Abstract
To identify the genetic basis of circulating concentrations of monocyte chemoattractant protein-1 (MCP-1), we conducted genome-wide association analyses for MCP-1 in 3 independent cohorts (n = 9598). The strongest association was for serum MCP-1 with a nonsynonymous polymorphism, rs12075 (Asp42Gly) in DARC, the gene for Duffy antigen receptor for chemokines, a known vascular reservoir of proinflammatory cytokines (minor allele frequency, 45.6%; P < 1.0 * 10−323). This association was supported by family-based genetic linkage at a locus encompassing the DARC gene (genome-wide P = 8.0 * 10−13). Asp42Gly accounted for approximately 20% of the variability in serum MCP-1 concentrations and also was associated with serum concentrations of interleukin-8 and RANTES. While exploring a lack of association between this polymorphism and EDTA plasma MCP-1 concentrations (P = .82), we determined that both clotting and exogenous heparan sulfate (unfractionated heparin) released substantial amounts of MCP-1 from Darc. Quantitative immunoflow cytometry failed to identify meaningful Asp42Gly-associated differences in Darc expression, suggesting that a functional change is responsible for the differential cytokine binding. We conclude that Asp42Gly is a major regulator of erythrocyte Darc-mediated cytokine binding and thereby the circulating concentrations of several proinflammatory cytokines. We have also identified for the first time 2 mechanisms for the release of reservoir chemokines with possible clinical implications.
Introduction
Chronic low-grade inflammation, often comprising activation of both the innate immune system and the coagulation/fibrinolysis pathways,1 is observed in cardiovascular diseases and several other chronic diseases of aging. It is characterized by elevated concentrations of circulating inflammatory biomarkers (eg, C-reactive protein), cytokine and chemokine mediators (eg, interleukin-6), and coagulation biomarkers (eg, fibrin degradation products such as D-dimer).
Monocyte chemoattractant protein-1 (MCP-1) is a member of the large family of human chemokines (subfamily CC chemokines) and is central to the inflammatory process. Circulating MCP-1 is produced mainly by endothelial and smooth muscle cells.2 Two predominant forms of 9 and 13 kDa (differing in degree of O-glycosylation)3 are induced by various inflammatory stimuli and act via the G protein–coupled receptor CCR2.4 Circulating MCP-1 is bound by the chemokine receptor Darc (Duffy antigen receptor for chemokines) that exhibits specific chemokine binding capacities for CC and CXC cytokines.5 MCP-1 is capable of gradient-induced leukocyte recruitment to sites of inflammation, and it promotes angiogenesis and the maturation of immune cells.6,7 In clinical studies MCP-1 concentrations are elevated in sepsis, autoimmune diseases, and cancer.8-10 Sound evidence from experimental data and initial clinical observations indicate a role for MCP-1 in insulin-resistance and atherosclerosis,11,12 underscoring the relevance of MCP-1 pathophysiology for a broad spectrum of chronic inflammatory diseases.
Common clinical characteristics explain only a small proportion of the variability of serum MCP-1 concentrations,13 and the contribution of genetic factors is incompletely understood. Multivariable-adjusted heritability of MCP-1 concentrations in whites of European descent has been reported to be 0.37 in plasma and 0.44 in serum.14,15 Initial linkage studies showed peaks of genome-wide significance on chromosome 1 and 3.14,15 Multiple single nucleotide polymorphisms (SNPs) in the MCP-1 gene (CCL2) have been reported to be related to blood concentrations of MCP-1, but to date, only rs1024611 (−2518 A/G) has been replicated.16,17 To identify the genetic basis of the proinflammatory cytokine MCP-1, we used family-based genetic linkage and genome-wide association (GWA) studies. Analyses were performed for serum and plasma concentrations separately. To understand the pathophysiology of discrepancies in our serum versus plasma GWA findings, we conducted a series of in vitro experiments.
Methods
Study cohorts
We examined persons of self-reported European descent from the community-based Atherosclerosis Risk in Communities (ARIC)18 (n = 729), Framingham Heart Study (FHS)19 (n = 6771), and Monitoring of Trends and Determinants in Cardiovascular Disease/Kooperative Gesundheitsforschung in der Region Augsburg (MONICA/KORA)20 (n = 1625).
Further details on the phenotype and genotype samples are provided in the supplemental data (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). MCP-1 and other biomarkers were measured by commercially available test kits (supplemental Table 2), MCP-1 serum concentrations were measured in FHS and MONICA/KORA and plasma MCP-1 in MONICA/KORA and ARIC. In the GWA samples, plasma RANTES measurements were available in ARIC; FHS had available serum concentrations of intercellular adhesion molecule-1, interleukin-6, and myeloperoxidase. In the above-mentioned subcohort of MONICA/KORA, data on additional inflammatory biomarkers (C-reactive protein, intercellular adhesion molecule-1, interferon-inducible protein-10, interleukin-6, interleukin-8, interleukin-18, macrophage migration inhibitory factor, myeloperoxidase, RANTES) were measured in serum. Two hundred four persons overlapped between the GWA and replication sample. Omitting those persons from the analysis did not change the results significantly.
Samples for biochemical analysis to examine the difference between serum and plasma were obtained from local volunteers at the Laboratory for Clinical Biochemistry Research in Vermont.
Study protocols were approved by all participating institutional review boards, and all participants gave written informed consent in accordance with the Declaration of Helsinki.
Genotyping
SNP genotyping in the different studies was performed with the use of the Affymetrix 6.0 gene chip in ARIC, the Affymetrix Human Mapping 500K Array Set and 50K Human Gene Focused Panel in FHS, and the Affymetrix Human Mapping 500K Array in MONICA/KORA.
Details on quality control used for genotyping are described in supplemental Table 1. For linkage analyses in the Offspring and Generation 3 cohorts of the FHS, a subset of 4157 SNPs with low levels of linkage disequilibrium were selected, covering the 22 autosomal chromosomes with an average SNP spacing of 0.84 cM or 669 Kb and an average information content of 86%.
Targeted genotyping was performed in a subgroup of FHS participants (n = 2666) for the imputed top association SNP (rs12075) in the DARC gene with the use of a Taqman assay according to the manufacturer's instructions. For replication, a subcohort of the MONICA/KORA (n = 1453) was genotyped with the 50K IBC gene chip (http://bioinf.itmat.upenn.edu/cvdsnp/).
Imputation
Imputation to the approximately 2.5 million CEU HapMap SNPs was performed in all 3 studies.21 All used the Markov chain–based method implemented in the program (Mach1 Version 1.0.1.5).22 For all imputed SNPs the reliability of imputation was assessed by comparing the observed dosage variance with the expected variance under Hardy-Weinberg equilibrium.
Statistical analysis
Continuous data are presented as mean and standard deviation (SD) for clinical characteristics; median and 25th to 75th percentiles for biomarkers; and percentages by cohort for categorical measures. Skewed distribution of MCP-1 values led to natural logarithmic transformation for further analyses. Mean and SD of phenotypes and covariates are provided in Table 1. Spearman correlation coefficients were calculated for serum and plasma MCP-1 concentrations in the MONICA/KORA cohort.
Linkage
In the FHS, families with 2 or more persons with both phenotype and genotype data were available for linkage analyses (n = 6539 participants in 878 families). A recently developed variance component model with a robust score statistic was applied.23 This approach is relatively robust to violations of the normality assumption, which may be a problem with MCP-1 residuals. We computed the robust statistic on the LOD scale to facilitate interpretation.
Genetic association analysis
All SNPs were tested for deviation from the Hardy-Weinberg equilibrium. Because serum and plasma MCP-1 represent different phenotypes, analyses were performed separately. Primary analyses were conducted for age- and sex-adjusted biomarker concentrations. We defined multivariable-adjusted models a priori as secondary analyses (see supplemental Table 1 for variable definitions by cohort). The FHS generated cohort-specific residuals adjusting for age and sex, and unstandardized residuals were used in tests for associations with SNPs. Linear mixed effect models were calculated, to account for familial correlation among FHS participants. Family-based association testing was additionally used in the FHS. All studies fitted an additive genetic model (1 degree of freedom trend test) relating MCP-1 to genotype (0, 1, 2 variant alleles). In ARIC and MONICA/KORA, the relation of the natural logarithm of MCP-1 concentrations to genotype was examined by linear regression. We report the mean of the log per copy of the minor allele (β), standard error [SE(β)], and corresponding P values.
Genome-wide significance
We defined genome-wide statistical significance at a conservative threshold of P less than 5 * 10−8 for testing common genetic variants across the genome, which approximates a Bonferroni correction for approximately 1 000 000 independent hypotheses tests. We also provide results for a threshold expecting 1 false positive P more than 1/number of SNPs (∼ 2.5 million), P less than 4 * 10−7.
Population stratification
We assessed population stratification by applying principal components analysis to the genotypes in ARIC and the FHS with the use of EIGENSTRAT (http://www.broad.mit.edu/tools/software.html).24 We found no significant association between the first 10 principal components and MCP-1 levels. Therefore, even if population stratification was present in our European descent samples, it was unlikely to alter our overall findings. For MCP-1 association results we provided the genomic inflation factor λ.
Meta-analysis
We examined results from individual studies with all imputed SNPs and combined individual estimates of genotype effect by using inverse-variance weighting in a fixed effects meta-analysis. In addition to the meta-analysis based on combining regression estimates, we applied an approach based on combining test statistics (such as signed z scores weighted by effective sample). Both approaches yielded similar results; the results of the fixed effects method are presented. Pre–meta-analysis exclusions were low minor allele frequency (MAF; < 5% MONICA/KORA, < 1% FHS and ARIC), call rate (< 95% MONICA/KORA and FHS, < 90% ARIC), or Hardy-Weinberg-equilibrium P less than 10−6 for genotyped SNPs and ratio of observed to expected genotype variance less than 0.3 for imputed SNPs. We used the software METAL (http://www.sph.umich.edu/csg/abecasis/Metal/index.html) for meta-analysis.
The distribution of the observed and expected P values is provided in quantile-quantile plots (supplemental Figure 1).
Determinants of MCP-1 concentrations in serum versus plasma
To test whether the difference between serum and EDTA plasma could be explained by the presence or lack of calcium or the differences in the number of freeze-thaw cycles or the plasma clotting, blood from 5 donors was collected into EDTA and heparin collection tubes, as well as prepared for serum. Samples were snap-frozen to −80°C, then thawed and assayed, as well as assayed fresh. MCP-1 was measured by a commercially available enzyme-linked immunoabsorbent assay (R&D Systems; http://www.rndsystems.com/). In addition, before diluting for the assay, EDTA plasma was recalcified to plasma calcium levels (∼ 3.0mM), and clotted; EDTA was added to serum (2.5mM final concentration) to chelate the calcium. To assess whether heparin had a direct effect on the assay, blood was collected into EDTA collection tubes, and plasma was prepared (n = 5). To this plasma we added the dissolved contents of a heparin collection tube to yield a final heparin concentration approximately the same as that in heparin plasma (14 U/mL).
We ran size exclusion chromatography columns for EDTA and heparin plasma and serum. Approximately 1 mL of each sample was applied to a Sephacryl S-200HR Hi-Prep 16/60 column (Pharmacia), 1 mL/min, 0.1M ammonium carbonate, and fractions were collected, read for protein concentration by absorbance at 280 nm, dried in a speed-vac (Savant), rehydrated in phosphate-buffered saline with 0.2% bovine serum albumin at a 10-fold concentration, and assayed for MCP-1 by ELISA.
To investigate whether heparin was more efficient at releasing MCP-1 than clotting, blood (n = 4) was collected into EDTA. To one aliquot we added calcium (∼ 5mM final concentration), and the sample was allowed to clot; MCP-1 was measured in the resulting “serum.” To another aliquot, heparin was added (14 U/mL final concentration) and incubated for 15 minutes at room temperature, after which time protamine (0.15 mg/mL final concentration) was added to neutralize the heparin. Calcium was added as above, and the sample ws allowed to clot, and MCP-1 was measured in the resulting serum.
To determine the effects of therapeutic heparin, we performed 2 experiments. First, blood was drawn into EDTA collection tubes (also containing antiproteases aprotinin [150 KIU/mL] and D-Phe-Pro-Arg CMK [45μM]; SCAT; Haematologic Technologies) as part of a clinical trial comparing heparin with bivalirudin (a direct thrombin inhibitor) as adjunctive therapy during coronary stent placement. Samples were collected at baseline and after 1 hour of infusion, and MCP-1 was measured. Second, we compared the effect of different concentrations of heparin on measured plasma MCP-1 concentrations. Blood was collected into EDTA collection tubes and mixed with a small volume of heparin, previously dissolved in phosphate-buffered saline and serially diluted by one-third. The first tube yielded a concentration of heparin in the EDTA blood equivalent to that in a heparin collection tube, approximately 14 U/mL. After incubation the samples were centrifuged, and the MCP-1 was assayed in the resulting plasma.
To examine the amount of MCP-1 that is released from platelets, blood was drawn from 2 donors into EDTA and citrate collection tubes; the latter was done to allow for maximal platelet activation during clotting. Platelet-rich plasma was prepared, and MCP-1 was assayed before and after recalcification and before and after clotting. To examine the amount of MCP-1 that is released from peripheral blood mononuclear cells (PBMCs), blood was drawn from 4 donors into citrated Cell Preparation Tubes (Becton Dickinson), and PBMCs were collected. Platelet-free citrated plasma was prepared and 2 aliquots were prepared; PBMCs were added back to 1 aliquot. Both aliquots were recalcified and allowed to clot. MCP-1 was measured on the resulting serum.
Darc expression
Finally to determine whether the Asp42Gly polymorphism was associated with differential Darc expression, we drew blood from 16 volunteers and genotyped them at rs12075 by real-time polymerase chain reaction with the use of TaqMan Assay By Design (Applied Biosystems) under standard conditions (TaqMan SNP Genotyping Assays Protocol, Rev. B, Part no. 4332856b; Applied Biosystems). TaqMan genotypes were verified by sequencing. For flow cytometry, antibodies were obtained from R&D Systems. Washed red cells were prepared from EDTA whole blood and incubated with allophycocyanin-conjugated monoclonal anti–human Darc (no. FAB4139A) or allophycocyanin-conjugated isotype control antibody (no. IC003A) according to the product insert. Cell surface expression of Darc was determined by flow cytometric analysis with the use of a BD LSRII (BD Biosciences). We confirmed the flow cytometric results by visual observation with the use of confocal microscopy. Blood was processed as for flow cytometry, and the fluorescently stained cells were placed suspended in PBS/0.5% BSA, placed onto the coverslip of a glass-bottom culture dish (MatTek), and 1 drop of Aqua-Poly/Mount (Polysciences Inc) was added. Photos were taken at 40× magnification with a Zeiss Laser Scanning Confocal Microscope (LSM 510; Carl Zeiss MicroImaging Inc) equipped with a 40× PlanNeofluar, 1.3 numerical aperture (NA) objective. Images were outputted directed as .lsm files, converted to TIFF and then to PowerPoint Version 2007 files.
Results
Description of the cohorts
A total of 9125 persons (51% women overall) with high-quality genome-wide marker data and MCP-1 values were available for analysis across 3 cohorts: the FHS, the ARIC study, and the MONICA/KORA study. MCP-1 was measured in serum (n = 8396; FHS and MONICA/KORA) and in EDTA plasma (n = 2354; ARIC and MONICA/KORA). The characteristics by cohort are provided in Table 1. The correlation between MCP-1 serum and EDTA plasma concentrations in a small sample of healthy persons was R2 = 0.568, with serum values being almost twice those in plasma (supplemental Table 3).
Study participants characteristics by analysis sample
| Characteristic . | ARIC (n = 729) . | FHS (n = 6771) . | MONICA/KORA (n = 1625) . |
|---|---|---|---|
| Age, y, mean ± SD | 56.5 ± 5.1 | 49.4 ± 13.8 | 52.5 ± 10.1 |
| Women, % | 34.8 | 53.4 | 50.7 |
| Current smoking, % | 30.3 | 15.5 | 15.4 |
| Body mass index, kg/m2, mean ± SD | 27.0 ± 4.6 | 27.4 ± 5.5 | 27.3 ± 4.1 |
| Waist, cm, mean ± SD | 97.5 ± 12.4 | 96.0 ± 15.0 | 90.3 ± 12.4 |
| Systolic blood pressure, mm Hg, mean ± SD | 122 ± 18 | 121 ± 17 | 133 ± 18 |
| Total/HDL cholesterol, ratio, mean ± SD | 5.0 ± 1.9 | 3.9 ± 1.4 | 4.7 ± 1.7 |
| Glucose, mg/dL, mean ± SD | 107 ± 33 | 99 ± 23 | 100 ± 34 |
| Triglycerides, mg/dL, mean ± SD | 143 ± 89 | 125 ± 91 | 180 ± 142 |
| Diabetes, % | 11.8 | 7.5 | 4.4 |
| Hypertension treatment, % | 20.5 | 19.5 | 16.9 |
| Lipid treatment, % | 3.0 | 13.4 | 4.2 |
| Aspirin intake, %* | — | 18.5 | 4.6 |
| Hormone replacement (women), % | 19.7 | 16.5 | 21.3 |
| Prevalent cardiovascular disease, % | 4.2 | 6.2 | 1.5 |
| MCP-1, pg/mL, median (25th-75th percentile) | |||
| Plasma | 392 (341-447) | — | 142 (119-170) |
| Serum | — | 323 (263-401) | 312 (252-384) |
| MCP-1, ln transformed mean ± SD† | |||
| Plasma | 5.97 ± 0.23 | — | 4.98 ± 0.35 |
| Serum | — | 5.78 ± 0.34 | 5.74 ± 0.35 |
| Characteristic . | ARIC (n = 729) . | FHS (n = 6771) . | MONICA/KORA (n = 1625) . |
|---|---|---|---|
| Age, y, mean ± SD | 56.5 ± 5.1 | 49.4 ± 13.8 | 52.5 ± 10.1 |
| Women, % | 34.8 | 53.4 | 50.7 |
| Current smoking, % | 30.3 | 15.5 | 15.4 |
| Body mass index, kg/m2, mean ± SD | 27.0 ± 4.6 | 27.4 ± 5.5 | 27.3 ± 4.1 |
| Waist, cm, mean ± SD | 97.5 ± 12.4 | 96.0 ± 15.0 | 90.3 ± 12.4 |
| Systolic blood pressure, mm Hg, mean ± SD | 122 ± 18 | 121 ± 17 | 133 ± 18 |
| Total/HDL cholesterol, ratio, mean ± SD | 5.0 ± 1.9 | 3.9 ± 1.4 | 4.7 ± 1.7 |
| Glucose, mg/dL, mean ± SD | 107 ± 33 | 99 ± 23 | 100 ± 34 |
| Triglycerides, mg/dL, mean ± SD | 143 ± 89 | 125 ± 91 | 180 ± 142 |
| Diabetes, % | 11.8 | 7.5 | 4.4 |
| Hypertension treatment, % | 20.5 | 19.5 | 16.9 |
| Lipid treatment, % | 3.0 | 13.4 | 4.2 |
| Aspirin intake, %* | — | 18.5 | 4.6 |
| Hormone replacement (women), % | 19.7 | 16.5 | 21.3 |
| Prevalent cardiovascular disease, % | 4.2 | 6.2 | 1.5 |
| MCP-1, pg/mL, median (25th-75th percentile) | |||
| Plasma | 392 (341-447) | — | 142 (119-170) |
| Serum | — | 323 (263-401) | 312 (252-384) |
| MCP-1, ln transformed mean ± SD† | |||
| Plasma | 5.97 ± 0.23 | — | 4.98 ± 0.35 |
| Serum | — | 5.78 ± 0.34 | 5.74 ± 0.35 |
HDL indicates high-density lipoprotein.
Intake at least 3 per week FHS, at least 1 per week ARIC.
SD is the standard deviation of transformed biomarker values.
GWA study identified 2 loci for MCP-1 concentrations
Two genetic loci reached the predefined threshold of genome-wide significance (5 * 10−8) in the meta-analysis for serum MCP-1 (Table 2), and a third locus showed suggestive evidence of an association for EDTA plasma MCP-1. The strongest association was found for an imputed SNP in the DARC gene (rs12075, A/G, Asp42Gly) and serum MCP-1 concentrations, with an average per minor allele log MCP-1 concentration decrease of 0.23 (95% confidence interval, 0.22-0.25; P < 1.0 * 10−323) with a minor allele frequency (G allele) of 45.6% (Figure 1). Rs12075 is known to be responsible for the 2 polymorphic blood group antigens Fyb (aspartate) and Fya (glycine). MCP-1 concentrations by rs12075 genotype are provided in Table 3.
Findings with genome-wide association
| . | Serum . | Plasma . | ||||
|---|---|---|---|---|---|---|
| FHS . | MONICA/KORA . | Meta-analysis . | ARIC . | MONICA/KORA . | Meta-analysis . | |
| Chromosome 1 locus (rs12075, missense (Asp42Gly), A/G, DARC, physical position 157441978) | ||||||
| Age- and sex-adjusted | ||||||
| β Estimate, SE | −0.23, 0.006 | −0.25, 0.014 | −0.23, 0.006 | −0.002, 0.013 | −0.002, 0.013 | −0.002, 0.009 |
| P | 4.5 * 10−321 | 1.5 * 10−73 | < 1.0 * 10−323 | .87 | .85 | .82 |
| MAF, % | 45.5 | 46.3 | 45.6 | 47.6 | 46.3 | 47.0 |
| Multivariable-adjusted | ||||||
| β Estimate, SE | −0.23, 0.006 | −0.25, 0.014 | 0.23, 0.006 | 0.001, 0.013 | 0.002, 0.013 | 0.001, 0.009 |
| P | < 1.0 * 10−323 | 1.53 * 10−73 | < 1.0 * 10−323 | .99 | .88 | .91 |
| Chromosome 3 locus (rs12495098, unknown function, G/T, CCR2; CCR3, physical position 46317506) | ||||||
| Age- and sex-adjusted | ||||||
| β Estimate, SE | 0.11, 0.013 | 0.12, 0.024 | 0.11, 0.012 | 0.06, 0.024 | 0.10, 0.022 | 0.08, 0.016 |
| P | 9.7 * 10−16 | 5.6 * 10−7 | 1.1 * 10−20 | .011 | 5.7 * 10−6 | 3.7 * 10−7 |
| MAF, % | 8.7 | 8.4 | 8.6 | 7.7 | 8.4 | 8.1 |
| Multivariable-adjusted | ||||||
| β Estimate, SE | 0.11, 0.013 | 0.12, 0.024 | 0.11, 0.012 | 0.06, 0.023 | 0.10, 0.022 | 0.08, 0.016 |
| P | 1.9 * 10−16 | 5.6 * 10−7 | 1.7 * 10−21 | .01 | 4.5 * 10−6 | 3.3 * 10−7 |
| Chromosome 3 locus (rs10510751, unknown function, G/T, CCR2; CCR3, physical position 46321029) | ||||||
| Age- and sex-adjusted | ||||||
| β Estimate, SE | 0.11, 0.013 | 0.12, 0.024 | −0.11, 0.012 | 0.06, 0.023 | 0.10, 0.022 | 0.08, 0.016 |
| P | 8.9 * 10−16 | 6.1 * 10−7 | 1.2 * 10−20 | .011 | 5.7 * 10−6 | 3.7 * 10−7 |
| MAF, % | 8.7 | 8.3 | 8.6 | 7.8 | 8.3 | 8.1 |
| Multivariable-adjusted | ||||||
| β Estimate, SE | 0.11, 0.013 | 0.12, 0.024 | 0.11, 0.012 | 0.06, 0.023 | 0.10, 0.022 | 0.08, 0.016 |
| P | 1.7 * 10−16 | 6.1 * 10−7 | 1.9 * 10−21 | .01 | 4.5 * 10−6 | 4.0 * 10−7 |
| Chromosome 2 locus (rs7601375, T/C, intronic, COBLL1, physical position 46317506) | ||||||
| Age- and sex-adjusted | ||||||
| β Estimate, SE | −0.01, 0.006 | −0.006, 0.013 | −0.01, 0.006 | −0.04, 0.012 | −0.04, 0.012 | −0.04, 0.009 |
| P | .07 | .65 | .07 | .0007 | .0004 | 7.8 * 10−7 |
| MAF, % | 40.8 | 40.0 | 40.1 | 39.7 | 40.0 | 39.9 |
| Multivariable-adjusted | ||||||
| β Estimate, SE | −0.01, 0.006 | −0.006, 0.013 | −0.01, 0.006 | −0.04, 0.012 | −0.04, 0.012 | −0.04, 0.009 |
| P | .07 | .65 | .07 | .0004 | .0006 | 9.6 * 10−7 |
| . | Serum . | Plasma . | ||||
|---|---|---|---|---|---|---|
| FHS . | MONICA/KORA . | Meta-analysis . | ARIC . | MONICA/KORA . | Meta-analysis . | |
| Chromosome 1 locus (rs12075, missense (Asp42Gly), A/G, DARC, physical position 157441978) | ||||||
| Age- and sex-adjusted | ||||||
| β Estimate, SE | −0.23, 0.006 | −0.25, 0.014 | −0.23, 0.006 | −0.002, 0.013 | −0.002, 0.013 | −0.002, 0.009 |
| P | 4.5 * 10−321 | 1.5 * 10−73 | < 1.0 * 10−323 | .87 | .85 | .82 |
| MAF, % | 45.5 | 46.3 | 45.6 | 47.6 | 46.3 | 47.0 |
| Multivariable-adjusted | ||||||
| β Estimate, SE | −0.23, 0.006 | −0.25, 0.014 | 0.23, 0.006 | 0.001, 0.013 | 0.002, 0.013 | 0.001, 0.009 |
| P | < 1.0 * 10−323 | 1.53 * 10−73 | < 1.0 * 10−323 | .99 | .88 | .91 |
| Chromosome 3 locus (rs12495098, unknown function, G/T, CCR2; CCR3, physical position 46317506) | ||||||
| Age- and sex-adjusted | ||||||
| β Estimate, SE | 0.11, 0.013 | 0.12, 0.024 | 0.11, 0.012 | 0.06, 0.024 | 0.10, 0.022 | 0.08, 0.016 |
| P | 9.7 * 10−16 | 5.6 * 10−7 | 1.1 * 10−20 | .011 | 5.7 * 10−6 | 3.7 * 10−7 |
| MAF, % | 8.7 | 8.4 | 8.6 | 7.7 | 8.4 | 8.1 |
| Multivariable-adjusted | ||||||
| β Estimate, SE | 0.11, 0.013 | 0.12, 0.024 | 0.11, 0.012 | 0.06, 0.023 | 0.10, 0.022 | 0.08, 0.016 |
| P | 1.9 * 10−16 | 5.6 * 10−7 | 1.7 * 10−21 | .01 | 4.5 * 10−6 | 3.3 * 10−7 |
| Chromosome 3 locus (rs10510751, unknown function, G/T, CCR2; CCR3, physical position 46321029) | ||||||
| Age- and sex-adjusted | ||||||
| β Estimate, SE | 0.11, 0.013 | 0.12, 0.024 | −0.11, 0.012 | 0.06, 0.023 | 0.10, 0.022 | 0.08, 0.016 |
| P | 8.9 * 10−16 | 6.1 * 10−7 | 1.2 * 10−20 | .011 | 5.7 * 10−6 | 3.7 * 10−7 |
| MAF, % | 8.7 | 8.3 | 8.6 | 7.8 | 8.3 | 8.1 |
| Multivariable-adjusted | ||||||
| β Estimate, SE | 0.11, 0.013 | 0.12, 0.024 | 0.11, 0.012 | 0.06, 0.023 | 0.10, 0.022 | 0.08, 0.016 |
| P | 1.7 * 10−16 | 6.1 * 10−7 | 1.9 * 10−21 | .01 | 4.5 * 10−6 | 4.0 * 10−7 |
| Chromosome 2 locus (rs7601375, T/C, intronic, COBLL1, physical position 46317506) | ||||||
| Age- and sex-adjusted | ||||||
| β Estimate, SE | −0.01, 0.006 | −0.006, 0.013 | −0.01, 0.006 | −0.04, 0.012 | −0.04, 0.012 | −0.04, 0.009 |
| P | .07 | .65 | .07 | .0007 | .0004 | 7.8 * 10−7 |
| MAF, % | 40.8 | 40.0 | 40.1 | 39.7 | 40.0 | 39.9 |
| Multivariable-adjusted | ||||||
| β Estimate, SE | −0.01, 0.006 | −0.006, 0.013 | −0.01, 0.006 | −0.04, 0.012 | −0.04, 0.012 | −0.04, 0.009 |
| P | .07 | .65 | .07 | .0004 | .0006 | 9.6 * 10−7 |
Data are provided for the strongest associations for the respective locus for SNPs available in both studies (genotyped or imputed).
dbSNP positions are from NCBI Build 36. For comparison the respective results for multivariable-adjusted associations (secondary analysis) are provided. The β values are for log-transformed MCP-1 residuals.
Plasma MCP-1 concentrations were not available in FHS, serum concentrations were not available in ARIC.
LD r2 rs12495098 and rs10510751 are 1.0.
GWA study of serum MCP-1. (A) Signal-intensity plot showing the age- and sex-adjusted association of SNPs across all autosomes with MCP-1 in the GWA analysis. The coordinate provides the number of the chromosome. Lines indicate P value thresholds for P = 5 * 10−8. (B) Regional plots of associations with lowest P value for the DARC gene. Based on HapMapCEU, NCBI Build 36. The color coding represents r2 with lead SNP (rs12075), white indicates r2 < 0.01; gray, 0.01 ≤ r2 < 0.1; yellow, 0.1 ≤ r2 < 0.2; orange, 0.2 ≤ r2 < 0.5. Signal intensity for multivariable-adjusted serum concentrations and results for plasma are presented in supplemental Figure 2.
GWA study of serum MCP-1. (A) Signal-intensity plot showing the age- and sex-adjusted association of SNPs across all autosomes with MCP-1 in the GWA analysis. The coordinate provides the number of the chromosome. Lines indicate P value thresholds for P = 5 * 10−8. (B) Regional plots of associations with lowest P value for the DARC gene. Based on HapMapCEU, NCBI Build 36. The color coding represents r2 with lead SNP (rs12075), white indicates r2 < 0.01; gray, 0.01 ≤ r2 < 0.1; yellow, 0.1 ≤ r2 < 0.2; orange, 0.2 ≤ r2 < 0.5. Signal intensity for multivariable-adjusted serum concentrations and results for plasma are presented in supplemental Figure 2.
MCP-1 concentrations by rs12075 (Asp42Gly) genotype
| . | ARIC (n = 729) . | FHS (n = 6771) . | MONICA/KORA (n = 1625) . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| A/A . | A/G . | G/G . | A/A . | A/G . | G/G . | A/A . | A/G . | G/G . | |
| MCP-1, pg/mL, median (25th-75th percentile) | |||||||||
| Plasma | 398 (337-462) | 388 (338-441) | 392 (347-447) | — | — | — | 142 (118-172) | 143 (119-171 | 143 (120-168) |
| Serum | — | — | — | 389 (324-466) | 318 (264-380) | 259 (214-310) | 376 (315-450) | 307 (256-365) | 240 (204-297) |
| MCP-1, ln transformed, mean ± SD | |||||||||
| Plasma | 5.98 ± 0.25 | 5.96 ± 0.23 | 5.98 ± 0.21 | — | — | — | 4.98 ± 0.35 | 4.98 ± 0.34 | 4.98 ± 0.32 |
| Serum | — | — | — | 5.97 ± 0.31 | 5.76 ± 0.32 | 5.55 ± 0.29 | 5.94 ± 0.29 | 5.72 ± 0.32 | 5.50 ± 0.32 |
| . | ARIC (n = 729) . | FHS (n = 6771) . | MONICA/KORA (n = 1625) . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| A/A . | A/G . | G/G . | A/A . | A/G . | G/G . | A/A . | A/G . | G/G . | |
| MCP-1, pg/mL, median (25th-75th percentile) | |||||||||
| Plasma | 398 (337-462) | 388 (338-441) | 392 (347-447) | — | — | — | 142 (118-172) | 143 (119-171 | 143 (120-168) |
| Serum | — | — | — | 389 (324-466) | 318 (264-380) | 259 (214-310) | 376 (315-450) | 307 (256-365) | 240 (204-297) |
| MCP-1, ln transformed, mean ± SD | |||||||||
| Plasma | 5.98 ± 0.25 | 5.96 ± 0.23 | 5.98 ± 0.21 | — | — | — | 4.98 ± 0.35 | 4.98 ± 0.34 | 4.98 ± 0.32 |
| Serum | — | — | — | 5.97 ± 0.31 | 5.76 ± 0.32 | 5.55 ± 0.29 | 5.94 ± 0.29 | 5.72 ± 0.32 | 5.50 ± 0.32 |
In MONICA/KORA, MCP-1 serum concentrations were available for 1626 persons. Some characteristics have missing values (indicated by —).
The genomic control λ in all cohorts was low (λGC ≤ 1.017). Signal-intensity plots for the association of SNPs across all autosomes with MCP-1 in the GWA analysis are provided in supplemental Figure 2.
Multivariable-adjusted results (see supplemental Table 1 for variables) were similar to the top findings derived from the primary analysis. Because rs12075 was imputed, we genotyped the SNP in a subset of FHS participants (n = 2666); these analyses showed an average per allele decrease of 0.217 (SE = 0.008, P = 1.3 * 10−136) compared with those obtained with the imputed SNP data (−0.224, SE = 0.009, P = 3.5 * 10−117) in the same subgroup.
For replication, rs12075 was genotyped in a different subcohort of the MONICA/KORA sample (n = 1424) and resulted in an average decrease of 0.28 (SE = 0.03, P = 2.2 * 10−16) for serum MCP-1 concentrations (Table 4). DARC Asp42Gly explained approximately 20% and 19% of the residual variability in MCP-1 serum concentrations in FHS and MONICA/KORA, respectively. Adding the SNP increased the total variance explained by the covariates in age- and sex-adjusted models from 4.9% to 18.5% in FHS and from 4.6% to 23.0% in MONICA/KORA; the comparable changes for multivariable models were 7.3% to 18.6% in FHS and 6.7% to 24.5% in MONICA/KORA.
Different inflammatory biomarkers in relation to SNP rs12075
| Biomarker . | n . | Median (25th to 75th percentile) . | β . | SE . | 95% CI . | P . |
|---|---|---|---|---|---|---|
| ARIC, plasma | ||||||
| RANTES, pg/mL | 1114 | 5996 (2006-10 259) | 0.039 | 0.073 | −0.10 to 0.18 | .59 |
| FHS, serum | ||||||
| Intercellular adhesion molecule-1* | 6897 | 241.0 (208.9-285.9) | 0.007 | 0.005 | −0.004 to 0.017 | .20 |
| Interleukin-6* | 3042 | 2.71 (1.82-4.31) | −0.011 | 0.020 | −0.050 to 0.028 | .59 |
| Myeloperoxidase* | 2941 | 39.95 (27.76-59.41) | −0.028 | 0.017 | −0.061 to 0.004 | .09 |
| MONICA/KORA subcohort, serum | ||||||
| C-reactive protein* | 1453 | 1.47 (0.66-3.21) | −0.014 | 0.044 | −0.101 to 0.073 | .76 |
| Intercellular adhesion molecule-1 | 1436 | 728.9 (590.4-902.4) | 13.11 | 11.43 | −9.29 to 35.51 | .25 |
| Interferon-inducible protein-10* | 1423 | 220.3 (144.4-357.0) | −0.024 | 0.032 | −0.086 to 0.039 | .46 |
| Interleukin-6* | 1432 | 2.35 (1.41-3.80) | −0.057 | 0.044 | −0.142 to 0.029 | .194 |
| Interleukin-8*† | 1424 | 6.9 (5.0-9.8) | −0.068 | 0.025 | −0.117 to −0.019 | .007 |
| Interleukin-18* | 1424 | 171.4 (113.2-243.3) | −0.034 | 0.031 | −0.095 to 0.028 | .284 |
| Macrophage migration inhibitory factor* | 1379 | 18.0 (14.5-22.8) | −0.018 | 0.015 | −0.047 to 0.010 | .21 |
| MCP-1*† | 1424 | 201.6 (126.1-312.1) | −0.28 | 0.03 | −0.35 to −0.22 | 2.2 * 10−16 |
| Myeloperoxidase* | 1268 | 120.0 (94.2-154.0) | −0.006 | 0.016 | −0.038 to 0.026 | .70 |
| RANTES*† | 1380 | 22 844 (14 938-35 604) | −0.068 | 0.022 | −0.111 to −0.025 | .002 |
| Biomarker . | n . | Median (25th to 75th percentile) . | β . | SE . | 95% CI . | P . |
|---|---|---|---|---|---|---|
| ARIC, plasma | ||||||
| RANTES, pg/mL | 1114 | 5996 (2006-10 259) | 0.039 | 0.073 | −0.10 to 0.18 | .59 |
| FHS, serum | ||||||
| Intercellular adhesion molecule-1* | 6897 | 241.0 (208.9-285.9) | 0.007 | 0.005 | −0.004 to 0.017 | .20 |
| Interleukin-6* | 3042 | 2.71 (1.82-4.31) | −0.011 | 0.020 | −0.050 to 0.028 | .59 |
| Myeloperoxidase* | 2941 | 39.95 (27.76-59.41) | −0.028 | 0.017 | −0.061 to 0.004 | .09 |
| MONICA/KORA subcohort, serum | ||||||
| C-reactive protein* | 1453 | 1.47 (0.66-3.21) | −0.014 | 0.044 | −0.101 to 0.073 | .76 |
| Intercellular adhesion molecule-1 | 1436 | 728.9 (590.4-902.4) | 13.11 | 11.43 | −9.29 to 35.51 | .25 |
| Interferon-inducible protein-10* | 1423 | 220.3 (144.4-357.0) | −0.024 | 0.032 | −0.086 to 0.039 | .46 |
| Interleukin-6* | 1432 | 2.35 (1.41-3.80) | −0.057 | 0.044 | −0.142 to 0.029 | .194 |
| Interleukin-8*† | 1424 | 6.9 (5.0-9.8) | −0.068 | 0.025 | −0.117 to −0.019 | .007 |
| Interleukin-18* | 1424 | 171.4 (113.2-243.3) | −0.034 | 0.031 | −0.095 to 0.028 | .284 |
| Macrophage migration inhibitory factor* | 1379 | 18.0 (14.5-22.8) | −0.018 | 0.015 | −0.047 to 0.010 | .21 |
| MCP-1*† | 1424 | 201.6 (126.1-312.1) | −0.28 | 0.03 | −0.35 to −0.22 | 2.2 * 10−16 |
| Myeloperoxidase* | 1268 | 120.0 (94.2-154.0) | −0.006 | 0.016 | −0.038 to 0.026 | .70 |
| RANTES*† | 1380 | 22 844 (14 938-35 604) | −0.068 | 0.022 | −0.111 to −0.025 | .002 |
Models were age- and sex-adjusted, MONICA/KORA additionally adjusted for survey.
Biomarkers were log-transformed for analysis of relation to rs12075.
Significant associations (P < .05) were identified for inflammatory biomarkers with a Darc strong-binding chemokine profile, but not for other serum inflammatory markers.
Notably, rs12075 was not associated with plasma MCP-1 concentrations in MONICA/KORA and ARIC (P = .82; Table 2). The lack of association was confirmed in a subset of FHS participants (n = 263; P = .24).
Because Darc is a chemokine receptor/reservoir with an established profile of chemokine binding,5 the discrepancy between serum and plasma findings led us to hypothesize that serum MCP-1 levels represented the combination of circulating plasma levels and additional MCP-1 liberated from the erythrocyte Darc reservoir by clotting.
A second locus on chromosome 3 was identified at the chemokine receptor gene cluster recently reported to be associated with EDTA plasma MCP-1 levels by linkage.14 Two SNPs, rs12495098 and rs10510751, in complete linkage disequilibrium (D′ = 1.0, r2 = 1.0) at the CCR2/CCR3 cytokine receptor gene cluster were consistently associated with MCP-1 concentrations in both serum and EDTA plasma, although slightly under the genomewide significance level for plasma, P = 3.7 * 10−7. There was no evidence for interaction ([non]–multiplicative) between the chromosome 1 and 3 locus.
A potential locus was identified on chromosome 2 with an intronic SNP in the COBLL1 (COBL-like 1) gene, associated with EDTA plasma MCP-1. Additional SNPs with a meta-analysis P value of less than 1 * 10−6 for serum or EDTA plasma MCP-1 are tabulated in the supplemental data. We examined SNPs in the previously reported gene coding for MCP-1, CCL2. The lowest P value for a SNP in the (candidate) cytokine cluster comprising the CCL2 gene was for rs9893096 (MAF, 47.6%), with an average per allele increase of 0.02 (SE, 0.006, P = 4.3 * 10−5). The meta-analysis for the previously replicated rs1024611, −2518 A/G (LD r2 = 0.41 with rs9893096) showed a P value of .003.
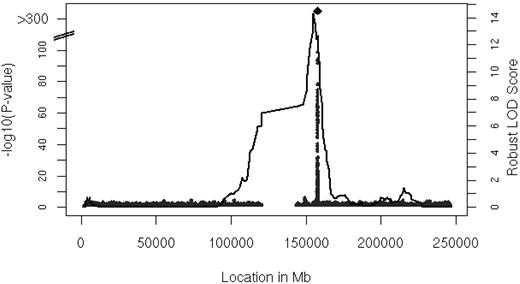
Linkage analysis identified a locus for serum MCP-1 levels on chromosome 1
We performed linkage analysis with families in the FHS with 2 or more persons that had both serum MCP-1 and genotype data available (n = 6539 participants in 878 families). The results of linkage analysis are presented in Figure 2. One convincing locus was evident on chromosome 1 at 154.8 Mb with a logarithm of odds (LOD) score of 14.3 (genome-wide P = 8.0 * 10−13). The linkage peak substantially decreased after adjustment for rs12075 to LOD of 2.47 (the multivariable-adjusted LOD = 15.96 decreased to LOD = 2.71 after inclusion of rs12075), which was imputed with a ratio of observed to expected variance of 0.8, indicating that the imputation was imperfect (ratio = 1 for good imputation). Family-based association testing applied to the genotyped SNPs showed significant association in the vicinity of rs12075 (minimum family-based association testing P = 9.0 * 10−26) in the FHS.
Linkage analysis of serum MCP-1. Age- and sex-adjusted multipoint linkage of MCP-1 on chromosome 1 (154.8 Mb) with an LOD score of 14.3; genome-wide P = 8.0 * 10−13. The DARC gene is located between 157 441 134 and 157 442 914 base pairs (bp) on chromosome 1.
Linkage analysis of serum MCP-1. Age- and sex-adjusted multipoint linkage of MCP-1 on chromosome 1 (154.8 Mb) with an LOD score of 14.3; genome-wide P = 8.0 * 10−13. The DARC gene is located between 157 441 134 and 157 442 914 base pairs (bp) on chromosome 1.
Erythrocyte Darc is a major source of MCP-1 in serum
To explore the hypothesis that erythrocyte Darc was the source of the additional MCP-1 in serum compared with EDTA plasma, we first asked whether serum levels, but not plasma levels, of other known Darc-binding chemokines showed an association with rs12075. Significant associations (P < .05) were identified for serum interleukin-8 and RANTES, but not for other serum inflammatory markers outside the known Darc strong-binding chemokine profile (Table 4), such as interferon-inducible protein-10. In addition, no association was observed between rs12075 and EDTA plasma concentrations of RANTES in the ARIC cohort.
We were able to rule out assay artifacts (presence or absence of calcium, freezing and thawing, or clotting plasma) as a source of the difference (n = 5 donors; Table 5). During these studies we observed approximately 2-fold increased values of MCP-1 in heparin collection tubes; we confirmed that, similar to EDTA, heparin had no direct effect on the MCP-1 assay (n = 5, 156.0 ± 13.2 pg/mL in EDTA versus 162.2 ± 18.3 pg/mL in EDTA plus heparin). In addition, because it had been proposed that platelets2 and monocytes and T cells3 might present MCP-1 under some conditions, we were able to exclude relevant release of MCP-1 from platelets and PBMCs during the clotting process (supplemental Table 4).
Effect of calcium or freezing and thawing on MCP-1 assay
| Sample . | None . | Additional additive . | |
|---|---|---|---|
| EDTA . | Calcium . | ||
| EDTA | 124.5 (14.7) | 123.9 (13.9) | |
| EDTA −80°C | 127.2 (10.7) | 121.9 (15.8) | |
| Serum | 161.3 (22.9) | 155.1 (24.0) | |
| Serum −80°C | 156.5 (23.0) | 161.3 (24.8) | |
| Heparin | 236.9 (22.7) | ||
| Heparin −80°C | 233.4 (25.2) | ||
| Sample . | None . | Additional additive . | |
|---|---|---|---|
| EDTA . | Calcium . | ||
| EDTA | 124.5 (14.7) | 123.9 (13.9) | |
| EDTA −80°C | 127.2 (10.7) | 121.9 (15.8) | |
| Serum | 161.3 (22.9) | 155.1 (24.0) | |
| Serum −80°C | 156.5 (23.0) | 161.3 (24.8) | |
| Heparin | 236.9 (22.7) | ||
| Heparin −80°C | 233.4 (25.2) | ||
Data represent the geometric mean (SEM) of 5 donors (in pg/mL).
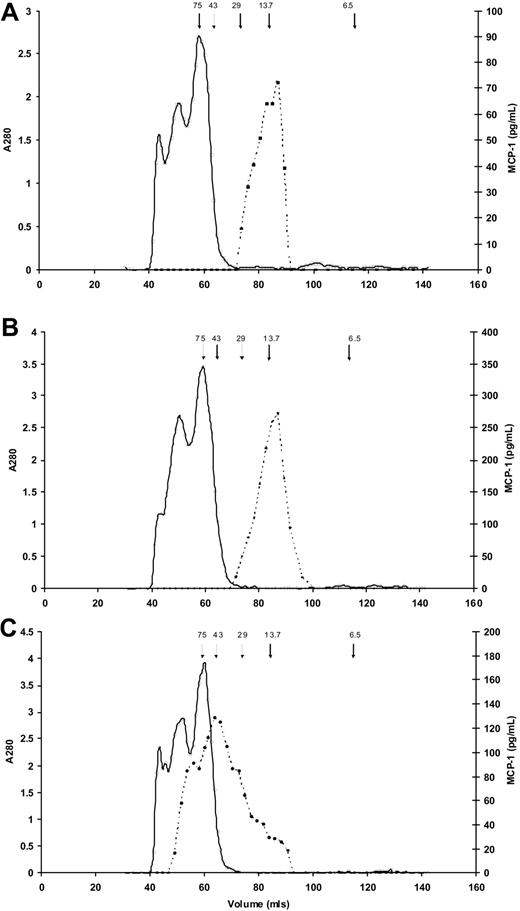
To confirm that enzymes activated during clotting/fibrinolysis did not have a proteolytic effect on MCP-1 resulting in altered immunogenicity, we used size exclusion chromatography to characterize serum and plasma MCP-1. In both EDTA plasma (166.9 pg/mL) and serum (377.4 pg/mL), MCP-1 predominantly eluted at an apparent molecular weight of approximately 13 500 kDa, the expected value for MCP-1 monomers (Figure 3A-B). In addition, and in contrast, Figure 3C shows a large molecular weight range for MCP-1 in heparin plasma (316.5 pg/mL), suggesting binding of the MCP-1 to glycosaminoglycans present in unfractionated heparin.
Molecular weight estimates of MCP-1 in serum and plasma. Results of size exclusion chromatography for MCP-1 (dashed lines) in (A) EDTA plasma, (B) serum, and (C) heparin plasma. Arrows indicate elution of molecular weight standards, in kDa; solid line indicates absorbance at 280 nm. (A) In EDTA plasma MCP-1 predominantly elutes from the gel filtration column at a molecular weight of approximately 13 500 kDa. The slight shoulder suggests possible dimerization. (B) MCP-1 in serum elutes predominantly as a single peak similar to EDTA plasma. (C) MCP-1 in heparin plasma elutes at a large molecular weight range, suggesting binding of the MCP-1 to the various molecular weight glycosaminoglycans present in unfractionated heparin.
Molecular weight estimates of MCP-1 in serum and plasma. Results of size exclusion chromatography for MCP-1 (dashed lines) in (A) EDTA plasma, (B) serum, and (C) heparin plasma. Arrows indicate elution of molecular weight standards, in kDa; solid line indicates absorbance at 280 nm. (A) In EDTA plasma MCP-1 predominantly elutes from the gel filtration column at a molecular weight of approximately 13 500 kDa. The slight shoulder suggests possible dimerization. (B) MCP-1 in serum elutes predominantly as a single peak similar to EDTA plasma. (C) MCP-1 in heparin plasma elutes at a large molecular weight range, suggesting binding of the MCP-1 to the various molecular weight glycosaminoglycans present in unfractionated heparin.
We next assessed whether therapeutic heparin increased MCP-1 concentrations with the use of data from a clinical trial comparing heparin with bivalirudin as adjunctive therapy during coronary stent placement. Concentrations of MCP-1 in peripheral blood at baseline (171.2 ± 37.6 pg/mL) and 1 hour after heparin application (167.2 ± 37.8 pg/mL) were similar (n = 6 patients in each arm; supplemental Table 5). Therapeutic heparin is commonly given to achieve systemic concentrations of approximately 0.3 to 0.7 U/mL25 ; in an experiment whereby we titrated blood with unfractionated heparin, we did not observe a marked increase in MCP-1 concentrations with heparin concentrations less than 1.6 U/mL (supplemental Table 6).
Taken together, these results provided convincing evidence that serum and heparin plasma contained MCP-1 freed from erythrocytes by blood clotting and competition with soluble heparin, respectively.
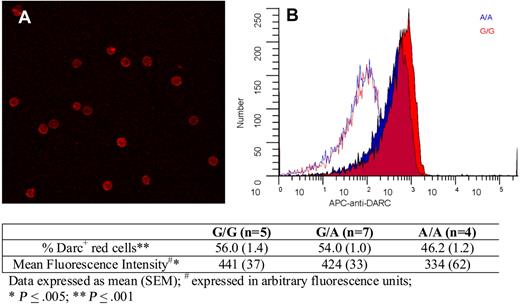
Finally, we explored whether there were any Asp42Gly-associated differences in Darc expression on red cells. Figure 4 shows erythrocyte Darc expression; quantitative immuno–flow cytometry indicated that compared with G/G persons, A/A persons exhibited an 18% decrease in the percentage of Darc+ cells and a 24% decrease in mean fluorescence intensity. Because these differences were relatively small and not in the expected direction, we conclude that it is unlikely that differential expression because of the Asp42Gly change explains increased Darc-bound MCP-1 on the erythrocytes of A/A persons and that differential chemokine binding is more likely explained by a change in Darc structure and function.
Darc expression on erythrocytes. (A) A representative field showing Darc expression on erythrocytes visualized by confocal microscopy (“Darc expression”). (B) Flow cytometric histograms of red cell Darc expression from representative persons with A/A (blue) and G/G (red) genotypes. The corresponding isotype controls are shown as red and blue lines. The table indicates the average values for the percentage of erythrocytes expressing Darc and the mean fluorescent intensity from persons with G/G, A/G, and A/A genotypes, as described in “Darc expression.”
Darc expression on erythrocytes. (A) A representative field showing Darc expression on erythrocytes visualized by confocal microscopy (“Darc expression”). (B) Flow cytometric histograms of red cell Darc expression from representative persons with A/A (blue) and G/G (red) genotypes. The corresponding isotype controls are shown as red and blue lines. The table indicates the average values for the percentage of erythrocytes expressing Darc and the mean fluorescent intensity from persons with G/G, A/G, and A/A genotypes, as described in “Darc expression.”
Discussion
The 3 major findings of this study are the following: first, in a meta-analysis of 3 community-based samples with persons of European descent, we identified 2 previously unknown genomic loci related to circulating MCP-1 concentrations. The strongest association was with serum MCP-1 concentrations and rs12075 a nonsynonymous SNP in the DARC gene (Asp42Gly). We identified a second locus consistently associated with both serum and EDTA plasma MCP-1 concentrations in a cluster of chemokine receptor genes, including the MCP-1 receptor, CCR2.
Second, we have identified an important mechanism for freeing chemokines from the Darc reservoir, coagulation. This mechanism was hypothesized from the differential association of rs12075 with serum versus EDTA plasma MCP-1 and was substantiated by determining that serum levels of other Darc-related cytokines also were associated with variation in rs12075, and that immunoassay artifacts because of differences in sample types or handling or in activation of coagulation enzymes were unlikely. Third, we established a second mechanism by which Darc-bound MCP-1 is released from erythrocytes by heparin if present at high enough concentrations.
Variation in Darc is associated with serum cytokine levels
Darc belongs to the blood group alloantigen system and serves for phenotyping Darc status of donor blood in blood banking. The Asp42Gly polymorphism determines the 2 polymorphic antigens Fya and Fyb.26 Differential binding capacities of chemokines related to rs12075 are novel. Functionally, Darc constitutes a 7 transmembrane domain multispecific chemotactic receptor that is unique in binding both CC (MCP-1, RANTES) and CXC (interleukin-8) cytokines.27 Darc does not participate in signal transduction directly because it lacks a necessary DRY motif. However, recent data indicate that cytokine binding to Darc leads to heterodimerization with CCR5 and inhibition of CCR5 signaling.28 In the vasculature, Darc is present on erythrocytes and venule endothelium. Endothelial Darc is required for proper chemokine-mediated recruitment of leukocytes and for transendothelial chemokine transport. Darc is unique as a chemokine receptor in that internalized chemokines remain fully active. Erythrocyte Darc has been proposed to act as a chemokine reservoir, allowing high circulating vascular concentrations without desensitization of circulating leukocytes.29 However, to date mechanisms for the release of Darc-bound chemokines have not been established. Although we have now established clotting as one such mechanism, the molecular processes and purpose (if any) for MCP-1 release during clotting are unknown. Although we excluded that relevant amounts of MCP-1 are set free from platelets and PBMCs, other Darc ligands such as the CXC chemokine neutrophil activating peptide 2 stored in platelets may be released and compete with MCP-1 for its binding site.
Findings in humans and animal models regarding the absence of Darc suggest physiologic importance for the regulation of Darc function. Duffy-negative persons (erythrocyte Darc−, endothelial Darc+) have a lower mean white blood cell count and neutrophil count30 and have delayed graft function and increased graft failure after kidney transplantation.31 We also note that complete loss of Darc is rare.27 Darc knockout mice lack protection from hyperacute inflammatory responses, probably mediated primarily by lack of erythrocyte Darc,32 and also exhibit rapid clearance of vascular chemokines after injection.33 Mice lacking only endothelial Darc exhibit deficient leukocyte emigration,27 and recent studies have identified DARC as a major regulator of murine bone mineral density through a role in cytokine-mediated osteoclastogenesis.34
Although in general recent GWA studies have reported small effect sizes, we found a single variant, DARC rs12075 (Asp42Gly), that explains approximately 20% of the variation in MCP-1 serum concentration, which is considerably greater than all previously known correlates combined.13 It is notable that DARC is a trans-acting variant and is not on the same chromosome as the gene coding for MCP-1 (CCL2). As observed before,14 family-based genetic linkage for serum MCP-1 confirms our GWA results with a convincingly high LOD score of greater than 14. The single SNP, rs12075, located under the peak explains most of this linkage signal. From the increased strength of association after genotyping, the evidence from the linkage results, and the fact that rs12075 is a nonsynonymous polymorphism in the cytokine binding region of Darc, we conclude that it is probable that rs12075 may fully explain the relation to serum MCP-1 concentrations at this locus.
Few SNPs in the genome show a population differentiation as marked as rs12075.35 The aspartate-to-glycine amino acid difference associated with rs12075 forms the basis for Duffy blood groups Fyb (Asp, A allele) and Fya (Gly, G allele). Several nonsynonymous changes have been identified on the Fyb background (Arg89Cys, Ala100Thr, GATA-1), whose effects are unlikely to explain our rs12075 observations.
Although the mechanism by which variance at this location causes a difference in chemokine binding needs definitive determination, our preliminary examination using immuno-flow cytometry suggests that a simple difference in erythrocyte Darc expression is unlikely to be the answer. Flow cytometric analysis with monoclonal anti-Darc antibody failed to show any significant increase in Darc expression in erythrocytes from A/A homozygote persons (in fact there appeared to be a modest decrease), suggesting a genotype-mediated change in function rather than a change in expression causing different levels of circulating MCP-1. The location of a necessary sulfation at Tyr4136 suggests the possibility of more efficient sulfation or augmentation of chemokine binding to the sulfated Tyr, in the presence of the negatively charged Asp, becauseAsp42 Darc was associated with greater erythrocyte-bound MCP-1.
We speculate that these findings may have clinical importance. First, the genetically determined capacity of Darc to bind chemokines, as estimated by accounting for 20% of the variability of serum MCP-1, may be important under multiple pathophysiologic conditions ranging from cancer and atherosclerosis to inflammatory diseases in general. There is evidence that Darc-mediated chemokine sequestration helps determine tumor microenvironment, angiogenesis, and metastasis.37,38 Under inflammatory stimuli, endothelial Darc is up-regulated and modulates the inflammatory response,28,39 and recent results suggest an important role for Darc-sequestered cytokines in the regulation of bone metabolism.34 Because erythrocyte Darc may act both to limit free chemokine activity and to present chemokine to key local environments when needed, genetic variation in these capacities may have significant implications.
Second, our finding of the release of chemokines such as MCP-1 from Darc during clotting (although the exact mechanisms remain to be elucidated) suggests localized increased chemokine levels in thrombotic states such as acute coronary syndromes,40 venous thromboembolism,41 or disseminated intravascular coagulation, as well as during physiologically necessary hemostasis. Because Darc is present on venules, microthrombosis may liberate chemokines from endothelial Darc as well. Whether increased local chemokine levels act to ameliorate or exacerbate clot formation and/or the inflammatory effects associated with hemostasis and thrombosis remains to be determined.
Third, supraphysiologic heparin concentration liberated relatively large quantities of MCP-1. We did not observe a systemic increase in Darc sequestered inflammatory biomarkers after therapeutic application of heparin. However, although it seems possible that concentrations of heparin sufficient to release MCP-1 from red cells are achieved at the wave front propagated by a start-up bolus of heparin (eg, 80-100 U/kg), the consequent effects of these hypothesized relatively large local chemokine concentrations are uncertain. We cannot exclude that our observation is an in vitro phenomenon.
Variation at the MCP-1 receptor is associated with MCP-1 concentrations in serum and plasma
We observed a second locus associated with both serum and EDTA plasma levels of MCP-1 in a chemokine receptor gene cluster region on chromosome 3. Our results are strengthened by a recent linkage scan that showed a peak for plasma MCP-1 concentrations on chromosome 3 with a LOD score of 3.5 at 78cM, which coincides with the same chemokine receptor cluster.14 In the FHS linkage study, we did not observe a relevant peak in this area even with a larger sample compared with the original publication, a finding which may be due to the small effect size.15 CCR2 resides in this region and codes for 2 isoforms of the MCP-1 receptor,4 which mediate the biologic effects of MCP-1; thus, CCR2 is a good candidate for further exploration. The interaction of MCP-1 and CCR2 are important in blood monocyte homeostasis and mobilization from the bone marrow.42 Furthermore, recent investigations showed that the signaling chemokine receptor CCR2 is responsible for homeostatic and clearance functions for chemokines in the circulation and tissues.43 As seen for other decoy receptors such as D6, a high-affinity receptor for CC chemokines,44 dysfunction of these scavengers leads to increased amounts of circulating chemokines.43 Although not consistently, CCR2 polymorphisms have been related to coronary atherosclerosis.45,46 Because the CCR2 association with MCP-1 concentration was at least as strong when considering the effects of Darc-bound MCP-1 (β estimate, SE = 0.11, 0.012 for serum; SE = 0.08, 0.016 for EDTA plasma) we hypothesize an important receptor-mediated physiologic activity associated with Darc-bound cytokine.
Prior investigations have shown associations between the candidate gene that codes for MCP-1, CCL2 (17q11.2-q21.1) and MCP-1 concentrations.13,16,17 The lowest P value in the present GWA study in the chemokine cluster that comprises CCL2 was found for rs9893096 (P = 4.3 * 10−5). The meta-analysis for the previously replicated rs1024611, −2518 A/G, did not reach genome-wide significance (P = .003), but this locus remains a reasonable candidate.
Limitations
We acknowledge we cannot be certain that the identified SNPs are the causal variants; follow-up functional studies will provide important insights. In addition, the meta-analysis was performed in persons of European descent. We suspect that associations will differ in racially diverse cohorts because of the known, marked frequency variation in the top SNP rs12075 by ethnicity and the effect of other SNPs such as the GATA-1 mutation (FY-) which results in the absence of Darc expression on erythrocytes.
We observed 3 new loci with small-to-moderate effect sizes, but we may not have identified variants that did not reach the a priori–defined significance threshold. Because of moderate sample size we were only able to identify strong signals, for example, the known candidate gene CCL2 did not meet the predefined significance criteria. Follow-up meta-analyses with larger datasets will be required to provide information on genetic variation too modest to achieve the genome-wide significance threshold in the current analysis.
Although we observed differential function of erythrocyte Darc based on the rs12075 genotype, we cannot be certain that this difference would be expressed by the Darc in other locations such as venule endothelium. However, because the difference seems mediated by an amino acid change, this is probable. Finally, because we used a monoclonal antibody to assess Darc expression on red cells, we cannot rule out possible effects of the Asp42Gly amino acid difference in immunoreactivity.
In conclusion, we report novel genetic loci with genome-wide significance in relation to circulating MCP-1 concentrations and pathophysiologic plausibility that have not been identified by candidate gene approaches. Further exploration and functional analyses of these loci will show whether they can serve as clinically relevant targets for a better understanding of the inflammatory process and associated diseases. We report for the first time 2 possible mechanisms for releasing erythrocyte Darc-sequestered proinflammatory cytokines, and our findings on the relation between DARC and circulating chemokine concentrations may have clinical implications in the pathophysiology of cancer, inflammation, thrombosis, and possibly the administration of heparin.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The persons and funding agencies that made this work possible are listed in the Acknowledgments in the supplemental material.
National Institutes of Health
Authorship
Contribution: C.M.B., J.C.B., E.J.B., S.B., A.D., J.D., P.D., P.T.E., C.G., C.H., T.I., J.F.K., W.K., A.P., B.M.P., R.B.S., R.P.T., R.S.V., and J.D.W. provided the study concept and design; C.M.B., J.C.B., E.J.B., J.D., M.D., P.T.E., C.G., G.H., C.H., N.S.J., J.F.K., N. Khuseyinova, N. Klopp, W.K., A.C.M., A.P., C.T., R.P.T., and J.D.W. acquired data; C.M.B., J.C.B., J.D., P.D., E.J.B., P.T.E., T.I., N.S.J., N. Klopp, W.K., A.P., B.M.P., R.B.S., R.P.T., J.F.Y., Y.Z., and E.B. analyzed and interpreted the data; J.C.B., E.J.B., J.D., P.T.E., W.K., A.P., B.M.P., R.B.S., and R.P.T. drafted the manuscript; C.M.B., J.C.B., E.J.B., S.B., A.D., J.D., P.D., P.T.E., C.G., G.H., R.C.H., T.I., N.S.J., J.F.K., N. Khuseyinova, W.K., A.P., B.M.P., R.B.S., R.P.T., R.S.V., J.D.W., J.C.M.W., Y.Z., and E.B. critically revised the manuscript for important intellectual content; J.C.B., M.B., A.D., J.D., M.G.L., R.P.T., J.F.Y., and Y.Z. analyzed statistics; E.J.B., P.T.E., R.C.H., W.K., B.M.P., R.B.S., R.P.T., and R.S.V. obtained funding; M.B., J.C.B., E.J.B., M.D., N.S.J., A.C.M., B.M.P., C.T., and R.P.T. provided administrative, technical, or material support; E.J.B., J.D., W.K., B.M.P., R.P.T., and J.C.M.W. supervised the study. M.B., J.C.B., E.M.B., J.D., P.T.E., R.C.H., W.K., B.M.P., R.B.S., and R.P.T. had full access to the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. All authors gave final approval of the version to be published. The corresponding authors certify that all authors have agreed to all the content in the manuscript, including the data as presented.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Emelia J. Benjamin, Boston University School of Medicine, Framingham Heart Study, 73 Mount Wayte Ave, Framingham, MA 01702-5827; e-mail: emelia@bu.edu; or Ron C. Hoogeveen, Baylor College of Medicine, Atherosclerosis Risk in Communities Study, Center for Cardiovascular Disease Prevention, Methodist DeBakey Heart Center, 6565 Fannin, MS F-701, Houston, TX 77030; e-mail: ronh@bcm.tmc.edu; or Wolfgang Koenig, University of Ulm, MONICA/KORA, Albert-Einstein-Allee 23, 89081 Ulm, Germany; e-mail: wolfgang.koenig@uniklinik-ulm.de; or Russell P. Tracy, University of Vermont, 208 S Park Dr, Ste 2, Colchester, VT 05446; e-mail: russell.tracy@uvm.edu.
References
Author notes
R.B.S., J.B., M.B., J.D., P.T.E., R.C.H., E.J.B., W.K., and R.P.T. contributed equally to this study.
A.D. and J.C.M.W. are members of The Netherlands Consortium on Healthy Aging (NCHA).


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal