SEM). **P < .01; ***P < .001.
Knockdown of Hipk1 or Hipk2 results in changes in expression of mostly dissimilar transcripts
To determine which genes were deregulated by Hipk2 depletion, and perhaps explain the subsequent defects in proliferation, enucleation, and hemoglobin accumulation, mRNA was isolated from fetal liver erythroid cultures 36 hours after shRNA infection, and processed and hybridized to Agilent mouse expression arrays along with the control. Figure 6A compares the log ratios between Hipk2 and Hipk1 knockdown cultures: most genes are unchanged and fall inside the lines representing 2-fold changes. Very few differentially expressed genes overlapped between Hipk1 and Hipk2 knockdown: specifically, only 49 genes were significantly reduced in both Hipk1 and Hipk2 knockdown samples and 10 genes were significantly induced in both samples (supplemental Table 2).
Knockdown of Hipk1 or Hipk2 results in changes in expression in related but nonidentical groups of genes. Total RNA was isolated from fetal liver erythroid cultures grown in Epo-containing media 36 hours after shRNA retroviral infection, and then reverse-transcribed, labeled, and hybridized onto Agilent mouse expression arrays. Each sample was isolated and processed in duplicate and compared with a luciferase control isolated during the same experiment. (A) Log2 ratios for each knockdown culture compared with control are plotted: sample shHK1 (Hipk1-KD) is on the abscissa and shHK2 (Hipk2-KD) on the ordinate axis. Dashed lines indicate ratios of 2-fold increase (1) and 2-fold decrease (−1) in expression compared with control. Note the paucity of genes falling in the top right or bottom left corners. (B) Functional ontology classifications for significantly changed genes (both increased and decreased) in the Hipk2-knockdown cells; only significantly enriched categories are listed.
Knockdown of Hipk1 or Hipk2 results in changes in expression in related but nonidentical groups of genes. Total RNA was isolated from fetal liver erythroid cultures grown in Epo-containing media 36 hours after shRNA retroviral infection, and then reverse-transcribed, labeled, and hybridized onto Agilent mouse expression arrays. Each sample was isolated and processed in duplicate and compared with a luciferase control isolated during the same experiment. (A) Log2 ratios for each knockdown culture compared with control are plotted: sample shHK1 (Hipk1-KD) is on the abscissa and shHK2 (Hipk2-KD) on the ordinate axis. Dashed lines indicate ratios of 2-fold increase (1) and 2-fold decrease (−1) in expression compared with control. Note the paucity of genes falling in the top right or bottom left corners. (B) Functional ontology classifications for significantly changed genes (both increased and decreased) in the Hipk2-knockdown cells; only significantly enriched categories are listed.
In Figure 6B, the transcripts whose expression was significantly changed after Hipk2 knockdown were categorized by gene ontology for their biologic process or molecular function using the Database for Annotation, Visualization and Integrated Discovery (http://david.abcc.ncifcrf.gov). Significantly changed genes after Hipk2 depletion fell into categories mostly involving the regulation of cell proliferation and apoptosis; the most significant category of genes affected by Hipk2 knockdown was cell-cycle control, which correlates with the S phase accumulation shown earlier (Figure 3A). Microarray results have been submitted to the Gene Expression Omnibus under accession number GSE17106.
shRNA knockdown of Hipk2 results in reductions of some, but not all, erythroid-specific transcripts and many genes involved in cell-cycle control
To elucidate mechanisms through which hemoglobin production might be inhibited by Hipk2 knockdown, quantitative RT-PCR analyses (Table 1) were performed on mRNA isolated for the microarray studies described in Figure 6, using primers against erythroid-specific genes, genes involved in heme synthesis and hemoglobin metabolism, as well as some genes involved in cell-cycle regulation. Hipk2 knockdown resulted in the decreased expression of many transcripts, such as erythroid-important genes, such as Klf1, Gata1, ankyrin, spectrin, band 3, and band 4.2, as well as several genes involved in the heme biosynthetic pathway and production of hemoglobin, such as mitoferrin 1, adult α-globin (Hba-a1), and embryonic β-like globin (Hbb-bh1). Genes involved in regulation of the cell cycle and proliferation, such as p21, Rb1, and cyclin E2, were also deregulated. Notably, some transcripts were decreased more than 80% (with significant P values), such as Band 3, Band 4.1, and mitoferrin 1.
mRNA expression changes in Hipk2-depleted fetal liver cultures as determined by quantitative PCR analysis
| RefSeq ID . | Gene symbol . | Gene name . | Fold change . | P . | Description . |
|---|---|---|---|---|---|
| Erythropoiesis | |||||
| Down-regulation | |||||
| NM_031158 | Ank1 | Ankyrin 1, erythroid | −0.77 | < .009 | Actin cytoskeleton organization |
| NM_019670 | Diap3 | Diaphenous homolog 3 (MDia2) | −0.50 | < .112 | Actin binding |
| NM_183428 | Ebp4.1 | Band 4.1 | −0.84 | < .001 | Actin cytoskeleton organization |
| NM_133245 | Edrf1 | α-Hemoglobin stabilizing protein | −0.59 | > .011 | Hemoglobin binding |
| NM_010149 | EpoR | Erythropoietin receptor | −0.57 | < .013 | Cytokine receptor activity |
| NM_008089 | Gata1 | GATA-binding protein 1 | −0.50 | < .001 | Regulation of transcription |
| NM_008090 | Gata2 | GATA-binding protein 2 | −0.36 | < .323 | Regulation of transcription |
| NM_010369 | Gypa1 | Glycophorin A | −0.55 | .084 | Cytoskeletal anchoring protein |
| NM_010635 | Klf1 | Kruppel-like factor 1, erythroid (EKLF) | −0.46 | < .001 | Regulation of transcription |
| NM_009007 | Rac1 | RAS-related C3 botulinum substrate 1 | −0.52 | < .066 | Small GTPase-mediated signal transduction |
| NM_009008 | Rac2 | RAS-related C3 botulinum substrate 2 | −0.36 | > .002 | Small GTPase-mediated signal transduction |
| NM_011403 | Slc4a1 | Band 3 | −0.81 | < .008 | Anion exchange; cytoskeleton |
| NM_013675 | Spnb1 | Spectrin 1, β | −0.61 | < .017 | Actin cytoskeleton organization |
| NM_011638 | Trfr2 | Transferrin receptor 2 | −0.10 | > .150 | Iron ion transport activity |
| NM_009569 | Zfpm1 | Friend-of-GATA1 (FOG1) | −0.56 | < .004 | Regulation of transcription |
| Up-regulation | |||||
| NM_019740 | Foxo3 | Forkhead box O3a (Foxo3a) | 0.04 | > .019 | Regulation of transcription |
| Heme biosynthesis and hemoglobin production | |||||
| Down-regulation | |||||
| NM_009653 | Alas2 | ALAS2 | −0.50 | .003 | Heme biosynthesis |
| NM_007998 | Fech | Ferrochelatase (Fech) | −0.16 | < .001 | Heme biosynthesis |
| NM_008218 | Hba-a1 | Hemoglobin-α adult chain 1 | −0.64 | > .039 | Heme binding, oxygen transport |
| NM_010405 | Hba-x | Hemoglobin X, α-like embryonic chain | −0.29 | < .074 | Heme binding, oxygen transport |
| NM_008220 | Hbb-b1 | Hemoglobin, β adult major chain | −0.57 | > .003 | Heme binding, oxygen transport |
| NM_008219 | Hbb-bh1 | Hemoglobin Z, β-like embryonic chain | −0.60 | > .012 | Heme binding, oxygen transport |
| NM_026331 | Slc25a37 | Mitoferrin 1 | −0.83 | < .011 | Mitochondrial iron ion transport |
| Cell cycle and proliferation | |||||
| Down-regulation | |||||
| NM_009743 | Bcl2l1 | Bcl-2 like 1 (Bcl-XL) | −0.61 | < .007 | Regulation of apoptosis |
| NM_009829 | Ccnd2 | Cyclin D2 | −0.58 | < .054 | Cell cycle control at G1/S transition |
| NM_007669 | Cdkn1a | Cyclin-dept kinase inhibitor 1a (p21, cip1) | −0.33 | < .001 | Cell cycle progression through G1 |
| NM_009875 | Cdkn1b | Cyclin-dept kinase inhibitor 1b (p27, kip1) | −0.56 | .004 | Cell cycle control at G1 |
| NM_010786 | Mdm2 | Murine double minute 2 | −0.82 | Negative regulator of p53 | |
| NM_009029 | Rb1 | Retinoblastoma 1 | −0.70 | .000 | Tumor suppressor, regulation of cell cycle |
| NM_011640 | Trp53 | p53 (transformation related protein 53) | −0.38 | .144 | Tumor suppressor, cell division regulation |
| Up-regulation | |||||
| NM_007628 | Ccna1 | Cyclin A1 | 0.33 | .339 | Cell cycle regulation |
| NM_007631 | Ccnd1 | Cyclin D1 | 0.13 | < .093 | Reentry into mitotic cell cycle |
| NM_001037134 | Ccne2 | Cyclin E2 | 1.31 | > .021 | Cell cycle regulation |
| NM_007658 | Cdc25a | Cell division cycle 25 homolog A | 0.18 | < .068 | Cell cycle regulation |
| NM_010848 | Myb | Myb proto-oncogene (C-myb) | 0.59 | > .040 | Transcription; cell cycle control |
| NM_007483 | RhoB | Ras homolog gene family, member B | 1.91 | < .081 | Negative regulation of cell cycle |
| Miscellaneous | |||||
| Down-regulation | |||||
| NM_007913 | Egr1 | Early growth response 1 | −0.06 | < .037 | Regulation of transcription |
| NM_001033228 | Itga1 | Integrin-α1 precursor (VLA-1; CD49a) | −0.68 | > .003 | Cell adhesion |
| NM_023045 | Xpo7 | Ran-binding protein 16 (Exportin 7) | −0.74 | > .007 | Protein export from nucleus |
| NM_010019 | Dapk | Death-associated protein kinase 2 | −0.58 | .000 | Positive regulator of apoptosis |
| Up-regulation | |||||
| NM_013768 | Prmt5 | Protein arginine N-methyltransferase 5 | 0.94 | < .799 | Methyltransferase activity |
| RefSeq ID . | Gene symbol . | Gene name . | Fold change . | P . | Description . |
|---|---|---|---|---|---|
| Erythropoiesis | |||||
| Down-regulation | |||||
| NM_031158 | Ank1 | Ankyrin 1, erythroid | −0.77 | < .009 | Actin cytoskeleton organization |
| NM_019670 | Diap3 | Diaphenous homolog 3 (MDia2) | −0.50 | < .112 | Actin binding |
| NM_183428 | Ebp4.1 | Band 4.1 | −0.84 | < .001 | Actin cytoskeleton organization |
| NM_133245 | Edrf1 | α-Hemoglobin stabilizing protein | −0.59 | > .011 | Hemoglobin binding |
| NM_010149 | EpoR | Erythropoietin receptor | −0.57 | < .013 | Cytokine receptor activity |
| NM_008089 | Gata1 | GATA-binding protein 1 | −0.50 | < .001 | Regulation of transcription |
| NM_008090 | Gata2 | GATA-binding protein 2 | −0.36 | < .323 | Regulation of transcription |
| NM_010369 | Gypa1 | Glycophorin A | −0.55 | .084 | Cytoskeletal anchoring protein |
| NM_010635 | Klf1 | Kruppel-like factor 1, erythroid (EKLF) | −0.46 | < .001 | Regulation of transcription |
| NM_009007 | Rac1 | RAS-related C3 botulinum substrate 1 | −0.52 | < .066 | Small GTPase-mediated signal transduction |
| NM_009008 | Rac2 | RAS-related C3 botulinum substrate 2 | −0.36 | > .002 | Small GTPase-mediated signal transduction |
| NM_011403 | Slc4a1 | Band 3 | −0.81 | < .008 | Anion exchange; cytoskeleton |
| NM_013675 | Spnb1 | Spectrin 1, β | −0.61 | < .017 | Actin cytoskeleton organization |
| NM_011638 | Trfr2 | Transferrin receptor 2 | −0.10 | > .150 | Iron ion transport activity |
| NM_009569 | Zfpm1 | Friend-of-GATA1 (FOG1) | −0.56 | < .004 | Regulation of transcription |
| Up-regulation | |||||
| NM_019740 | Foxo3 | Forkhead box O3a (Foxo3a) | 0.04 | > .019 | Regulation of transcription |
| Heme biosynthesis and hemoglobin production | |||||
| Down-regulation | |||||
| NM_009653 | Alas2 | ALAS2 | −0.50 | .003 | Heme biosynthesis |
| NM_007998 | Fech | Ferrochelatase (Fech) | −0.16 | < .001 | Heme biosynthesis |
| NM_008218 | Hba-a1 | Hemoglobin-α adult chain 1 | −0.64 | > .039 | Heme binding, oxygen transport |
| NM_010405 | Hba-x | Hemoglobin X, α-like embryonic chain | −0.29 | < .074 | Heme binding, oxygen transport |
| NM_008220 | Hbb-b1 | Hemoglobin, β adult major chain | −0.57 | > .003 | Heme binding, oxygen transport |
| NM_008219 | Hbb-bh1 | Hemoglobin Z, β-like embryonic chain | −0.60 | > .012 | Heme binding, oxygen transport |
| NM_026331 | Slc25a37 | Mitoferrin 1 | −0.83 | < .011 | Mitochondrial iron ion transport |
| Cell cycle and proliferation | |||||
| Down-regulation | |||||
| NM_009743 | Bcl2l1 | Bcl-2 like 1 (Bcl-XL) | −0.61 | < .007 | Regulation of apoptosis |
| NM_009829 | Ccnd2 | Cyclin D2 | −0.58 | < .054 | Cell cycle control at G1/S transition |
| NM_007669 | Cdkn1a | Cyclin-dept kinase inhibitor 1a (p21, cip1) | −0.33 | < .001 | Cell cycle progression through G1 |
| NM_009875 | Cdkn1b | Cyclin-dept kinase inhibitor 1b (p27, kip1) | −0.56 | .004 | Cell cycle control at G1 |
| NM_010786 | Mdm2 | Murine double minute 2 | −0.82 | Negative regulator of p53 | |
| NM_009029 | Rb1 | Retinoblastoma 1 | −0.70 | .000 | Tumor suppressor, regulation of cell cycle |
| NM_011640 | Trp53 | p53 (transformation related protein 53) | −0.38 | .144 | Tumor suppressor, cell division regulation |
| Up-regulation | |||||
| NM_007628 | Ccna1 | Cyclin A1 | 0.33 | .339 | Cell cycle regulation |
| NM_007631 | Ccnd1 | Cyclin D1 | 0.13 | < .093 | Reentry into mitotic cell cycle |
| NM_001037134 | Ccne2 | Cyclin E2 | 1.31 | > .021 | Cell cycle regulation |
| NM_007658 | Cdc25a | Cell division cycle 25 homolog A | 0.18 | < .068 | Cell cycle regulation |
| NM_010848 | Myb | Myb proto-oncogene (C-myb) | 0.59 | > .040 | Transcription; cell cycle control |
| NM_007483 | RhoB | Ras homolog gene family, member B | 1.91 | < .081 | Negative regulation of cell cycle |
| Miscellaneous | |||||
| Down-regulation | |||||
| NM_007913 | Egr1 | Early growth response 1 | −0.06 | < .037 | Regulation of transcription |
| NM_001033228 | Itga1 | Integrin-α1 precursor (VLA-1; CD49a) | −0.68 | > .003 | Cell adhesion |
| NM_023045 | Xpo7 | Ran-binding protein 16 (Exportin 7) | −0.74 | > .007 | Protein export from nucleus |
| NM_010019 | Dapk | Death-associated protein kinase 2 | −0.58 | .000 | Positive regulator of apoptosis |
| Up-regulation | |||||
| NM_013768 | Prmt5 | Protein arginine N-methyltransferase 5 | 0.94 | < .799 | Methyltransferase activity |
Discussion
Hipk2 has been shown to be indispensable for proliferation, cell survival, and terminal differentiation in several cell types; Hipk1 and Hipk2 are thought to be functionally redundant in hematopoietic development because of gross defects in hematopoiesis in Hipk1/2 double-deficient but not single-knockout mice. The roles of either Hipk1 or Hipk2 in erythropoiesis are unknown. Here, we demonstrate the novel and significant role that Hipk2 plays in the complex regulation of terminal erythropoiesis, involving both terminal proliferation and some essential aspects of terminal differentiation.
We found that the significant induction of Hipk1 and Hipk2 during terminal erythroid differentiation occurs during the same stage when several other erythroid-important regulators, such as Foxo3, Klf1 (EKLF), and Gata1, and erythroid-specific genes, such as erythroid membrane proteins, heme biosynthetic enzymes, and α- and β-globins are also induced (Figure 1). Through shRNA knockdown experiments, we showed that Hipk2 is essential for normal terminal erythroid proliferation, maximal expression of hemoglobin and other erythroid-specific proteins, as well as for normal enucleation. Hipk2 is not critical for cell-surface expression of erythroid-important markers TER119 or CD71, however. In contrast, Hipk1 may not play a significant role in terminal erythroid proliferation or differentiation, but we cannot make firm conclusions about these findings because we were not able to achieve more than partial HIPK1 knockdown at the protein level. These data demonstrate that Hipk2 plays a clear and novel role in the regulation of terminal erythroid differentiation and only suggest that Hipk1 and Hipk2 may have only partially redundant functions during erythropoietic development.
A novel role for Hipk2 in terminal erythroid differentiation
Normal levels of Hipk2 appear to be critical for some aspects of terminal erythroid differentiation, such as enucleation and maximal hemoglobin production. Although there was no effect of either Hipk1 or Hipk2 knockdown on measures of differentiation, such as cell size or TER119 and CD71 expression (Figure 4A-B), Hipk2 knockdown did drastically reduce the percentage of enucleated cells (Figure 4C). There were more undifferentiated, unhemoglobinized cells after Hipk2 knockdown than Hipk1 knockdown or control. Hemoglobin quantification in the knockdown cultures also showed that there was also significantly less hemoglobin per cell after Hipk2 depletion than Hipk1 depletion or control.
Evidence from microarray and quantitative PCR analysis of mRNA from each knockdown culture also supports the fact that Hipk2 regulates hemoglobin accumulation: many transcripts of genes involved in hemoglobin production, such as α- and β-globins, and heme biosynthetic enzymes, such as mitoferrin 1 (Slc25a37), were reduced significantly after Hipk2 knockdown. The common expression pattern between Hipk1/Hipk2 and GATA1, FoxO3, and EKLF (Figure 1B-C) supports the possibility that Hipk2 may play a role in these canonical regulatory pathways of hemoglobin production.
Hipk2's role in terminal erythroid proliferation may provide another mechanism explaining “ineffective erythropoiesis”
Hipk2 appears to be necessary for normal terminal erythroid proliferation, and the inhibition of cell growth observed after Hipk2 knockdown (Figure 2D) is consistent with an increase in apoptotic and dead cells (Figure 3C-D) and a greater accumulation of cells in S phase at 48 hours over that seen in Hipk1 knockdown or control cells (Figure 3A). These findings are consistent with other studies implicating the role of Hipk2 in regulation of cell survival and proliferation7,11,14,18,22 but are the first involving erythropoiesis specifically.
The specific combination of accumulation of erythroblasts in S phase, increased apoptosis of precursors, and defective terminal erythroid differentiation is termed “ineffective erythropoiesis.” This phenotype also occurs in conditions affecting DNA synthesis, such as megaloblastic anemia resulting from folate or cobalamin deficiency,23 or defects in cell-cycle control of erythropoiesis, such as depletion of E2F proteins.24,25 In the latter study, using mice deficient in E2F4, Kinross et al24 showed that the promotion of cellular proliferation and cell-cycle progression impaired some aspects of terminal erythroid differentiation but did not result in a complete block in erythropoiesis. This, along with our findings of deregulation of many cell cycle– as well as erythroid-specific genes after Hipk2 knockdown, supports the hypothesis that cell-cycle progression may be coupled directly to some, but not all, aspects of terminal erythroid differentiation, such as induction of important erythroid-specific genes, and that the phenotype of “ineffective erythropoiesis” may be a block in the regulation of erythroid gene expression by cell-cycle control. Hipk2 likely plays a role in this regulatory pathway.
To further examine the regulatory role of Hipk2 in erythropoiesis, it will be important to determine exactly which regulators HIPK2 phosphorylates during erythropoiesis. Through computational analysis, we determined which transcription factor-binding consensus sequences were enriched in the promoter regions of sets of genes that were differentially expressed in the Hipk2 knockdown cells, compared with random promoter regions in the remainder of the genome. To begin with, we analyzed the set of genes that had functions related to proliferation or cell-cycle regulation and found that c-MYB-binding sites were the most enriched in this set of promoters (P < .001). Binding sites for c-MYB were also enriched in the promoters of the globin genes and heme biosynthetic enzymes that specifically decreased after Hipk2 knockdown (P < .001). Given evidence in the literature that degradation of c-MYB is necessary for terminal differentiation in many hematopoietic cell types (eg, high expression of c-Myb in immature cells promotes proliferation and inhibits differentiation26 ), and HIPK2 is known to inhibit c-MYB transcriptional activity (after Wnt1 stimulation) by promoting its phosphorylation and subsequent proteosomal degradation,12 it is possible that one of the pathways through which Hipk2 may regulate terminal erythroid differentiation may involve the stabilization of c-MYB. Further experiments concerning the phosphorylation status of c-MYB and its aberrant accumulation in Hipk2 knockdown cultures will be necessary to confirm this hypothesis. We have performed chromatin immunoprecipitation on wild-type cells and confirmed that c-MYB is bound to some of these promoters (data not shown).
In conclusion, our examination of Hipk1 and Hipk2 knockdown in fetal liver erythroid differentiation has uncovered the clear role that Hipk2 plays in terminal erythroid proliferation, hemoglobin production, and enucleation. The combination of increased apoptosis, accumulation in S phase, and impaired proliferation in erythroid progenitors is consistent with conditions defined as “ineffective erythropoiesis,” and our findings on the novel role of Hipk2 in erythropoiesis may lead to further understanding of the coupling of cell-cycle control to the regulation of some aspects of terminal erythroid differentiation, such as normal cell growth and erythroid-specific gene expression.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Deepa Mokshagundam for cloning the shRNA retroviral constructs.
This work was supported in part by grants from the National Institutes of Health (P01 HL 32262; K08 DK076848-03 to S.M.H.) and Amgen (H.F.L.).
National Institutes of Health
Authorship
Contribution: S.M.H. designed experiments, performed research, analyzed data, and wrote the manuscript; K.A.B. performed experiments and wrote the manuscript; and H.F.L. designed experiments and reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Harvey F. Lodish, Whitehead Institute for Biomedical Research, 9 Cambridge Center, Cambridge, MA 02142; e-mail: lodish@wi.mit.edu.
References
Supplemental data
Forward and reverse sequences are listed.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal