Abstract
Inhibiting the expression of the HIV-1 coreceptor CCR5 holds great promise for controlling HIV-1 infection in patients. Here we report stable knockdown of human CCR5 by a short hairpin RNA (shRNA) in a humanized bone marrow/liver/thymus (BLT) mouse model. We delivered a potent shRNA against CCR5 into human fetal liver-derived CD34+ hematopoietic progenitor/stem cells (HPSCs) by lentiviral vector transduction. We transplanted vector-transduced HPSCs solidified with Matrigel and a thymus segment under the mouse kidney capsule. Vector-transduced autologous CD34+ cells were subsequently injected in the irradiated mouse, intended to create systemic reconstitution. CCR5 expression was down-regulated in human T cells and monocytes/macrophages in systemic lymphoid tissues, including gut-associated lymphoid tissue, the major site of HIV-1 replication. The shRNA-mediated CCR5 knockdown had no apparent adverse effects on T-cell development as assessed by polyclonal T-cell receptor Vβ family development and naive/memory T-cell differentiation. CCR5 knockdown in the secondary transplanted mice suggested the potential of long-term hematopoietic reconstitution by the shRNA-transduced HPSCs. CCR5 tropic HIV-1 infection was effectively inhibited in mouse-derived human splenocytes ex vivo. These results demonstrate that lentiviral vector delivery of shRNA into human HPSCs could stably down-regulate CCR5 in systemic lymphoid organs in vivo.
Introduction
Chemokine receptor CCR5 is an attractive therapeutic target for inhibiting HIV-1, as it serves as a HIV-1 coreceptor and is essential for CCR5 tropic HIV-1 infection.1-4 Blocking CCR5 expression should prevent HIV-1 infection at the initial stage of the viral life cycle. Individuals with a Δ32/Δ32 homozygous mutation in the CCR5 gene do not express CCR5, are highly protected from HIV-1, and are apparently normal.5-7 Recently, an HIV+ acute myelogenous leukemia patient was treated for leukemia and HIV infection by bone marrow transplantation using donated CCR5 Δ32/Δ32 marrow. After the transplantation, nearly 100% of the patient's blood cells were replaced with donor cells. HIV DNA and RNA were undetectable at 20 months, even after the discontinuation of highly active antiretroviral therapy.8 This evidence supports that long-term and stable reduction of CCR5 is a promising strategy for treating HIV-infected patients. The major limitation of this strategy is the difficulty of identifying human leukocyte antigen–matched CCR5 Δ32/Δ32 homozygous donors as the mutation exists in approximately 1% of white populations and is rare in other ethnic populations.9
Small interfering RNAs (siRNAs) induce sequence-specific degradation of mRNAs by RNA interference.10 Many forms of siRNA have been used to inhibit HIV coreceptors and HIV-1 gene expression in in vitro and in vivo experimental settings.11-18 To stably inhibit HIV replication, we and others developed lentiviral vectors that are capable of stably delivering short hairpin RNA (shRNA) in mammalian cells.19-25 We demonstrated that expression of CCR5-specific shRNA in human primary T lymphocytes results in efficient CCR5-knockdown and protection of cells from HIV-1 infection in vitro.22 However, we and others recognized that a high level of sustained shRNA expression may be toxic to cells because of competition with endogenous micro-RNA biogenesis, induction of interferon responses, and/or off-targeting effects.23,26-33 To stably reduce CCR5 expression without cytotoxicity, we identified a highly efficient shRNA (shRNA 1005) directed to human CCR5 mRNA using the enzymatic production of RNAi libraries (EPRIL) screening technique.21,34 We expressed shRNA 1005 using the transcriptionally weak H1 promoter to stably reduce CCR5 expression without inducing cytotoxicity in human primary peripheral blood lymphocytes in vitro.21,34 To test stable CCR5 reduction in vivo, we used a nonhuman primate hematopoietic stem cell transplantation model in which we were able to demonstrate stable reduction of CCR5 expression in peripheral blood lymphocytes in shRNA-transduced CD34+ cell-transplanted rhesus macaques.21 Because of a single nucleotide mismatch in the shRNA 1005 target sequence between human and rhesus macaque CCR5 mRNA, we mutated the human CCR5 shRNA 1005 so that it would be 100% homologous to the corresponding rhesus macaque CCR5 mRNA target sequence. This rhesus macaque-specific shRNA 1005 inhibited rhesus macaque CCR5 expression but not human CCR5 expression.21
In this study, we used a recently developed humanized bone marrow/liver/thymus (hu-BLT) mouse model to examine the down-regulation of human CCR5 expression using shRNA 1005 against human CCR5 mRNA.35,36 Unlike other humanized mouse models, this model allows us to examine the effects of shRNA expression during T-cell differentiation in the transplanted tissue thymus and liver (thy/liv). We found that differentiated T cells were able to migrate systemically and develop functional primary and secondary lymphoid organs. We demonstrated here that an implant of lentiviral vector–mediated CCR5 shRNA-transduced CD34+ cells did result in efficient and stable CCR5-knockdown in multiple lymphoid organs, including in gut-associated mucosal lymphoid tissues, without causing apparent adverse effects. We found that the CCR5 knockdown was sufficient to inhibit CCR5 tropic HIV-1 infection in isolated splenocytes ex vivo.
Methods
Lentiviral vector construction and production
The construction of lentiviral vector FG12 H1shRNACCR5 (1005) was previously described.21,37 Briefly, we generated a random shRNA library directed to human CCR5 mRNA sequences by adapting the EPRIL method, which were then expressed using a lentiviral vector with an H1 promoter. A total of 400 clones of a vesicular stomatitis virus-G pseudotyped lentiviral vector were individually produced and screened for efficient CCR5 knockdown using CEM.NKR-CCR5 cells in 96-well plates. The most effective clone (human CCR5-shRNA(1005)) sequence consisted of 5′ sense siRNA-GAGCAAGCTCAGUUUACACC-loop-UUGUCCGAC-antisense siRNA-GGUGUAAACUGAGCUUGCUC-UU3′. To express the mCherry fluorescent protein from a lentiviral vector, mCherry cDNA was extracted from pmCherry (Clontech) using the BamHI and EcoRI sites and cloned into the corresponding sites of the FG11F lentiviral vector.22 High titer (> 3 × 108 enhanced green fluorescent protein [EGFP]+ or mCherry+ units/mL) vesicular stomatitis virus-G pseudotyped lentiviral vectors were prepared by calcium phosphate plasmid DNA transfection in 293T cells as previously described.22 The concentrated vector stocks were titered on 293T cells based on EGFP or mCherry expression.
Human fetal thymus and CD34+ and CD34− cell isolation from a fetal liver
Human fetal liver and thymus were obtained without identification information under federal and state regulations from the University of California, Los Angeles (UCLA) CFAR Gene and Cellular Therapy Core Laboratory and UCLA OB-GYN. Human fetal liver was digested with 1 mg/mL hyaluronidase (Sigma-Aldrich), 1 mg/mL collagenase type IV (Sigma-Aldrich), and 2 units/mL DNase I (Roche Diagnostics) containing AIM-V medium (Invitrogen) for 2 hours. To avoid bacterial and fungal contamination, human fetal tissues and cells were cultured with 450 μg/mL Zosyn (Wyeth) and 2.5 μg/mL amphotericin B (Sigma-Aldrich) in appropriate medium. Fetal liver mononuclear cells were separated by a Ficoll-Hypaque (GE Healthcare) density gradient. CD34+ and CD34− cells were separated using the magnetic-activated cell sorting Direct CD34 Progenitor Cell Isolation Kit (Miltenyi Biotec) according to the manufacturer's instructions. The purity of CD34+ cells was more than 97% as evaluated by fluorescence-activated cell sorter (FACS).
Lentiviral vector transduction
CD34+ (0.5 × 106) and CD34− cells (4.5 × 106) from a single fetal liver were seeded into RetroNectin (Takara)-coated plates with 2% bovine serum albumin in Yssel's medium (Gemini). After a 1-hour incubation, cells were infected with either the FG12 H1shRNACCR5 (1005) or FG11FmCherry lentiviral vector at multiplicity of infection (MOI) of 0.2 to 3 for overnight.
Generation of hu-BLT mice
NOD.CB17-Prkdcscid/J and NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice were purchased from The Jackson Laboratory and maintained in the animal facilities at UCLA in accordance with protocols approved by the UCLA animal research committee. hu-BLT mice were prepared as previously described35 with modifications for the implantation of the vector-transduced thy/liv organoid under the kidney capsule. Six- to 8-week-old male mice were implanted with a portion of human fetal thymus and Matrigel-solidified lentivirally transduced CD34+ (0.5 × 106) and CD34− cells (4.5 × 106) within 5 μL of Matrigel (BD Biosciences) under the kidney capsule. Three weeks after implantation, CD34+/CD34−/thymus-implanted mice were irradiated (325 cGy by 60Co irradiation) and transplanted with lentivirus vector–transduced autologous human CD34+ cells (1 × 106) using a 26-gauge needle through the tail vein or retro-orbital vein.
Serial transplantation of bone marrow–derived cells in the hu-BLT mouse model
Recipient mice (in duplicate) were prepared in an identical fashion using nontransduced thy/liv tissue. BLT mouse donor bone marrow cells were isolated from femurs of a vector-transduced BLT donor at 14 weeks after CD34+ cell transplantation. The BLT donor bone marrow cell suspension (volume 575 μL) was both directly injected into the thy/liv implant (volume 25 μL) and intravenously (volume 50 μL) into the irradiated recipient mice at 18 weeks after thy/liv transplantation.
Cell isolation from peripheral blood, bone marrow, thy/liv, lymph node, spleen, lung, liver, and intestines
Single-cell suspensions were prepared from peripheral blood, bone marrow, thy/liv, lymph nodes, spleen, lung, liver, and intestines, as previously described with modifications.35,38 Mouse peripheral blood was stained with antibodies for 30 minutes, treated with red blood cell lysis (RBCL; 4.15 g of NH4Cl, 0.5 g of KHCO3, and 0.019 g of ethylenediaminetetraacetic acid in 500 mL of H2O) buffer for 10 minutes and washed with FACS buffer. Bone marrow, thy/liv implant, lymph nodes, and spleen mononuclear cells were finely minced into small fragments and resuspended in 5 mL of RPMI 1640. The supernatant was filtered through a 70-μm cell strainer. The pellet was washed in FACS buffer (2% fetal calf serum in phosphate-buffered saline [PBS]) and resuspended in RBCL buffer for 10 minutes. The cells were centrifuged at 1500g for 2 minutes and washed with FACS buffer. Lung lobes were minced into small pieces with scissors, treated with collagenase type IV, passed through a 16-gauge needle, and filtered through a 70-μm cell strainer. Cells were resuspended in RPMI 1640, laid over the 70% of Percoll/PBS, and centrifuged at 2000g for 20 minutes. The pellet was washed in FACS buffer. The liver was minced into small fragments and passed through a 16-gauge needle and filtered through a 70-μm cell strainer. Cells were resuspended in RPMI 1640, laid over the 70% of Percoll/PBS, and centrifuged at 2000g for 20 minutes. Cells were treated with RBCL buffer and filtered through a 70-μm cell strainer and washed with FACS buffer. The small intestine was flushed in ice-cold PBS, cut into small fragments, and inverted to expose the intraepithelial surface. These fragments were shaken gently for 30 minutes at 37°C in buffer (10mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid; Invitrogen), 5mM ethylenediaminetetraacetic acid (Nippon Gene), and 3% fetal calf serum in Hanks balanced salt solution (Mediatech Inc). The fragments were incubated with lamina propria lymphocyte (LPL) digestion medium (500 μg/mL collagenase type II, Sigma-Aldrich; and 10 units/mL DNase I in RPMI 1640) for 60 minutes at 37°C. The supernatants were collected through a 70-μm cell strainer. Isolated LPL cells were suspended in 40% of Percoll/Dulbecco modified Eagle medium. Cell suspension was laid over the 70% of Percoll/PBS and centrifuged at 2000g for 20 minutes. LPL cells were washed with FACS buffer.
Flow cytometry
All isolated tissue mononuclear cells were stained with monoclonal antibodies to human CCR5 conjugated with PE-Cy5 (2D7; BD Biosciences), CD45 conjugated with biotin or APC (HI30; eBioscience), CD4 conjugated with PE-Cy7 (SK3; BD Biosciences) or APC-Cy7 (RPA-T4; eBioscience), CD8 conjugated with APC (RPA-T8; BD Biosciences), PerCP or PE-Cy5 (HIT8a; BD Biosciences), CD45RA conjugated with PE-Cy7 (L48; BD Biosciences), CD27 conjugated with PE (M-T271; BD Biosciences), CD19 conjugated with PerCP (SJ25C1; BD Biosciences), CD33 conjugated with PE-Cy7 (P67.6; eBioscience), CD14 conjugated with PE (M5E2; BD Biosciences), and CD3 conjugated with PerCP (SK7; BD Biosciences) according to the manufacturer's instructions. The cells were then washed in FACS buffer twice, stained with Streptavidin AlexaFluor 350 (Invitrogen), washed again with FACS buffer, and fixed with 2% formaldehyde in PBS. EGFP, mCherry, and antibody tagged receptor expressions were examined on the FC500 (Beckman Coulter), LSRII, or FACS Vantage (BD Biosciences). The data were analyzed by FlowJo (TreeStar) software.
HIV-1 production and infection ex vivo
HIV-1NL4-339 and HIV-1NFNSX SL940,41 were produced by calcium phosphate transfection in 293T cells. The virus supernatants were filtered with 0.22-μm filters and stored at −80°C. Infectious titers were determined by infecting phytohemagglutinin (PHA)/interleukin-2–activated human peripheral blood mononuclear cells using serially diluted viruses. For the ex vivo infection, CD8-depleted PHA/interleukin-2–activated splenocytes were infected with HIV-1 at an MOI of 2.5 for 2 hours.
TCR repertoire analysis
Lentiviral vector–transduced EGFP and mCherry+ cells were flow-sorted using FACS vantage from the bulk mononuclear cells separated from thy/liv tissues. The total RNA was isolated from these sorted cells using Trizol-LS reagent (Invitrogen) per the manufacturer's recommendations, and cDNA was prepared using 20 μL of this total RNA by a high-capacity cDNA reverse transcription kit (Applied Biosystems). To define the T-cell receptor (TCR) repertoire diversity, TCR-Vβ spectratyping was performed as described earlier,42 using primers flanking to complementarity determining region-3. To explain briefly, we performed independent TCR-Vβ-specific amplifications for all 24 TCR-Vβ families using one Vβ specific forward primer along with the common fluorescent labeled (6-FAM, VIC, or NED) TCR-β constant gene-specific reverse primer. A fraction of these amplified products, with fluorescent dye on each fragment, were then resolved through the ABI-3130 capillary electrophoresis system, and the fragment length distribution was analyzed based on obtained fluorescent intensity and its electrophoretic mobility using Genemapper software (Applied Biosystems). The statistical significance of the Vβ-repertoire distribution was assessed between the EGFP and mCherry+ samples by Mann-Whitney testing using Graphpad Prism, Version 3 software.
Results
A novel method of generating a lentiviral vector–transduced hematopoietic progenitor/stem cell (HPSC)–transplanted hu-BLT mouse model
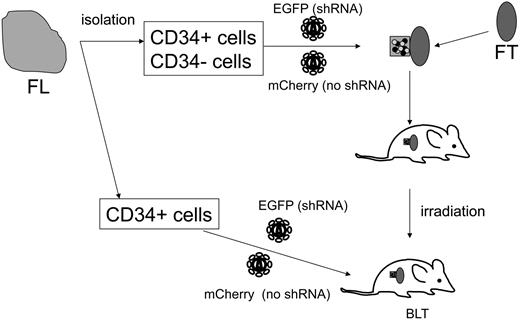
Systemic reconstitution of human hematopoietic cells in the hu-BLT mouse model requires a thy/liv tissue transplantation followed by total body irradiation and subsequent CD34+ cell transplantation.35,36 In a previous report, lentiviral vector transduction of CD34+ cells, followed by implantation into irradiated thy/liv-implanted hu-BLT mice, resulted in EGFP expression in human lymphocytes residing in lymphoid tissues, albeit at low efficiency (1%-4%).35 Given that efficient reconstitution of CCR5 shRNA-expressing human T lymphocytes is critical for analyzing CCR5 knockdown, we examined a novel method of generating a vector-transduced thy/liv implant (Figure 1). We solidified vector-transduced fetal liver-derived CD34+ cells and CD34− cells with Matrigel and transplanted it with a thymus segment under the kidney capsule. This method allows vector-transduced HPSCs to migrate into the adjacent thymus segment and produce a vector-transduced thy/liv organoid. We included CD34− cells as our pilot study determined that the transplantation of a thymus segment, along with CD34+ and CD34− cells, increased the efficiency of thymus engraftment 1.7-fold compared with the transplantation of a thymus segment, along with CD34+ cells (data not shown). The function of CD34− cells for thymus organoid development is not clear at this time. CD34− cells may function as stromal cells to support hematopoietic stem cell engraftment.
Diagram of generating a lentiviral vector–transduced HPSC-transplanted hu-BLT mouse. CD34+ and CD34− cells isolated from a human fetal liver were transduced by either shRNA (EGFP+) or no shRNA (mCherry+) vectors. The transduced cells were solidified with Matrigel and implanted under a kidney capsule with a piece of human fetal thymus. Three weeks after the implantation, the mouse was irradiated and intravenously injected with vector-transduced autologous CD34+ cells. FL indicates human fetal liver segment; and FT, human fetal thymus segment.
Diagram of generating a lentiviral vector–transduced HPSC-transplanted hu-BLT mouse. CD34+ and CD34− cells isolated from a human fetal liver were transduced by either shRNA (EGFP+) or no shRNA (mCherry+) vectors. The transduced cells were solidified with Matrigel and implanted under a kidney capsule with a piece of human fetal thymus. Three weeks after the implantation, the mouse was irradiated and intravenously injected with vector-transduced autologous CD34+ cells. FL indicates human fetal liver segment; and FT, human fetal thymus segment.
To effectively control shRNA-transduced human CD34+ cell engraftment and differentiation, we used 2 lentiviral vectors expressing different fluorescent protein markers. The CCR5 shRNA vector expressed EGFP, and the non-shRNA control vector expressed mCherry, a variant of red fluorescent protein. This 2-fluorescent reporter system allows us to simultaneously detect CCR5 shRNA-expressing cells (EGFP+) and nonexpressing cells (mCherry+) to determine whether CCR5 shRNA vector-transduced cells differ from control vector-transduced cells in regards to levels of stability and specificity of CCR5 reduction within the same animal. We transduced CD34+ and CD34− cells with each vector, mixed cells after transduction, and transplanted them under the kidney capsule with a thymus segment. Vector transduction efficiencies in the CD34+ and CD34− cells used for the thy/CD34+/− transplantations were mean = 50.8% (n = 10) and mean = 15.4% (n = 7), respectively, for EGFP vectors and mean = 42.9% (n = 10) and mean = 17.9% (n = 7), respectively, for mCherry vectors. Three weeks after thymus and vector-transduced CD34+/CD34− cell transplantation under the kidney capsule, we intravenously injected vector-transduced autologous CD34+ cells into a sublethally irradiated mouse for systemic hematopoietic cell reconstitution. Vector transduction efficiencies for the intravenously injected CD34+ cells were mean = 45.0% (n = 9) for EGFP vectors and mean = 28.3% (n = 9) for mCherry vectors.
Engraftment and differentiation of shRNA 1005-transduced HPSCs in systemic lymphoid organs of the hu-BLT mouse model
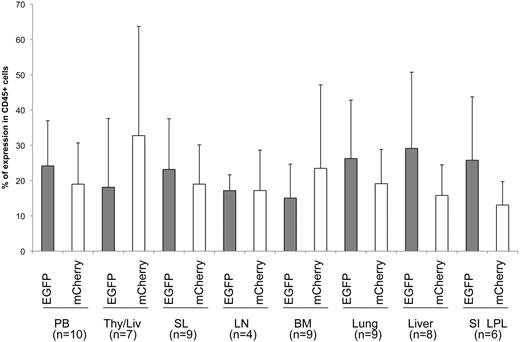
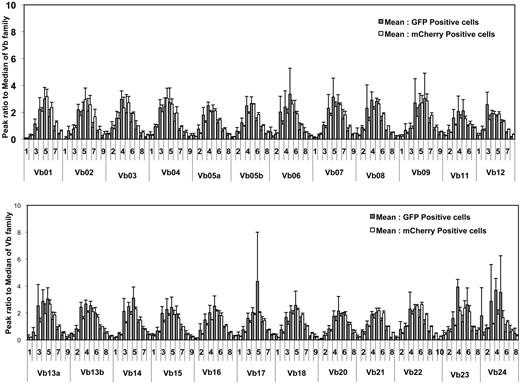
We examined human cell engraftment in transplanted mice between 13 and 20 weeks after CD34+ cell intravenous injection. Human leukocyte CD45+ cells were detected in a lymphocyte-gated population in multiple lymphoid organs from transplanted mice by flow cytometric analysis. Human CD45+ cells were under the detection limit in control nontransplanted mice (data not shown). Comparable levels of EGFP and mCherry expression were found in this human CD45+ population in multiple lymphoid organs in transplanted mice (Figure 2), suggesting that shRNA 1005 expression did not affect human HPSC differentiation and migration. To examine the effect of shRNA expression on TCR rearrangement, we examined TCR vβ rearrangement using a quantitative polymerase chain reaction (PCR)–based TCR spectratyping assay in FACS-purified EGFP+ and mCherry+ thymocytes from thy/liv organoids (n = 3; Figure 3). The profile and peak distribution of each TCRvβ showed normal Gaussian distribution in both EGFP+ and mCherry+ sorted populations. These data suggest that shRNA 1005 expression did not affect polyclonal human TCR development.
Human hematopoietic differentiation of shRNA 1005-transduced HPSCs in multiple lymphoid organs. EGFP and mCherry reporter gene expression was examined in human CD45+ cells in gated lymphocyte population in multiple tissues. Samples were analyzed between 14 and 20 weeks after intravenous CD34+ cell injection. Mean percentage EGFP and percentage mCherry expression are shown in each organ. No significant difference was found (P > .05) between percentage EGFP+ and percentage mCherry+ cells in various tissues by Student t test. Data were generated from n = 4 to 10 individual animals from an aggregate of 8 donors. Error bar represents SD. PB indicates peripheral blood; Thy/Liv, transplanted human thy/liv organoid; BM, bone marrow; SL, spleen; LN, lymph nodes; SI LPL, small intestine lamina propria lymphocytes; and n, number of samples.
Human hematopoietic differentiation of shRNA 1005-transduced HPSCs in multiple lymphoid organs. EGFP and mCherry reporter gene expression was examined in human CD45+ cells in gated lymphocyte population in multiple tissues. Samples were analyzed between 14 and 20 weeks after intravenous CD34+ cell injection. Mean percentage EGFP and percentage mCherry expression are shown in each organ. No significant difference was found (P > .05) between percentage EGFP+ and percentage mCherry+ cells in various tissues by Student t test. Data were generated from n = 4 to 10 individual animals from an aggregate of 8 donors. Error bar represents SD. PB indicates peripheral blood; Thy/Liv, transplanted human thy/liv organoid; BM, bone marrow; SL, spleen; LN, lymph nodes; SI LPL, small intestine lamina propria lymphocytes; and n, number of samples.
Polyclonal human TCRvβ development in thymocytes differentiated from shRNA 1005-transduced HPSCs. Total RNA isolated from FACS-purified EGFP+ or mCherry+ thymocytes taken from thy/liv organoids of 3 reconstituted BLT mice were subjected to a quantitative PCR-based TCR spectratyping analysis. Peak ratio was compared with the median of each TCR Vβ family between EGFP+ and mCherry+ thymocytes. There was no significant difference between the 2 groups (P = .668, by Mann-Whitney test). TCRs are made from splicing of V-D-J regions, and the complementarity determining region-3 region is therefore random in size. Thus, the PCR output bands corresponding to different sized splice products, and the intensity of these bands should be Gaussian in distribution if T-cell repertoires are random.
Polyclonal human TCRvβ development in thymocytes differentiated from shRNA 1005-transduced HPSCs. Total RNA isolated from FACS-purified EGFP+ or mCherry+ thymocytes taken from thy/liv organoids of 3 reconstituted BLT mice were subjected to a quantitative PCR-based TCR spectratyping analysis. Peak ratio was compared with the median of each TCR Vβ family between EGFP+ and mCherry+ thymocytes. There was no significant difference between the 2 groups (P = .668, by Mann-Whitney test). TCRs are made from splicing of V-D-J regions, and the complementarity determining region-3 region is therefore random in size. Thus, the PCR output bands corresponding to different sized splice products, and the intensity of these bands should be Gaussian in distribution if T-cell repertoires are random.
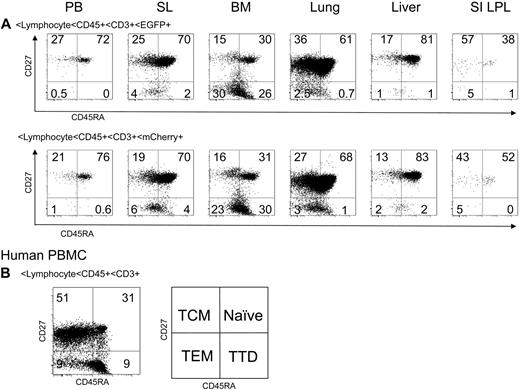
To examine the effect of shRNA 1005 expression on naive and memory T-cell differentiation, we examined CD27 and CD45RA expression on CD3+ T lymphocytes in multiple lymphoid organs. Profiles of CD27 and CD45RA expression were similar between EGFP+ and mCherry+ gated CD3+ T-cell population in spleen, lung, and liver (n = 3; Figure 4; supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article), suggesting shRNA 1005 expression did not affect naive and memory T-cell differentiation. Taken together, these results suggest that shRNA 1005-transduced human HPSCs could differentiate into T lymphocytes in primary and secondary lymphoid organs with no apparent shRNA-induced cytotoxicity.
Naive and memory T-cell differentiation of shRNA 1005-expressing T cells. (A) Naive and memory T-cell differentiation was analyzed by CD27 and CD45RA cell surface expression in gated EGFP+ and mCherry+ CD3+ T lymphocytes from multiple lymphoid organs. (B) A normal human peripheral blood mononuclear cell staining control is shown as a control. TCM indicates central memory T cells; TEM, effector memory T cells; and TTD, terminally differentiated cells.
Naive and memory T-cell differentiation of shRNA 1005-expressing T cells. (A) Naive and memory T-cell differentiation was analyzed by CD27 and CD45RA cell surface expression in gated EGFP+ and mCherry+ CD3+ T lymphocytes from multiple lymphoid organs. (B) A normal human peripheral blood mononuclear cell staining control is shown as a control. TCM indicates central memory T cells; TEM, effector memory T cells; and TTD, terminally differentiated cells.
shRNA 1005 down-regulates CCR5 expression in systemic lymphoid organs
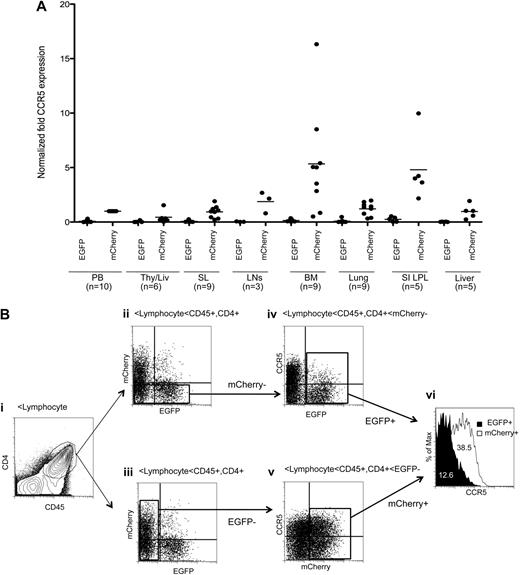
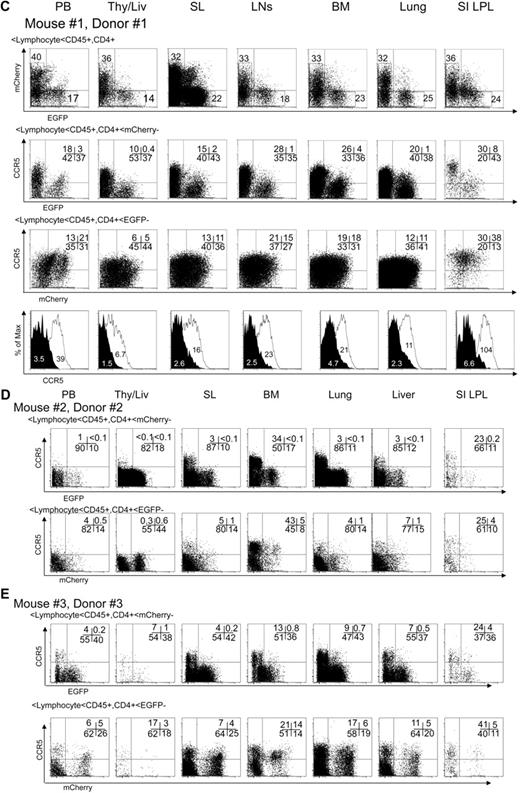
We examined CCR5 expression in human CD4+/CD45+ T lymphocytes in multiple lymphoid tissues in reconstituted mice at 14 to 20 weeks after intravenous CD34+ cell injection (Figure 5A). CCR5 expression was efficiently reduced in the EGFP+ population relative to the mCherry+ population in all tissues analyzed. Notably, CCR5 reduction was efficient, even in highly CCR5-expressing tissues, such as the bone marrow and gut. Basal CCR5 expression levels in each transplanted mouse varied as previously reported in the hu-BLT mouse model.38,43 Variation of CCR5 expression has been documented in humans.44 We have shown 3 representative datasets from 3 mice transplanted with different human HPSC donors (Figure 5B-D). CCR5 expression was efficiently down-regulated, even in the animal with the highest CCR5 basal expression level (Figure 5C).
Efficient CCR5 down-regulation in human CD4+ T lymphocytes in multiple lymphoid organs. (A) The level of CCR5 expression was compared in EGFP+ and mCherry+ human CD4+/CD45+ lymphocytes in lymphoid tissues from multiple transplanted mice. We normalized CCR5 expression level using the mean CCR5 expression in mCherry+ cells from peripheral blood (PB) as 1. Samples were obtained between 14 and 20 weeks after intravenous CD34+ cell injection. Bar represents mean value; n indicates number of samples. Aggregate difference, comparing CCR5 expression in EGFP+ versus mCherry+ cells in all tissues, was statistically significant by Student t test (P < .001). (B) A representative gating scheme. (i) The human CD4+/CD45+ population was identified in the gated lymphocyte population in spleen. (ii-iii) EGFP and mCherry expression was identified in the CD4+/CD45+ gated population. (iv) mCherry− population was further gated to analyze CCR5 expression in EGFP+ and EGFP− population. (v) EGFP− population was further gated to analyze CCR5 expression in mCherry+ and mCherry− population. (vi) The mean fluorescent intensity of CCR5 expression was compared in EGFP+ and mCherry+ population. (C-D) Representative data showing CCR5 down-regulation in multiple lymphoid tissues from a mouse with the highest CCR5 basal expression. (E) Additional dataset from mouse reconstituted with different donor. Data were analyzed as shown in panel B.
Efficient CCR5 down-regulation in human CD4+ T lymphocytes in multiple lymphoid organs. (A) The level of CCR5 expression was compared in EGFP+ and mCherry+ human CD4+/CD45+ lymphocytes in lymphoid tissues from multiple transplanted mice. We normalized CCR5 expression level using the mean CCR5 expression in mCherry+ cells from peripheral blood (PB) as 1. Samples were obtained between 14 and 20 weeks after intravenous CD34+ cell injection. Bar represents mean value; n indicates number of samples. Aggregate difference, comparing CCR5 expression in EGFP+ versus mCherry+ cells in all tissues, was statistically significant by Student t test (P < .001). (B) A representative gating scheme. (i) The human CD4+/CD45+ population was identified in the gated lymphocyte population in spleen. (ii-iii) EGFP and mCherry expression was identified in the CD4+/CD45+ gated population. (iv) mCherry− population was further gated to analyze CCR5 expression in EGFP+ and EGFP− population. (v) EGFP− population was further gated to analyze CCR5 expression in mCherry+ and mCherry− population. (vi) The mean fluorescent intensity of CCR5 expression was compared in EGFP+ and mCherry+ population. (C-D) Representative data showing CCR5 down-regulation in multiple lymphoid tissues from a mouse with the highest CCR5 basal expression. (E) Additional dataset from mouse reconstituted with different donor. Data were analyzed as shown in panel B.
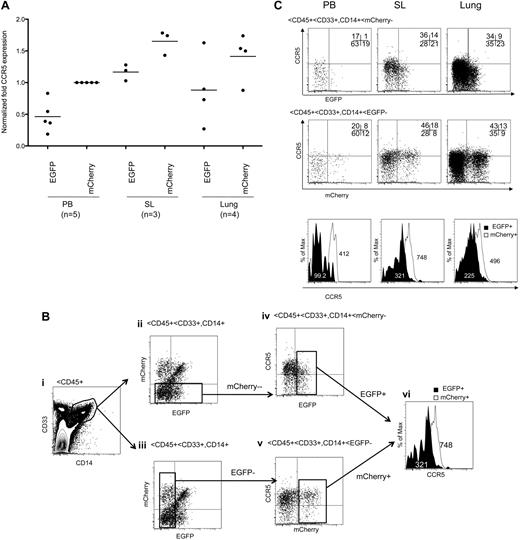
We next examined CCR5 down-regulation in the monocyte/macrophage populations, which are another major target of HIV-1 infection. CCR5 expression was reduced in the EGFP+ CD14+/CD33+ monocyte/macrophage population relative to the EGFP− population in peripheral blood, spleen, and lung (Figure 6A-B). In conclusion, shRNA 1005 successfully down-regulated CCR5 expression in CD4+ T lymphocytes and monocyte/macrophage populations in systemic lymphoid organs in vivo.
CCR5 down-regulation in EGFP-expressing human monocyte/macrophage population in multiple lymphoid organs. (A) Normalized mean CCR5 expression was compared in EGFP+ and mCherry+ human CD14+/CD33+ monocyte/macrophage population in multiple tissues. Bar represents mean value. Samples were analyzed between 14 and 20 weeks after intravenous CD34+ cell injection. Aggregate difference, comparing CCR5 expression in EGFP+ versus mCherry+ cells in all tissues, was statistically significant by Student t test (P < .001). (B) Gating strategies are the same as shown in Figure 5B except for the use of CD14 and CD33 markers. (C) Representative data showing CCR5 expression in CD14+/CD33+ monocyte/macrophage population.
CCR5 down-regulation in EGFP-expressing human monocyte/macrophage population in multiple lymphoid organs. (A) Normalized mean CCR5 expression was compared in EGFP+ and mCherry+ human CD14+/CD33+ monocyte/macrophage population in multiple tissues. Bar represents mean value. Samples were analyzed between 14 and 20 weeks after intravenous CD34+ cell injection. Aggregate difference, comparing CCR5 expression in EGFP+ versus mCherry+ cells in all tissues, was statistically significant by Student t test (P < .001). (B) Gating strategies are the same as shown in Figure 5B except for the use of CD14 and CD33 markers. (C) Representative data showing CCR5 expression in CD14+/CD33+ monocyte/macrophage population.
CCR5 was effectively down-regulated in CD4+ T cells in secondary transplanted animals
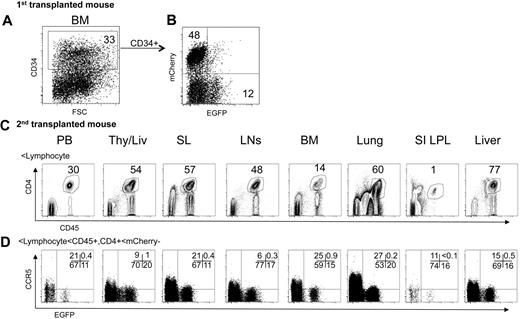
We performed a serial transplantation experiment to examine the long-term hematopoietic repopulation of shRNA 1005-transduced HPSCs and their subsequent CCR5 down-regulation. Bone marrow cells were isolated from the femurs of an EGFP and mCherry-expressing BLT mouse at 14 weeks after CD34+ cell transplantation. A total of 33% of the cells were CD34+ in isolated bone marrow cells (Figure 7A). Within the CD34+ cells, 12% were EGFP+ and 48% were mCherry+ (Figure 7B). The BM cells were directly injected into the thy/liv organoid and intravenously in irradiated recipient mice (n = 2). Human hematopoietic cell reconstitution was examined at 14 weeks after bone marrow cell transplantation. We detected EGFP+ human CD4+/CD45+ T cells in multiple tissues of the secondary transplanted mice (Figure 7C). mCherry+ cells were less than 1% positive; thus, we did not perform further analysis. CCR5 expression was efficiently down-regulated in EGFP+ cells relative to EGFP− cells in the CD45+/CD4+/mCherry− gated population (Figure 7D). These results demonstrated that lentiviral vector transduction of HPSCs and shRNA 1005 expression are capable of supporting long-term repopulation of the hematopoietic system and stable CCR5 knockdown in the BLT mouse model.
CCR5 down-regulation in second transplanted mice. The bone marrow cells from an EGFP- and mCherry-expressing transplanted donor mouse were isolated and analyzed for CD34 (A) and EGFP and mCherry expression (B). (C-D) Bone marrow cells were directly injected into a thy/liv tissue and intravenously in irradiated recipient mice (n = 2). (C) Human CD4+/CD45+ population was identified in gated lymphocyte population in multiple tissues 14 weeks after bone marrow cell injection. (D) CCR5 expression in EGFP+ and EGFP− population was examined in the gated mCherry−/CD4+/CD45+ population. Data from a representative mouse are shown.
CCR5 down-regulation in second transplanted mice. The bone marrow cells from an EGFP- and mCherry-expressing transplanted donor mouse were isolated and analyzed for CD34 (A) and EGFP and mCherry expression (B). (C-D) Bone marrow cells were directly injected into a thy/liv tissue and intravenously in irradiated recipient mice (n = 2). (C) Human CD4+/CD45+ population was identified in gated lymphocyte population in multiple tissues 14 weeks after bone marrow cell injection. (D) CCR5 expression in EGFP+ and EGFP− population was examined in the gated mCherry−/CD4+/CD45+ population. Data from a representative mouse are shown.
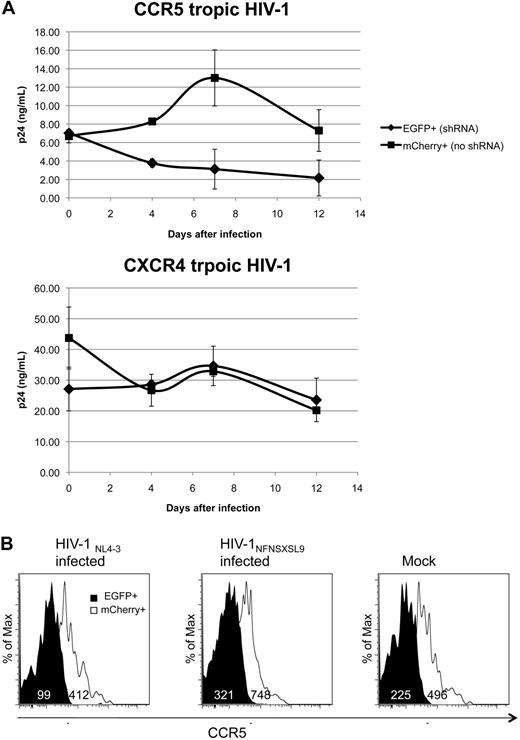
CCR5 tropic HIV is inhibited in ex vivo isolated CCR5 down-regulated splenocytes
To examine HIV susceptibility in CCR5 down-regulated cells, we FACS isolated EGFP+ and mCherry+ splenocytes from an animal. We depleted CD8+ cells from splenocytes activated ex vivo with PHA and interleukin-2. The cells (4 × 104) were infected with either CCR5 tropic HIV-1NFNSX SL9 or CXCR4 tropic HIV-1NL4-3 at an MOI of 2.5 in triplicate. There was no increase in p24 HIV gag capsid protein production in the culture supernatant of EGFP+ splenocytes over the 12-day culture period (Figure 8A). In contrast, mCherry+ splenocytes were susceptible to CCR5 tropic HIV-1NFNSX SL9 and produced approximately 4-fold higher levels of p24 in the culture supernatant compared with the EGFP+ splenocyte supernatants on day 7 and day 12, indicating that CCR5 tropic HIV-1 infection was effectively inhibited in CCR5 down-regulated cells. In contrast to the CCR5 HIV-1 infection, CXCR4 tropic HIV-1NL4-3 infection produced comparable amounts of p24 in both EGFP+ and mCherry+ splenocyte culture supernatants, confirming the specificity of the inhibition. CCR5 down-regulation was maintained in EGFP+ cells (Figure 8B). These results demonstrated that down-regulation of CCR5 by shRNA 1005 was sufficient to inhibit CCR5 tropic HIV-1 infection in ex vivo– stimulated cells.
CCR5 tropic HIV-1 inhibition ex vivo. (A) Splenocytes were isolated from a transplanted mouse at 20 weeks after CD34+ cell injection. Cells were activated with PHA for 2 days and interleukin-2 for 5 days. CD8+ cells were depleted and sorted for EGFP+ and mCherry+ cells at 99.6% purities. Sorted cells (4 × 104) were infected with CCR5 tropic HIV-1NFNSX SL9 or CXCR4 tropic HIV-1NL4-3 for 2 hours at MOI of 2.5 in parallel and in triplicate. Cells were washed 3 times after the infection. The amount of remaining input HIV-1 particles in culture supernatant was monitored 1 hour after infection by HIV p24 enzyme-linked immunosorbent assay. The amount of HIV production in culture supernatant was monitored by HIV p24 enzyme-linked immunosorbent assay at days 4, 7, and 12 after infection during the culture. The average p24 production in culture supernatant. Error bar represents SD. shRNA significantly affected HIV growth curve of HIV-1NFNSX SL9 but not HIV-1NL4-3 (P < .001 and P = .38, respectively, 2-way analysis of variance). (B) CCR5 expression in EGFP+ and mCherry+ cells at 12 days after HIV-1 infection.
CCR5 tropic HIV-1 inhibition ex vivo. (A) Splenocytes were isolated from a transplanted mouse at 20 weeks after CD34+ cell injection. Cells were activated with PHA for 2 days and interleukin-2 for 5 days. CD8+ cells were depleted and sorted for EGFP+ and mCherry+ cells at 99.6% purities. Sorted cells (4 × 104) were infected with CCR5 tropic HIV-1NFNSX SL9 or CXCR4 tropic HIV-1NL4-3 for 2 hours at MOI of 2.5 in parallel and in triplicate. Cells were washed 3 times after the infection. The amount of remaining input HIV-1 particles in culture supernatant was monitored 1 hour after infection by HIV p24 enzyme-linked immunosorbent assay. The amount of HIV production in culture supernatant was monitored by HIV p24 enzyme-linked immunosorbent assay at days 4, 7, and 12 after infection during the culture. The average p24 production in culture supernatant. Error bar represents SD. shRNA significantly affected HIV growth curve of HIV-1NFNSX SL9 but not HIV-1NL4-3 (P < .001 and P = .38, respectively, 2-way analysis of variance). (B) CCR5 expression in EGFP+ and mCherry+ cells at 12 days after HIV-1 infection.
Discussion
We examined our library-selected highly efficient shRNA 1005 for human CCR5 down-regulation in the hu-BLT mouse model. We chose the hu-BLT mouse model because this model allows us to examine the differentiation of shRNA 1005-transduced HPSCs as well as CCR5 down-regulation in systemic lymphoid organs. shRNA 1005-transduced human HPSCs differentiate into thymocytes in the transplanted human thy/liv and migrate and differentiate into naive and memory T lymphocytes in systemic lymphoid tissues in the BLT mouse model. It provides distinct advantages over other humanized mouse models where human T cells develop in the mouse thymus, raising concerns that the mouse thymic stroma may alter the TCR rearrangement. Our results showed that shRNA 1005-transduced HPSCs differentiated into T cells with a polyclonal TCR Vβ family repertoire, suggesting that TCR rearrangements were not affected by the shRNA 1005 expression. We observed stable down-regulation of human CCR5 expression in CD4+ T lymphocytes and monocytes/macrophages in primary and secondary lymphoid organs, including mucosal tissues, such as lung and gut-associated mucosal lymphoid tissues. Effective CCR5 down-regulation in gut-associated T cells is of particular importance for our study because these cells express a higher level of CCR5 than peripheral blood T cells and are the primary target of CCR5 tropic HIV-1 in patients.45 Bone marrow also contained relatively high levels of CCR5 expressing T lymphocytes in our study. The relatively high level of CCR5 expression in bone marrow was previously observed in the BLT mouse model.43 In addition, previous reports showed a high proportion of human bone marrow T cells displaying memory phenotypes, suggesting bone marrow as a preferential homing site for memory T cells in humans.46 These highly CCR5-expressing bone marrow CD4+ T cells could be also a primary target of CCR5 tropic HIV infection. Our data demonstrate efficient CCR5 down-regulation in CD4+ T cells in bone marrow. Our strategy may possibly protect these highly CCR5-expressing cells from HIV infection.
Our current study demonstrates that CCR5 down-regulation by shRNA 1005 effectively inhibited CCR5 tropic HIV-1 infection in isolated splenocytes ex vivo. Our next goal is to inhibit HIV infection in systemic lymphoid tissues in vivo. Unlike conventional drugs, gene therapy strategies have the potential to stably control HIV infection with a single treatment. For the treatment of chronic diseases, such as HIV-1, therapeutic genes must persist for years without causing adverse effects. The hu-BLT mouse model is an appropriate animal model system to investigate the effects of therapeutic gene expression, anti-HIV efficacy, and safety before human clinical trials. Our current results and future HIV challenge experiments could provide further evidence that it may be possible to create a single administration reagent, using a lentiviral vector–expressing CCR5 shRNA through hematopoietic stem cell transduction and transplantation, to stably control HIV-1 infection in patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Victor Garcia for valuable information and Jennifer Fulcher, Alvin Welch, Ana Beatriz Ruiz, Stephanie Matyas, Min Zhou, Patrick Kim, Ruth Cortado, Encarnacion Montecino-Rodriguez, Eun Mi Hur, Sonal Patel, Parvataneni Ram, and Broad Stem Cell Research Center flow core facility at UCLA for their reagents and technical support.
This work was supported by the Rheumatology Fellowship Training Grant T32 AR053463, UCLA AIDS Institute, UCLA Center for AIDS Research (CFAR), National Institute of Allergy and Infectious Diseases (AI028697), National Heart, Lung, and Blood Institute (1R01HL086409), and the National Cancer Institute (CA086306).
National Institutes of Health
Authorship
Contribution: S.S., B.L., I.S.Y.C., and D.S.A. designed the research; S.S., D.S.A., P.H., L.P., J.B., N.K., G.B., and P.K. performed the research and analyzed data; A.C. established a tissue procurement procedure; Z.G. and J.A.Z. developed CD34+/CD34−/thy transplantation experimental strategies; B.A. and O.Y. performed TCR spectratyping assay; and S.S., L.P., and D.S.A. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dong Sung An, UCLA David Geffen School of Medicine, Division of Hematology-Oncology, UCLA AIDS Institute, 188 BSRB, 615 Charles E. Young Dr South, Los Angeles, CA 90095; e-mail: an@ucla.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal