Abstract
To analyze prostaglandin E2 (PGE2) signaling in lymphoid cells, we introduce a multipronged strategy, combining temporal quantitative phosphoproteomics and phospho flow cytometry. We describe the PGE2-induced phosphoproteome by simultaneous monitoring of approximately 250 regulated phospho-epitopes, which, according to kinase prediction algorithms, originate from a limited number of kinase networks. Assessing these signaling pathways by phospho flow cytometry provided higher temporal resolution at various PGE2 concentrations in multiple lymphoid cell subsets. This showed elevated levels of protein kinase A (PKA) signaling in unstimulated CD8+CD45RO+ T cells, which correlated with suppressed proximal T-cell receptor signaling, indicating that PKA sets the threshold for activation. The combination of phosphoproteomics and high throughput phospho flow cytometry applied here provides a comprehensive generic framework for the analysis of signaling networks in mixed cell populations.
Introduction
Prostaglandin E2 (PGE2) plays a crucial role in the normal physiologic control of immune homeostasis, as well as in inflammation and cancer immune evasion, and pathophysiologic conditions1 such as acute inflammation,2 pain sensation,3 fever,4 and arthritis.5,6 PGE2 is known to signal through 4 G protein–coupled transmembrane receptors, EP1 to EP4.7 Although EP2 and EP4 are coupled to the G protein Gαs, leading to cyclic adenosine monophosphate (cAMP) production, EP1 is coupled to Gαq, resulting in inositol 1,4,5-trisphosphate generation and increased intracellular calcium [Ca2+]. In contrast, EP3 signals through Gαi that activates potassium channels and inhibits cAMP generation.8
In the immune system, PGE2 produced by innate immune cells such as monocytes and macrophages,9 as well as by regulatory T cells10 and endothelial cells, acts through EP1 to EP4 on lymphocytes to modulate immune functions. In T cells, PGE2 can inhibit T-cell receptor (TCR) signaling by the cAMP–protein kinase A (PKA)–Csk-Lck signaling cascade.11
In colorectal carcinoma, cyclooxygenase-2 (COX-2) expression has been shown to be 80% higher than in normal tissue.12 Administration of nonsteroidal anti-inflammatory drugs that inhibit COX-1 and COX-2 has been shown to be chemopreventive, reducing the relative risk of cancer, as well as reducing tumor size in patients with colorectal carcinoma.13 The antitumor effects of COX inhibitors result partly from their antiangiogenic effects.14,15 Furthermore, patients with colorectal carcinoma display elevated blood plasma levels of PGE2. COX-2 inhibition in these patients also leads to an increased antitumor immune response in vitro.16 This shows a role of PGE2 in inhibiting T-cell function and in cancer immune evasion that could be clinically important. It is therefore important to investigate the integrated signaling map, which results from the exposure of T cells to this inflammatory signal. Here, we aimed to generate a global understanding of phosphorylation-based signaling events in a variety of immune cells in response to PGE2.
In recent years significant progress has been made in the large-scale profiling of in vitro and in vivo protein phosphorylation with the use of a combination of specific phosphoprotein and phosphopeptide enrichment techniques (such as the here used online RP-TiO2-RP phosphopeptide enrichment) and high-resolution mass spectrometry, making it at present the method of choice to investigate temporally regulated phosphorylation-based signaling networks.17-20 In such an approach dynamic changes in protein phosphorylation may be globally investigated, unraveling thousands of phosphorylation sites on hundreds of different proteins. To assess dynamic regulation of these phosphorylation events, for instance on selective inhibition of kinases, stable isotope labeling is often implemented either by the use of amino acids in cell culture21 or by chemical peptide labeling of the proteolytic digests.22-25 In this study we use a highly efficient and cost-effective triple stable isotope-labeling approach at the peptide level18,22,26 to quantitatively assess temporal changes in phosphorylation events in lymphoid cells in response to PGE2 treatment.
Mass spectrometry–based phosphoproteomic analysis offers great depth about the number of phosphorylated sites that can be charted in an unbiased manner, allowing also the identification of many previously unidentified phosphopeptides. However, the technology is somewhat limited in the number of individual samples, which can be processed. On the basis of the outcome of our phosphoproteomics study and signaling networks derived from kinase prediction, we used a selected panel of phospho-specific antibodies in a high-throughput multisample, multiple time point phospho flow cytometric based profiling approach.27 This enabled us to assess PGE2-induced signaling in high-temporal resolution, at different concentrations of PGE2, with perturbation of signaling pathways, and in lymphocyte subpopulations by isolating peripheral blood mononuclear cells (PBMCs) and staining for 3 cell type–specific surface markers. Although studying signaling networks in isolated cell populations provides insight into stimulus-specific events under controlled in vitro conditions, phospho flow cytometry provides the opportunity to study signaling responses also in complicated cell mixtures, which allows intercellular secondary signaling events between different cell types. This approach provides experimental conditions, which are more physiologically relevant, but require that intercellular signaling is taken into account.
The combined phosphoproteomic mass spectrometry, phospho flow, and kinase prediction analyses yielded high-resolution information about the PGE2 signaling network. Furthermore, this combinational approach allowed the mapping of temporal signaling events in several cell types simultaneously, which is important because PGE2 signaling in healthy persons provides the basis to understand aberrant PGE2 signaling in diseased cells.
Methods
Cell isolation
Human peripheral blood was obtained from healthy volunteers (Ullevaal University Hospital Blood Center), and PBMCs were isolated with the use of gradient centrifugation (Lymphoprep; Axis-Shield PoC AS). For a detailed description, see supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Isolation of CD8+ T cells
CD8+ T cells were isolated with the use of a kit for the negative selection of CD8+ T cells (StemCell Technologies) followed by gradient centrifugation (Lymphoprep; Axis-Shield PoC AS).
Sample preparation for mass spectrometry
CD3+ lymphocytes, from a single donor, were stimulated with PGE2 (10μM) for 0, 10, or 60 minutes and then lysed and trypsin digested. For detailed description, see supplemental Methods.
Stable isotope labeling by amination of tryptic peptides
Mass spectrometry
For a detailed description of the mass spectrometer setup, and mass spectrometry data analysis, see supplemental Methods.
Bioinformatic handling of mass spectrometry data
The average ratio and SD of all quantified nonphosphorylated peptides was used to specify significant regulation. The average ratio of 3203 nonphosphorylated peptides between 0 and 10 minutes was calculated to be 1.022, with an SD of 0.23. For significant up-regulation or down-regulation everything outside the 95% confidence interval was considered to be up-regulated or down-regulated, that is, up-regulation for ratios greater than 1.48 and down-regulation for ratios less than 0.56. However, to increase stringency, up-regulation or down-regulation was defined as an increase in ratio of 2 or greater or decrease in ratio of 0.5 or less. All regulated phosphopeptides were searched against the PhosphoSitePlus database28 for previous identification and 3 common but independent kinase prediction algorithms (http://scansite.mit.edu29 ; www.networkin.info30 ; www.phosida.com31 ) to predict possible upstream kinases. For a full description of all algorithms used, see supplemental Methods.
For further analysis, only kinases that were predicted by at least 2 of the algorithms for the same phosphosite were considered. In addition, in vitro or in vivo determined upstream kinases of identified phosphopeptides were included in the kinase prediction dataset.
Sample preparation for phospho flow cytometry
PBMCs were stimulated at 37°C with PGE2 as indicated in the presence and absence of the PKA inhibitor H89 (20μM; 30 minutes preincubation at 37°C; Calbiochem), the cAMP antagonist Rp-8-Br-cAMPS (1mM; 30 minutes preincubation at 37°C; BioLog), or the cell permeable PKA inhibitor peptide myristoylated protein kinase inhibitor (PKI; 10μM; 45 minutes of preincubation at 37°C; Calbiochem).
Fluorescent cell barcoding and phospho flow staining
Three-dimensional fluorescent cell barcoding (FCB) was carried out as described elsewhere.27 For a full description, see supplemental Methods, supplemental Table 1, and supplemental Figure 5.
Flow cytometry
Flow cytometry was performed on a BD FACSCanto II (4-2-2) equipped with 405, 488, and 633 nm lasers. For analysis and visualization of flow cytometric results, the analysis software Cytobank was used (https://cytobank.stanford.edu).
Results
Investigation of the PGE2-stimulated phosphoproteome
Because of the coupling of the PGE2 receptors EP2 and EP4 to adenylyl cyclase, the activation of PKA was used as an expected primary marker for PGE2 signaling. To investigate PGE2 signaling kinetics, CD3+ T cells were stimulated with PGE2 (10μM) over a 60-minute time period, and cell lysates were immunoblotted with the use of an antiphospho PKA-substrate antibody (Figure 1A). Analysis of 4 independent experiments showed a rapid increase in PKA substrate phosphorylation after 1 minute, which continued to increase to maximal phosphorylation after 10 minutes. Phosphorylation reached near maximal levels at 5 minutes and remained high up to 15 minutes before most substrates returned to near basal after 60 minutes of PGE2 stimulation (Figure 1B). On the basis of these results we took time points of 0, 10, and 60 minutes for the subsequent temporal phosphoproteomics profiling experiments.
PGE2 signaling kinetics. (A) Immunoblot of CD3+ T lymphocytes stimulated with PGE2 (10μM) over a 60-minute time course with the use of a phospho-PKA substrate antibody (RRXpS/pT). (B) Relative density of 4 independent immunoblots plus SEM as depicted in panel A. (C) Mass spectrometry work flow. Three samples from 1 blood donor (CT, control; 10 and 60 minutes of PGE2 [10μM] stimulated) were lysed separately and in-solution trypsin digested. The resulting peptides were labeled with stable isotope labeling by reductive amination. Subsequently, the 3 samples of the 3 different time points were mixed at a ratio of 1:1:1. After SCX fractionation, phosphopeptides were subjected to online TiO2-LC enrichment, coupled to a LTQ Orbitrap mass spectrometer. Data analysis was conducted as described in the “Bioinformatic handling of mass spectrometry data.” LC-MS/MS indicates liquid chromatography tandem mass spectrometry.
PGE2 signaling kinetics. (A) Immunoblot of CD3+ T lymphocytes stimulated with PGE2 (10μM) over a 60-minute time course with the use of a phospho-PKA substrate antibody (RRXpS/pT). (B) Relative density of 4 independent immunoblots plus SEM as depicted in panel A. (C) Mass spectrometry work flow. Three samples from 1 blood donor (CT, control; 10 and 60 minutes of PGE2 [10μM] stimulated) were lysed separately and in-solution trypsin digested. The resulting peptides were labeled with stable isotope labeling by reductive amination. Subsequently, the 3 samples of the 3 different time points were mixed at a ratio of 1:1:1. After SCX fractionation, phosphopeptides were subjected to online TiO2-LC enrichment, coupled to a LTQ Orbitrap mass spectrometer. Data analysis was conducted as described in the “Bioinformatic handling of mass spectrometry data.” LC-MS/MS indicates liquid chromatography tandem mass spectrometry.
To investigate the temporal regulation of the PGE2-stimulated phosphoproteome, primary human CD3+ T cells were stimulated for 10 and 60 minutes with PGE2 (10μM) and compared with nonstimulated control cells (Figure 1C). To analyze peptides that originated from the 3 different stimulation time points simultaneously, we used stable isotope chemical labeling by peptide dimethylation.22 With the use of different isoforms of formaldehyde, peptides from each sample exhibited a minimum of 4 Da mass difference. Samples for the 3 different time points were labeled with heavy, intermediate, or light isoforms of formaldehyde and mixed at a total peptide ratio of 1:1:1. The resulting peptide mixture was then fractionated into 25 fractions with the use of the SCX chromatography to increase sensitivity in the subsequent online analysis of RP-TiO2-RP liquid chromatography tandem mass spectrometry.20 The eluted peptides were analyzed and identified by MS2 with the use of a LTQ-Orbitrap mass spectrometer (see supplemental Methods for detailed settings).
A total of 796 phosphopeptide isotopomer triplets (light, intermediate, and heavy) could be confidently identified, whereby each isotopomer peptide was individually sequenced to improve confidence of identification (Table 1; supplemental Table 4). Approximately one-third of these sites were found to be regulated in response to PGE2, whereby regulation was defined by an increase in ratio of 2 or greater or decrease in ratio of 0.5 or less at 10 and/or 60 minutes (see “Quantitation” in supplemental Methods). An overview of the temporal changes of these 247 unique, regulated phosphopeptides (from 207 unique phosphoproteins) is given in Table 1 and supplemental Table 2. A complete list of all identified phosphopeptides, their change in phosphorylation after PGE2 treatment, and their tandem mass spectrometry spectra can be viewed in supplemental data. To establish reproducibility of the detected changes in phosphorylation ratios, an additional experiment was conducted, and it showed high correlation in phosphorylation ratios with the original dataset, using linear regression analysis (supplemental Figure 3; supplemental Table 6). Furthermore, a total of 40 proteins were phosphorylated at multiple sites in response to PGE2. With the exception of only 4, all multiple phosphorylated proteins showed correlating changes in phosphorylation status at all detected phosphosites within each protein (see supplemental Table 7; supplemental data).
Summary of phosphoproteomic data
| Phosphoproteomic summary . | No. . |
|---|---|
| Identified phosphoproteins | 500 |
| Regulated phosphoproteins | 207 |
| Identified phosphopeptides | 796 |
| Regulated phosphopeptides | 247 |
| Kinase predicted phosphoproteins | 78 |
| Kinase predicted phosphopeptides | 103 |
| Kinase predicted phosphosites | |
| PKA | 41 |
| CAMKII | 18 |
| PKB/Akt | 14 |
| GSK3 | 15 |
| CK2 | 14 |
| CK1 | 11 |
| ERK | 9 |
| PKC | 9 |
| Cdk5 | 5 |
| PAK1 | 4 |
| MAPKAPK2 | 3 |
| p38 | 3 |
| PKG | 3 |
| AurB | 1 |
| Cdc2 | 1 |
| Clk2 | 1 |
| MKK3 | 1 |
| MKK6 | 1 |
| Mnk1 | 1 |
| PKD | 1 |
| PLK1 | 1 |
| Phosphoproteomic summary . | No. . |
|---|---|
| Identified phosphoproteins | 500 |
| Regulated phosphoproteins | 207 |
| Identified phosphopeptides | 796 |
| Regulated phosphopeptides | 247 |
| Kinase predicted phosphoproteins | 78 |
| Kinase predicted phosphopeptides | 103 |
| Kinase predicted phosphosites | |
| PKA | 41 |
| CAMKII | 18 |
| PKB/Akt | 14 |
| GSK3 | 15 |
| CK2 | 14 |
| CK1 | 11 |
| ERK | 9 |
| PKC | 9 |
| Cdk5 | 5 |
| PAK1 | 4 |
| MAPKAPK2 | 3 |
| p38 | 3 |
| PKG | 3 |
| AurB | 1 |
| Cdc2 | 1 |
| Clk2 | 1 |
| MKK3 | 1 |
| MKK6 | 1 |
| Mnk1 | 1 |
| PKD | 1 |
| PLK1 | 1 |
All regulated phosphosites were subjected to kinase prediction algorithms, and kinases that had been predicted for the same phosphosite by at least 2 kinase prediction algorithms were considered positive. Furthermore, kinases that have been shown to phosphorylate a specific phosphosite in vivo or in vitro were included in the predicted kinase dataset (Kinase prediction algorithms used http://scansite.mit.edu; www.networkin.info; www.phosida.com).
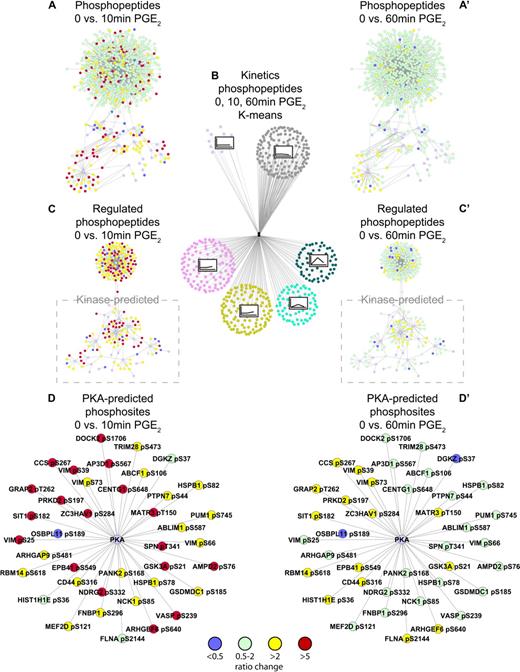
Analysis of the changes in phosphorylation of individual phosphopeptides in response to PGE2 treatment showed kinetic patterns, which were clustered according to their k-means (supplemental Figure 2). Phosphoproteins were also clustered, and a complete listing of kinetic clusters is presented in supplemental Table 5. Several phosphopeptides could be grouped into clusters, which increased significantly in phosphorylation after 10 minutes before decreasing to basal after 60 minutes (Figure 2A), as well as clusters, which gradually increased in phosphorylation over the 60-minute time period (Figure 2B). We color-coded the phosphopeptides according to their ratio change after 10 and 60 minutes of PGE2 treatment (Figure 3A,C,D and A',C',D', respectively). As can be observed in Figure 3A, C, and D, phosphorylation increases significantly after 10 minutes of PGE2 treatment with a large number of peptides exhibiting a ratio greater than 5. In comparison, most phosphopeptides exhibited little regulation after 60 minutes (Figure 3A and A' depicting all detected phosphopeptides and C and C' depicting regulated phosphopeptides only).
Identification and relative quantitation of detected phosphopeptides. (Top) Extracted ion chromatograms (XICs) of monoisotopic peaks were used to calculate peptide ratios (blue indicates control sample; purple, 10 minutes of PGE2 [10μM]; green, 60 minutes of PGE2 [10μM]). (Bottom) Tandem mass spectrometry spectra of detected phosphopeptides. (A) Phosphopeptide from AP-3 complex subunit δ-1 protein with phosphorylation at Ser567. (B) Phosphopeptide from Histone H1.4 protein with phosphorylation at Ser35. For complete mass spectrometry results, please refer to supplemental Table 4 and supplemental data.
Identification and relative quantitation of detected phosphopeptides. (Top) Extracted ion chromatograms (XICs) of monoisotopic peaks were used to calculate peptide ratios (blue indicates control sample; purple, 10 minutes of PGE2 [10μM]; green, 60 minutes of PGE2 [10μM]). (Bottom) Tandem mass spectrometry spectra of detected phosphopeptides. (A) Phosphopeptide from AP-3 complex subunit δ-1 protein with phosphorylation at Ser567. (B) Phosphopeptide from Histone H1.4 protein with phosphorylation at Ser35. For complete mass spectrometry results, please refer to supplemental Table 4 and supplemental data.
Quantitative phosphoproteomic mass spectrometry identifies the global PGE2-regulated phosphoproteome. K-means clustering analysis grouping phosphopeptides with similar phosphorylation profiles after 10 and 60 minutes of PGE2 (10μM) stimulation (B). All detected phosphopeptides were depicted and color coded according to the temporal change in phosphorylation ratio in response to PGE2 (10μM) after 10 and 60 minutes, A, C, D, and A', C', D', respectively; green indicates nonregulated phosphopeptide (ratio > 0.5 and < 2); yellow, ratio > 2 regulated phosphopeptide; red, ratio > 5 regulated phosphopeptide; blue, ratio < 0.5 regulated phosphopeptide; purple, predicted kinase. Individual networks were produced showing all identified phosphopeptides (A,A'), regulated phosphopeptides only (C,C') with kinase-predicted phosphopeptides highlighted, and PKA-predicted substrates (D,D'). All networks were visualized with the use of Cytoscape (www.cytoscape.org).
Quantitative phosphoproteomic mass spectrometry identifies the global PGE2-regulated phosphoproteome. K-means clustering analysis grouping phosphopeptides with similar phosphorylation profiles after 10 and 60 minutes of PGE2 (10μM) stimulation (B). All detected phosphopeptides were depicted and color coded according to the temporal change in phosphorylation ratio in response to PGE2 (10μM) after 10 and 60 minutes, A, C, D, and A', C', D', respectively; green indicates nonregulated phosphopeptide (ratio > 0.5 and < 2); yellow, ratio > 2 regulated phosphopeptide; red, ratio > 5 regulated phosphopeptide; blue, ratio < 0.5 regulated phosphopeptide; purple, predicted kinase. Individual networks were produced showing all identified phosphopeptides (A,A'), regulated phosphopeptides only (C,C') with kinase-predicted phosphopeptides highlighted, and PKA-predicted substrates (D,D'). All networks were visualized with the use of Cytoscape (www.cytoscape.org).
Predicted kinome
Aiming to deduce more detailed information about the PGE2-induced signaling network, all regulated phosphosites were subjected to 3 independent kinase prediction algorithms: NetworKIN,30 Scansite,29 and PHOSIDA.31 Although kinase prediction algorithm data at present cannot replace an experimental approach, it can be used as indicators for activated kinases in an experimental dataset.32 As we observed quite a few differences in the outcome between these 3 algorithms (supplemental Table 3), kinases were only considered as a possible upstream kinase of a specific phosphosite when at least 2 of the used algorithms predicted the same kinase. In addition, kinases that have been shown to phosphorylate a specific phosphosite in vitro or in vivo were included in the overview of the possible PGE2-induced kinome (supplemental Table 3). In summary, kinase prediction showed, as expected, an overrepresentation of PKA as a possible activated kinase with 41 phosphosites being predicted as possible PKA substrates (Table 2; Figure 3D and D'). Other predicted kinases include CAMKII with 18, PKB/Akt and glycogen synthase kinase 3 (GSK3) with 14, and CK2 with 14 substrates (Table 1). Importantly, according to the prediction algorithms, several of these kinases are able to phosphorylate an identical or similar motif, which complicates the identification of the kinase(s) responsible for the phosphorylation in question. For example, PKA and CAMKII are both able to phosphorylate the serine in an RXXS motif, whereas PKB/Akt is able to phosphorylate the serine in an RXRXXS motif.33 Although the PKA sites were uniquely predicted in 73% of the predictions, PKB/Akt and CAMKII were uniquely predicted only in 43% and 60% of the sites, respectively (Table 3).
Predicted PKA substrates
| IPI . | Protein . | Site1 . | Site2 . | Site3 . | 10 minutes . | 60 minutes . | ||
|---|---|---|---|---|---|---|---|---|
| Ratio . | STDEV . | Ratio . | STDEV . | |||||
| IPI00289608 | AP3D1, Isoform 2 of AP-3 complex subunit δ-1 | S567 | 16.87 | 4.08 | 1.84 | 0.58 | ||
| IPI00418471 | VIM, Vimentin | S39 | 12.63 | 3.75 | 2.06 | 0.61 | ||
| IPI00102670 | FNBP1, Isoform 2 of Formin-binding protein 1 | S296 | S301 | 11.79 | 1.47 | 1.13 | 0.04 | |
| IPI00009334 | PRKD2, Serine/threonine-protein kinase D2 | S197 | S198 | 10.43 | NaN | 2.37 | NaN | |
| IPI00021389 | CCS, Copper chaperone for superoxide dismutase | S267 | 9.63 | NaN | 2.68 | NaN | ||
| IPI00004407 | SIT1, Signaling threshold-regulating transmembrane adapter 1 | S182 | 9.21 | NaN | 2.24 | NaN | ||
| IPI00791144 | EPB41, EPB41 protein | S549 | 8.66 | NaN | 2.52 | NaN | ||
| IPI00008994 | NDRG2, Isoform 1 of protein NDRG2 | S332 | 8.38 | 0.90 | 1.95 | 0.24 | ||
| IPI00418471 | VIM, Vimentin | S25 | 8.31 | NaN | 1.83 | NaN | ||
| IPI00292228 | GSK3A, Glycogen synthase kinase-3 α | S21 | 7.95 | NaN | 2.08 | NaN | ||
| IPI00789551 | MATR3, Uncharacterized protein MATR3 | T150 | 7.47 | NaN | 4.00 | NaN | ||
| IPI00410067 | ZC3HAV1, Isoform 1 of zinc finger CCCH-type antiviral protein 1 | S284 | 7.41 | NaN | 3.91 | NaN | ||
| IPI00410067 | ZC3HAV1, Isoform 1 of zinc finger CCCH-type antiviral protein 1 | S284 | 7.30 | NaN | 3.31 | NaN | ||
| IPI00014256 | ARHGEF6, Isoform 1 of Rho guanine nucleotide exchange factor 6 | S640 | 7.06 | NaN | 2.35 | NaN | ||
| IPI00027430 | SPN, Leukosialin precursor | T341 | 6.93 | NaN | 1.94 | NaN | ||
| IPI00007722 | AMPD2, Adenosine monophosphate deaminase 2 | S76 | 6.10 | NaN | 1.91 | NaN | ||
| IPI00301058 | VASP, Vasodilator-stimulated phosphoprotein | S239 | 6.03 | 0.39 | 1.84 | 0.18 | ||
| IPI00022449 | DOCK2, Isoform 1 of dedicator of cytokinesis protein 2 | S1706 | 5.78 | NaN | 1.63 | NaN | ||
| IPI00217393 | CENTG1, Isoform 1 of centaurin-γ-1 | S648 | 5.72 | 0.42 | 1.81 | 0.10 | ||
| IPI00026928 | GRAP2, GRB2-related adapter protein 2 | T262 | 5.29 | 1.87 | 2.61 | 0.83 | ||
| IPI00418471 | VIM, Vimentin | S73 | 4.54 | 1.28 | 2.68 | 0.90 | ||
| IPI00442053 | ARHGAP9, Isoform 2 of Rho GTPase-activating protein 9 | S481 | 4.47 | 0.24 | 1.61 | 0.09 | ||
| IPI00013174 | RBM14, Isoform 1 of RNA-binding protein 14 | S618 | 4.26 | NaN | 2.66 | NaN | ||
| IPI00028065 | NCK1, Cytoplasmic protein NCK1 | S85 | 4.24 | NaN | 1.85 | NaN | ||
| IPI00329495 | ABLIM1, Isoform 1 of actin-binding LIM protein 1 | S587 | 4.05 | 1.28 | 1.57 | 0.43 | ||
| IPI00785100 | MEF2D, Myocyte enhancer factor 2D | S121 | 4.01 | NaN | 1.86 | NaN | ||
| IPI00032355 | PUM1, Pumilio homolog 1 | S745 | 3.68 | NaN | 1.61 | NaN | ||
| IPI00171176 | PANK2, Isoform 1 of pantothenate kinase 2, mitochondrial prec. | S168 | 3.26 | NaN | 1.36 | NaN | ||
| IPI00297160 | CD44, Isoform 12 of CD44 antigen precursor | S316 | 2.90 | NaN | 2.66 | NaN | ||
| IPI00028027 | GSDMDC1, Gasdermin domain-containing protein 1 | S185 | 2.88 | NaN | 1.52 | NaN | ||
| IPI00418471 | VIM, Vimentin | S66 | 2.84 | 0.21 | 1.15 | 0.10 | ||
| IPI00792186 | ABCF1, ATP-binding cassette, sub-family F (GCN20), member 1 | S106 | T109 | S110 | 2.73 | NaN | 1.20 | NaN |
| IPI00025512 | HSPB1, Heat shock protein β-1 | S78 | S82 | 2.58 | 0.21 | 1.20 | 0.03 | |
| IPI00438229 | TRIM28, Isoform 1 of transcription intermediary factor 1-β | S473 | 2.53 | 0.51 | 1.20 | 0.30 | ||
| IPI00102670 | FNBP1, Isoform 2 of formin-binding protein 1 | S296 | 2.42 | 0.20 | 0.77 | 0.13 | ||
| IPI00017338 | PTPN7, Isoform 1 of tyrosine-protein phosphatase non-receptor | S44 | 2.31 | NaN | 1.11 | NaN | ||
| IPI00025512 | HSPB1, Heat shock protein β-1 | S82 | 2.08 | 0.22 | 1.14 | 0.08 | ||
| IPI00302592 | FLNA, Filamin A, α isoform 1 | S2144 | 1.92 | NaN | 2.11 | NaN | ||
| IPI00217467 | HIST1H1E, Histone H1.4 | S36 | 1.90 | 0.05 | 3.10 | 0.02 | ||
| IPI00013835 | DGKZ, Isoform 1 of diacylglycerol kinase ζ | S37 | 0.86 | NaN | 0.40 | NaN | ||
| IPI00032970 | OSBPL11, Oxysterol-binding protein-related protein 11 | S189 | 0.09 | NaN | 0.02 | NaN | ||
| IPI . | Protein . | Site1 . | Site2 . | Site3 . | 10 minutes . | 60 minutes . | ||
|---|---|---|---|---|---|---|---|---|
| Ratio . | STDEV . | Ratio . | STDEV . | |||||
| IPI00289608 | AP3D1, Isoform 2 of AP-3 complex subunit δ-1 | S567 | 16.87 | 4.08 | 1.84 | 0.58 | ||
| IPI00418471 | VIM, Vimentin | S39 | 12.63 | 3.75 | 2.06 | 0.61 | ||
| IPI00102670 | FNBP1, Isoform 2 of Formin-binding protein 1 | S296 | S301 | 11.79 | 1.47 | 1.13 | 0.04 | |
| IPI00009334 | PRKD2, Serine/threonine-protein kinase D2 | S197 | S198 | 10.43 | NaN | 2.37 | NaN | |
| IPI00021389 | CCS, Copper chaperone for superoxide dismutase | S267 | 9.63 | NaN | 2.68 | NaN | ||
| IPI00004407 | SIT1, Signaling threshold-regulating transmembrane adapter 1 | S182 | 9.21 | NaN | 2.24 | NaN | ||
| IPI00791144 | EPB41, EPB41 protein | S549 | 8.66 | NaN | 2.52 | NaN | ||
| IPI00008994 | NDRG2, Isoform 1 of protein NDRG2 | S332 | 8.38 | 0.90 | 1.95 | 0.24 | ||
| IPI00418471 | VIM, Vimentin | S25 | 8.31 | NaN | 1.83 | NaN | ||
| IPI00292228 | GSK3A, Glycogen synthase kinase-3 α | S21 | 7.95 | NaN | 2.08 | NaN | ||
| IPI00789551 | MATR3, Uncharacterized protein MATR3 | T150 | 7.47 | NaN | 4.00 | NaN | ||
| IPI00410067 | ZC3HAV1, Isoform 1 of zinc finger CCCH-type antiviral protein 1 | S284 | 7.41 | NaN | 3.91 | NaN | ||
| IPI00410067 | ZC3HAV1, Isoform 1 of zinc finger CCCH-type antiviral protein 1 | S284 | 7.30 | NaN | 3.31 | NaN | ||
| IPI00014256 | ARHGEF6, Isoform 1 of Rho guanine nucleotide exchange factor 6 | S640 | 7.06 | NaN | 2.35 | NaN | ||
| IPI00027430 | SPN, Leukosialin precursor | T341 | 6.93 | NaN | 1.94 | NaN | ||
| IPI00007722 | AMPD2, Adenosine monophosphate deaminase 2 | S76 | 6.10 | NaN | 1.91 | NaN | ||
| IPI00301058 | VASP, Vasodilator-stimulated phosphoprotein | S239 | 6.03 | 0.39 | 1.84 | 0.18 | ||
| IPI00022449 | DOCK2, Isoform 1 of dedicator of cytokinesis protein 2 | S1706 | 5.78 | NaN | 1.63 | NaN | ||
| IPI00217393 | CENTG1, Isoform 1 of centaurin-γ-1 | S648 | 5.72 | 0.42 | 1.81 | 0.10 | ||
| IPI00026928 | GRAP2, GRB2-related adapter protein 2 | T262 | 5.29 | 1.87 | 2.61 | 0.83 | ||
| IPI00418471 | VIM, Vimentin | S73 | 4.54 | 1.28 | 2.68 | 0.90 | ||
| IPI00442053 | ARHGAP9, Isoform 2 of Rho GTPase-activating protein 9 | S481 | 4.47 | 0.24 | 1.61 | 0.09 | ||
| IPI00013174 | RBM14, Isoform 1 of RNA-binding protein 14 | S618 | 4.26 | NaN | 2.66 | NaN | ||
| IPI00028065 | NCK1, Cytoplasmic protein NCK1 | S85 | 4.24 | NaN | 1.85 | NaN | ||
| IPI00329495 | ABLIM1, Isoform 1 of actin-binding LIM protein 1 | S587 | 4.05 | 1.28 | 1.57 | 0.43 | ||
| IPI00785100 | MEF2D, Myocyte enhancer factor 2D | S121 | 4.01 | NaN | 1.86 | NaN | ||
| IPI00032355 | PUM1, Pumilio homolog 1 | S745 | 3.68 | NaN | 1.61 | NaN | ||
| IPI00171176 | PANK2, Isoform 1 of pantothenate kinase 2, mitochondrial prec. | S168 | 3.26 | NaN | 1.36 | NaN | ||
| IPI00297160 | CD44, Isoform 12 of CD44 antigen precursor | S316 | 2.90 | NaN | 2.66 | NaN | ||
| IPI00028027 | GSDMDC1, Gasdermin domain-containing protein 1 | S185 | 2.88 | NaN | 1.52 | NaN | ||
| IPI00418471 | VIM, Vimentin | S66 | 2.84 | 0.21 | 1.15 | 0.10 | ||
| IPI00792186 | ABCF1, ATP-binding cassette, sub-family F (GCN20), member 1 | S106 | T109 | S110 | 2.73 | NaN | 1.20 | NaN |
| IPI00025512 | HSPB1, Heat shock protein β-1 | S78 | S82 | 2.58 | 0.21 | 1.20 | 0.03 | |
| IPI00438229 | TRIM28, Isoform 1 of transcription intermediary factor 1-β | S473 | 2.53 | 0.51 | 1.20 | 0.30 | ||
| IPI00102670 | FNBP1, Isoform 2 of formin-binding protein 1 | S296 | 2.42 | 0.20 | 0.77 | 0.13 | ||
| IPI00017338 | PTPN7, Isoform 1 of tyrosine-protein phosphatase non-receptor | S44 | 2.31 | NaN | 1.11 | NaN | ||
| IPI00025512 | HSPB1, Heat shock protein β-1 | S82 | 2.08 | 0.22 | 1.14 | 0.08 | ||
| IPI00302592 | FLNA, Filamin A, α isoform 1 | S2144 | 1.92 | NaN | 2.11 | NaN | ||
| IPI00217467 | HIST1H1E, Histone H1.4 | S36 | 1.90 | 0.05 | 3.10 | 0.02 | ||
| IPI00013835 | DGKZ, Isoform 1 of diacylglycerol kinase ζ | S37 | 0.86 | NaN | 0.40 | NaN | ||
| IPI00032970 | OSBPL11, Oxysterol-binding protein-related protein 11 | S189 | 0.09 | NaN | 0.02 | NaN | ||
Phosphopeptides predicted as possible PKA substrates by 2 or more kinase prediction algorithms or by in vitro experimentation. Ratios of change in phosphorylation are shown for both 10- and 60-minute time points together with the SD when applicable.
Summary of peptides predicted for one or more of the kinases PKA, PKB/Akt, and CAMKII
| Kinase(s) . | No. of predicted peptides . | No. of in vitro substrates . | No. of total predictions . | Unique predictions for each kinase, % . |
|---|---|---|---|---|
| PKA | 30 | 3 | 41 | 73.2 |
| PKB/Akt | 6 | 2 | 14 | 42.9 |
| CAMKII | 9 | 0 | 15 | 60.0 |
| PKA and PKB/Akt | 3 | |||
| PKA and CAMKII | 3 | |||
| PKB/Akt and CAMKII | 1 | |||
| PKA and PKB/Akt and CAMKII | 2 |
| Kinase(s) . | No. of predicted peptides . | No. of in vitro substrates . | No. of total predictions . | Unique predictions for each kinase, % . |
|---|---|---|---|---|
| PKA | 30 | 3 | 41 | 73.2 |
| PKB/Akt | 6 | 2 | 14 | 42.9 |
| CAMKII | 9 | 0 | 15 | 60.0 |
| PKA and PKB/Akt | 3 | |||
| PKA and CAMKII | 3 | |||
| PKB/Akt and CAMKII | 1 | |||
| PKA and PKB/Akt and CAMKII | 2 |
The number of phosphopeptides predicted as substrates for 1 or more of the 3 kinases are listed to display the specificity of the prediction between the kinases. Unique prediction describes the fraction of the total number of predicted substrate peptides when only one specific kinase is implicated. For full details of all phosphoproteomic results and kinase prediction data, please refer to supplemental Table 3 and other supplemental data.
FCB combined with phospho flow analysis to study the PGE2-induced phosphoproteome
To multiplex the analysis of PGE2-induced phosphorylation-based signaling events under a variety of experimental conditions and to further validate the predicted activation of kinases by our phosphoproteomics dataset, we used phospho flow cytometry27 (Figures 4A and 5A). FCB combined with phospho flow allows the simultaneous analysis of a large number of samples with the capacity to assess selected phospho-epitope status at single cell resolution with the use of phospho-epitope–specific antibodies. In addition, signaling profiles of different cell populations in a complicated cell mixture can be assessed without extensive cell purification procedures.
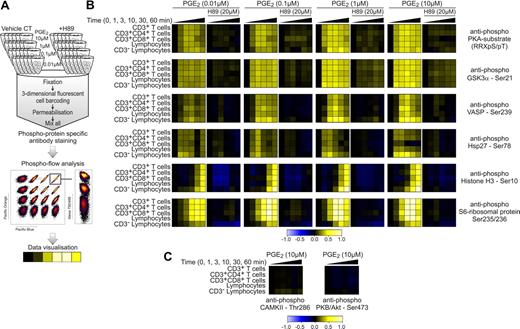
Phospho flow cytometry provides detailed insight into signaling events. (A) Phospho flow cytometry workflow overview. PBMCs were stimulated with PGE2 (0.01, 0.1, 1, and 10μM) over a 60-minute time period (0, 1, 3, 10, 30, and 60 minutes) in the absence and presence of the PKA inhibitor H89 (20μM). All samples were then fixed, subjected to fluorescent cell bar coding (FCB), and combined in 1 sample tube. Cells were then permeabilized and labeled with phospho-epitope–specific antibodies and cell type-specific surface marker antibodies before flow cytometric analysis and data analysis with the use of Cytobank (https://cytobank.stanford.edu). (B-C) Phospho flow cytometry results from 1 representative donor of 3 depicted as a heatmap. Warmer colors (yellow) indicate an increase in phospho-epitope–specific antibody signal, whereas colder colors (blue) indicate a decrease in signal. Differences in antibody signal were calculated with the first column of each lane as reference.
Phospho flow cytometry provides detailed insight into signaling events. (A) Phospho flow cytometry workflow overview. PBMCs were stimulated with PGE2 (0.01, 0.1, 1, and 10μM) over a 60-minute time period (0, 1, 3, 10, 30, and 60 minutes) in the absence and presence of the PKA inhibitor H89 (20μM). All samples were then fixed, subjected to fluorescent cell bar coding (FCB), and combined in 1 sample tube. Cells were then permeabilized and labeled with phospho-epitope–specific antibodies and cell type-specific surface marker antibodies before flow cytometric analysis and data analysis with the use of Cytobank (https://cytobank.stanford.edu). (B-C) Phospho flow cytometry results from 1 representative donor of 3 depicted as a heatmap. Warmer colors (yellow) indicate an increase in phospho-epitope–specific antibody signal, whereas colder colors (blue) indicate a decrease in signal. Differences in antibody signal were calculated with the first column of each lane as reference.
A combinational approach provides high-resolution signaling data. (A) Schematic overview of the combined phosphoproteomic and FCB-phospho flow methods that provide a high-resolution map of intracellular signaling and stimulus-specific signaling networks. (B) Summary depicting results obtained from kinase prediction, phosphoproteomics, and FCB-phospho flow (from left to right). Kinase prediction shows a probable connection between PKA and its substrates. All 3 depicted substrates (VASP, GSK3α, and heat shock protein 27 [Hsp27]) have been detected by mass spectrometry, and the relative changes in phospho-status at the specified sites have been determined. In addition, the detected phosphosites were studied with the use of FCB-phospho flow to show high-resolution kinetic information over extensive time courses in 5 lymphocyte cell populations, both in the absence and presence of a PKA inhibitor (H89). Color coding of histograms is according to a scale of 2 for VASP and GSK3α, and to a scale of 1 for Hsp27.
A combinational approach provides high-resolution signaling data. (A) Schematic overview of the combined phosphoproteomic and FCB-phospho flow methods that provide a high-resolution map of intracellular signaling and stimulus-specific signaling networks. (B) Summary depicting results obtained from kinase prediction, phosphoproteomics, and FCB-phospho flow (from left to right). Kinase prediction shows a probable connection between PKA and its substrates. All 3 depicted substrates (VASP, GSK3α, and heat shock protein 27 [Hsp27]) have been detected by mass spectrometry, and the relative changes in phospho-status at the specified sites have been determined. In addition, the detected phosphosites were studied with the use of FCB-phospho flow to show high-resolution kinetic information over extensive time courses in 5 lymphocyte cell populations, both in the absence and presence of a PKA inhibitor (H89). Color coding of histograms is according to a scale of 2 for VASP and GSK3α, and to a scale of 1 for Hsp27.
To characterize PGE2 kinetics in greater temporal detail, increasing concentrations of PGE2 (0.01, 0.1, 1, and 10μM PGE2) were investigated at time points of 0, 1, 3, 10, 30, and 60 minutes. Further, the effect of the PKA-inhibitor H89 (20μM) on PGE2-induced signaling events was evaluated (Figure 4B). By the use of cell surface markers, signaling events in the PBMC subpopulations CD3+ T cells, CD3+CD4+ T cells, CD3+CD8+ T cells, lymphocytes, and CD3− lymphocytes (natural killer cells and B cells) were investigated in parallel.
The antiphospho PKA-substrate antibody recognizes phosphorylated PKA substrates (RRXpS/pT). RRXpS/pT-phospho flow analysis was used to track PKA activation in response to PGE2 treatment, which indicated a gradual increase in the activation of PKA with a maximum at 10 minutes, decreasing again thereafter. Looking next at specific phosphosites, we observed that the kinetic profile of phospho-GSK3α Ser21 was similar to that of RRXpS/pT. In this context it is interesting to note that GSK3α Ser21 is a possible substrate for PKA,34 and phosphorylation at this site is known to inhibit kinase activity of GSK3α.35 Furthermore, phosphosites that were predicted substrates for GSK3α in our mass spectrometry dataset decreased in phosphorylation after PGE2 treatment, suggesting a direct link between PGE2, PKA, and GSK3α (supplemental Table 3). Phosphorylation of the actin regulatory protein vasodilator-stimulated phosphoprotein (VASP) has been shown to inhibit cytoskeletal rearrangement, affecting physiologically important processes such as T-cell polarization.36 Our data show that VASP Ser239 phosphorylation required higher concentrations of PGE2 than GSK3α Ser21, and VASP phosphorylation appears to be more transient. Heat shock protein 27 Ser78 phosphorylation, however, was observed to be maximal at 3 minutes after PGE2 stimulation (Figure 4B). The phosphosites investigated by these 3 antibodies were also detected in our phosphoproteomic mass spectrometry study and showed comparable profiles at 0, 10, and 60 minutes (Figure 5B). In addition, the phosphoproteomic dataset showed a steadily increasing phosphorylation of Histone H1.4 at Ser35 (Figure 2B). Because no phospho-epitope–specific antibody was available for this site, we decided to investigate Histone H3, which also has an N-terminal positioned phosphorylation motif for PKA (XRXS) at Ser10. With the use of the antiphospho Histone H3 antibody, comparable kinetic profiles were identified with a maximal phosphorylation after 60 minutes (Figure 4B). Histone phosphorylation is known to regulate gene transcription,37 establishing a link between PGE2 and transcriptional regulation. Phosphorylation of the S6-ribosomal protein at Ser235/236 showed a peak at 30 minutes before decreasing marginally at 60 minutes, indicating that PGE2 not only regulates transcription but also translation.38
When signaling events in distinct lymphocyte cell subsets were assessed in Figure 4B, differences in PGE2-induced signaling responses could be observed. In the case of VASP Ser239 phosphorylation, CD8+ T cells and CD3− lymphocytes (natural killer cells and B cells) showed a higher sensitivity and more prolonged response to PGE2 that with CD4+ T cells. Similarly, Histone H3 phosphorylation appeared earlier in CD3− lymphocytes. In contrast, S6-ribosomal protein phosphorylation in CD3− lymphocytes displayed a drastically delayed and less-sensitive kinetic profile in comparison to other lymphocyte cell subtypes (Figure 4B). However, at this point, the possibility remained that by investigating large subset classes of T cells such as CD4+ and CD8+ T cells, which can be further separated into functionally distinct cell types, clear differences in signaling profiles may remain hidden. This could occur because of opposing signaling responses in different cell populations, which potentially average out the detectable signal.
Although expected from the kinase prediction algorithms, no change in phosphorylation could be detected over a 60-minute time course with the use of a phospho-epitope–specific antibody against the autophosphorylation site of CAMKII (Thr286). Moreover, PKB/Akt phosphorylation at its activator site Ser473 did not increase with PGE2 (Figure 4C), indicating low or no activity change in response to PGE2 treatment in these 2 kinases. In support of this notion, measurements of Ca2+ levels in CD3+ lymphocytes showed little or no change in Ca2+ in response to 10 minutes of PGE2 (10μM) treatment (supplemental Figure 4A). This further suggests that CAMKII activity is low in response to PGE2 in CD3+ lymphocytes. Further, an in vitro kinase activity assay confirmed the algorithm-predicted activation of PKA in response to PGE2 treatment (supplemental Figure 4B), and the PKA inhibitor H89 consistently inhibited protein phosphorylation at all phospho-epitopes tested, irrespective of PGE2 concentration (Figure 4B). In addition, we investigated the mitogen-activated protein kinase signaling pathways and found no effect of PGE2 treatment, indicating that PGE2 does not significantly signal through the EP1 pathway in the absence of other stimuli, which is in line with our phosphoproteomics data. PGE2 alone did not directly affect the phosphorylation status of TCR signaling molecules investigated by phospho flow (supplemental Figure 6). However, our set of phosphoproteomic data identifies PGE2-induced phosphorylation changes at previously uncharacterized sites on a series of proteins in the TCR signaling cascade, confirming the inhibitory potential of PGE2 in respect to TCR signaling.39 Proteins that are involved in the regulation of enzyme activity as well as protein complex formation downstream of TCR (reviewed in Torgersen et al40 ) include CARMA1 (Ser441; 11-fold increase), phospholipase C γ (Ser1233; 10-fold increase), Wiskott-Aldrich syndrome protein–interacting protein (Ser340; 7-fold increase), VASP (Ser239; 6-fold increase), GADS (Thr262; 5-fold increase), nuclear factor of activated T cells cytoplasmic 2 (Ser236/243; 5-fold increase), FYN binding protein/adhesion and degranulation promoting adapter protein (Ser46; 4-fold increase), and Nck (Ser85; 4-fold increase) all of which constitute previously unnoticed sites of action in the PGE2 signaling network (supplemental Tables 2-4; supplemental data).
These data clearly show the advantages of the here described multipronged approach, whereby the regulation detected by phosphoproteomics in the whole assembly of cells can be used to subsequently monitor signaling events in distinct cell subsets in complicated cell mixtures, showing with high sensitivity the differences in signaling kinetics (Figure 5A-B).
Detailed signaling analysis of CD3+ T-cell subsets
Equipped with the data described earlier, we decided to investigate signaling profiles of functionally distinct CD3+ T-cell populations. We aimed to distinguish between the signaling profiles of CD4+ and CD8+ naive T cells (CD45RA) and CD4+ and CD8+ effector/memory T cells (CD45RO+). To allow a maximal number of channels available for perturbations of PGE2 signaling in PBMC cell mixtures, we identified CD4 and CD8 subsets of CD3+ T cells by the presence or absence of a CD4 marker and naive and effector/memory T cells by the presence or absence of a CD45RO marker. As can be seen in Figure 6A and B, CD8 effector/memory T cells (CD3+CD4−CD45RO+) show a low PKA-mediated signaling response after PGE2 treatment. Intriguingly, treatment with cAMP-elevating agents, such as the cAMP analog 8-CPT-cAMP, the broad-spectrum phosphodiesterase (PDE) inhibitor IBMX (3-isobutyl-1-methylxanthine), or the PDE4-specific inhibitor rolipram were not able to increase PKA-mediated signaling responses to levels observed in other investigated cell populations (Figure 6A-B). Nonetheless, both IBMX and 8-CPT-cAMP caused marginal, yet significant increases in PKA-mediated signaling, whereas PDE4-specific inhibition by rolipram showed no significant effect (Figure 6B). This indicates that, although described to play a crucial role in regulating cAMP levels in CD4+ T cells, PDE4 might not be involved in regulating cAMP levels in CD8 effector/memory T cells.41,42 We went on to investigate total signaling profiles in the described cell subsets and discovered that the basal levels of PKA signaling were significantly higher in unstimulated CD4+CD45RO+ and most evidently in CD4−CD45RO+ populations (Figure 6C-D). In contrast, the lowest level of basal PKA signaling could be seen in CD4 naive T cells when using the total CD3+ population as a reference (Figure 6C-D).
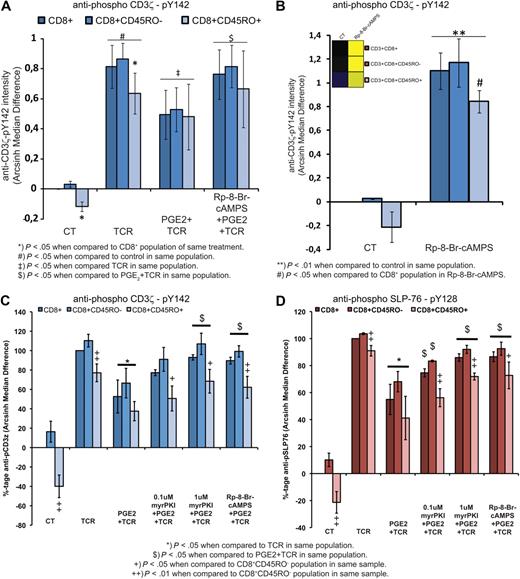
CD3+ T-cell subset analysis shows a highly active PKA signaling node in CD8+CD45RO+ T cells. (A) Heatmap depicting differences in Arcsinh median of the antiphospho PKA substrate (RRXpS/pT) antibody in CD3+ T-cell subsets after 10 minutes of PGE2 (10μM) alone or with IBMX (3-isobutyl-1-methylxanthine) or rolipram (5mM and 20μM, respectively; 30 minutes) preincubation, or 8-CPT-cAMP (300μM; 5 minutes). Heatmap depicts net signaling profiles from 1 representative blood donor with the first column in each row set as reference. (B) Statistical analysis of PKA signaling in selected T-cell subsets from 3 individual blood donors analyzed independently as in panel A (mean ± SD; n = 3). (C) Heatmap depicting total signaling dynamics of the same samples as in panel A with the control sample of the CD3+ T-cell population set as reference. (D) Statistical analysis from 3 individual blood donors analyzed independently, using the difference in basal PKA signaling of C (mean ± SD; n = 3). (E) TCR (anti-CD3 [OKT3] 1 μg/mL; anti-CD28 [CD28.2] 5 μg/mL) stimulation for the indicated time period with and without PGE2 (10μM; 10 minutes preincubation) measured by Arcsinh median differences in CD3ζ-chain phosphorylation at Tyr142. Heatmap depicts the relative level of signaling with the control sample of the CD3+ T-cell population set as reference from 1 representative blood donor. (F) Statistical analysis from 5 individual blood donors analyzed independently (mean ± SD; n = 5) of samples as in panel E. To account for large differences in TCR responses between different donors, data are normalized to percentages with TCR stimulation (1 minute) representing 100%. (G-H) Heatmaps depicting total signaling dynamics for the indicated phospho-epitope–specific antibodies in CD8+ T cells from 1 representative blood donor with the control sample of the CD8+ T-cell population as reference. One minute of TCR (anti-CD3 [OKT3] 1 μg/mL; anti-CD28 [CD28.2] 5 μg/mL) stimulation and PGE2 (10μM) preincubation for 10 minutes before TCR stimulation. In all heatmaps, warmer colors (yellow) indicate increases and cooler colors (blue) indicate decreases in Arcsinh median differences displayed on a scale of 1.
CD3+ T-cell subset analysis shows a highly active PKA signaling node in CD8+CD45RO+ T cells. (A) Heatmap depicting differences in Arcsinh median of the antiphospho PKA substrate (RRXpS/pT) antibody in CD3+ T-cell subsets after 10 minutes of PGE2 (10μM) alone or with IBMX (3-isobutyl-1-methylxanthine) or rolipram (5mM and 20μM, respectively; 30 minutes) preincubation, or 8-CPT-cAMP (300μM; 5 minutes). Heatmap depicts net signaling profiles from 1 representative blood donor with the first column in each row set as reference. (B) Statistical analysis of PKA signaling in selected T-cell subsets from 3 individual blood donors analyzed independently as in panel A (mean ± SD; n = 3). (C) Heatmap depicting total signaling dynamics of the same samples as in panel A with the control sample of the CD3+ T-cell population set as reference. (D) Statistical analysis from 3 individual blood donors analyzed independently, using the difference in basal PKA signaling of C (mean ± SD; n = 3). (E) TCR (anti-CD3 [OKT3] 1 μg/mL; anti-CD28 [CD28.2] 5 μg/mL) stimulation for the indicated time period with and without PGE2 (10μM; 10 minutes preincubation) measured by Arcsinh median differences in CD3ζ-chain phosphorylation at Tyr142. Heatmap depicts the relative level of signaling with the control sample of the CD3+ T-cell population set as reference from 1 representative blood donor. (F) Statistical analysis from 5 individual blood donors analyzed independently (mean ± SD; n = 5) of samples as in panel E. To account for large differences in TCR responses between different donors, data are normalized to percentages with TCR stimulation (1 minute) representing 100%. (G-H) Heatmaps depicting total signaling dynamics for the indicated phospho-epitope–specific antibodies in CD8+ T cells from 1 representative blood donor with the control sample of the CD8+ T-cell population as reference. One minute of TCR (anti-CD3 [OKT3] 1 μg/mL; anti-CD28 [CD28.2] 5 μg/mL) stimulation and PGE2 (10μM) preincubation for 10 minutes before TCR stimulation. In all heatmaps, warmer colors (yellow) indicate increases and cooler colors (blue) indicate decreases in Arcsinh median differences displayed on a scale of 1.
Interestingly, increased PKA-mediated signaling in effector/memory cells correlated with reduced proximal TCR signaling at the level of phosphorylation at the CD3ζ chain Tyr142 (Figure 6E-F). On TCR stimulation, CD3ζ phosphorylation after 1 minute reflects the preactivation environment, because TCR activation is blunted in populations with high basal PKA signaling (Figure 6F). Regardless of PKA signaling in the unstimulated state, when cells were preincubated with PGE2 before TCR activation, CD3ζ phosphorylation could be significantly inhibited (Figure 6F). These observations confirm previous publications that describe an inhibitory role of PKA in TCR signaling.11 For an overview of the basal phosphorylation status of all investigated epitopes, please refer to supplemental Figure 7.
To ensure that the obtained results from CD3+CD4− cells hold true for CD3+CD8+ cells, we repeated the above experiments on directly isolated CD3+CD8+ T cells (Figure 6G). Here, we also identified high basal phosphorylation of GSK3α at Ser21 and Histone H3 at Ser10 (Figure 6H). Both epitopes were identified as being up-regulated in our global PGE2-induced phospho signaling analysis and displayed sensitivity to the PKA inhibitor H89 (Figure 4B). Taken together, these results indicate that the high basal phosphorylation of GSK3α and Histone H3 is a direct consequence of high basal PKA activity observed in CD8+CD45RO+ T cells.
To examine in more detail the importance of PKA as a proximal gatekeeper for TCR activation, we investigated CD3ζ phosphorylation in response to TCR triggering. Here, we show that CD3ζ phosphorylation at Tyr142 in response to TCR stimulation can be significantly inhibited by PGE2 in all CD8+ T-cell populations, which in turn could be reversed to near maximal CD3ζ phosphorylation levels by inhibiting PKA type I activity (Figure 7A). Furthermore, incubation of unstimulated CD8 T cells with Rp-8-Br-cAMPS alone significantly increased the basal phosphorylation of CD3ζ in both CD8+ T-cell populations (Figure 7B). Importantly, the elevated levels of PKA signaling in CD8+CD45RO+ cells leading to CD3ζ hypophosphorylation could be reversed by the PKA type I antagonist (Figure 7B). The results obtained by antagonizing PKA type I in unstimulated CD8+ T cells show that basal PKA activity controls TCR activation at the level of CD3ζ phosphorylation and thereby provides a threshold that needs to be overcome to trigger TCR-induced downstream signaling events. This finding could be confirmed with the use of the myristoylated PKA inhibitor peptide myrPKI (Figure 7C-D). Preincubation of CD8+ T cells with PKI blocked PGE2-mediated inhibition of CD3ζ phosphorylation in response to TCR triggering (Figure 7C). When investigating signaling events further downstream of TCR triggering, SLP-76 phosphorylation was found to mirror the effects observed for CD3ζ phosphorylation, indicating that PKA regulates signaling events downstream of the TCR (Figure 7D).
Perturbation of the PKA signaling node leads to release from inhibition of T-cell activation. (A) CD8 T-cell populations were TCR stimulated (OKT3, 1 μg/mL; CD28.2, 5 μg/mL) for 1 minute with or without PGE2 (10μM; 10 minutes of preincubation) and with or without pretreatment with the PKA-RI antagonist Rp-8-Br-cAMPS (1mM; 30 minutes of preincubation). Cells were analyzed for CD3ζ phosphorylation at Tyr142. Data represent 4 independent experiments with individual blood donors expressed as mean ± SD; n = 4 of arcsinh median differences. (B) Effect of Rp-8-Br-cAMPS (1mM; 30 minutes of incubation) on basal CD3ζ phosphorylation at Tyr142. Warmer colors (yellow) in the heatmap representation (insert in B) indicate increases and cooler colors (blue) indicate decreases in Arcsinh median differences displayed on a scale of 1. Data represent 3 independent experiments with individual blood donors expressed as mean ± SD; n = 3 of arcsinh median differences. (C) CD8 T-cell populations were TCR stimulated (OKT3, 1 μg/mL; CD28, 5 μg/mL) for 1 minute with or without PGE2 (10μM; 10 minutes of preincubation) and with or without pretreatment with the PKA-RI antagonist Rp-8-Br-cAMPS (1mM; 30 minutes of preincubation) and the myristoylated PKA inhibitor peptide PKI (45 minutes of preincubation). Cells were analyzed for CD3ζ phosphorylation at Tyr142. Data represent 3 independent experiments from 3 individual blood donors expressed as mean ± SD of arcsinh median differences. (D) As in panel C, but SLP-76 phosphorylation at Tyr128 was analyzed. Data represent 3 independent experiments from 3 independent blood donors expressed as mean ± SD of arcsinh median differences.
Perturbation of the PKA signaling node leads to release from inhibition of T-cell activation. (A) CD8 T-cell populations were TCR stimulated (OKT3, 1 μg/mL; CD28.2, 5 μg/mL) for 1 minute with or without PGE2 (10μM; 10 minutes of preincubation) and with or without pretreatment with the PKA-RI antagonist Rp-8-Br-cAMPS (1mM; 30 minutes of preincubation). Cells were analyzed for CD3ζ phosphorylation at Tyr142. Data represent 4 independent experiments with individual blood donors expressed as mean ± SD; n = 4 of arcsinh median differences. (B) Effect of Rp-8-Br-cAMPS (1mM; 30 minutes of incubation) on basal CD3ζ phosphorylation at Tyr142. Warmer colors (yellow) in the heatmap representation (insert in B) indicate increases and cooler colors (blue) indicate decreases in Arcsinh median differences displayed on a scale of 1. Data represent 3 independent experiments with individual blood donors expressed as mean ± SD; n = 3 of arcsinh median differences. (C) CD8 T-cell populations were TCR stimulated (OKT3, 1 μg/mL; CD28, 5 μg/mL) for 1 minute with or without PGE2 (10μM; 10 minutes of preincubation) and with or without pretreatment with the PKA-RI antagonist Rp-8-Br-cAMPS (1mM; 30 minutes of preincubation) and the myristoylated PKA inhibitor peptide PKI (45 minutes of preincubation). Cells were analyzed for CD3ζ phosphorylation at Tyr142. Data represent 3 independent experiments from 3 individual blood donors expressed as mean ± SD of arcsinh median differences. (D) As in panel C, but SLP-76 phosphorylation at Tyr128 was analyzed. Data represent 3 independent experiments from 3 independent blood donors expressed as mean ± SD of arcsinh median differences.
The results presented here identify that elevated PKA type I signaling in unstimulated CD8+CD45RO+ T cells controls T-cell activation at the level of CD3ζ phosphorylation. In addition, our findings warrant caution to consider the averaging effect of opposing signaling states in different cell populations, when signaling profiles in complicated cell mixtures are to be interpreted and highlight the importance of investigating signaling events in functionally different cell populations individually.
Discussion
We describe a multipronged strategy to investigate signaling networks. An initial unbiased, quantitative phosphoproteomic mass spectrometry approach provides the basis for a high stringency kinase prediction of possible upstream kinases involved in the phosphorylation-based signaling network being investigated. Data from both the mass spectrometry as well as the kinase prediction is subsequently used for the selection of phospho-epitopes suitable for phospho flow cytometry–based monitoring of signaling pathway activity for several different cell types in parallel under a variety of experimental conditions. This novel approach leads to the identification of fundamental differences in signaling states in different T-cell subsets and highlights the importance of investigating signaling profiles in functionally distinct cell subpopulations.
The quantitative phosphoproteomic mass spectrometry data of PGE2-treated CD3+ T cells provides new insight into the complexity of PGE2-induced signaling events by assessing the temporal behavior of 247 regulated phosphosites on 207 phosphoproteins. In our larger full dataset of approximately 750 phosphosites, we identify a large number of previously unknown phosphosites (supplemental Table 4; supplemental data), of which many are regulated in response to PGE2 treatment. We next charted our phosphoproteomic dataset to select significantly regulated phosphosites for which antibodies were available, to be able to link our phosphoproteomic dataset to phospho flow cytometry. However, at present the number of phospho-epitope–specific antibodies available for a proteomic dataset of this size is limited. To circumvent this, we subjected all regulated phosphosites to stringent kinase prediction with the use of 3 independent kinase prediction algorithms.29-31 However, because these algorithms are at present based on limited datasets and because kinases are known to be able to phosphorylate similar or even identical phosphorylation motifs, false-positive identification of kinases is probable. Therefore, we included phospho-epitope–specific antibodies in our phospho flow antibody panel, which target phospho-epitopes that indicate increased activity of predicted kinases as well as established substrates of kinases of interest.
Using the selected antibody panel, we found that the CAMKII and PKB/Akt kinases were not significantly autophosphorylated in response to PGE2 stimulation in any of the investigated cell types, although their activation was strongly suggested by the performed kinase prediction algorithms (Figure 4C). The absence of an increase in Ca2+ on PGE2 stimulation (supplemental Figure 4) is also in agreement with the lack of autophosphorylation of CAMKII. These results indicate that CAMKII and PKB/Akt may be overrepresented in the kinase prediction datasets. This overrepresentation is most probably because both kinases share overlapping phosphorylation motifs with PKA. In addition, the sensitivity of investigated phospho-epitopes to the PKA inhibitor H89 indicates that the central node of the PGE2-signaling network is in fact PKA and that most phosphorylation events may result directly or indirectly from PKA activation. However, the identification of 6 unique predicted phosphorylation sites for PKB/Akt and 9 for CAMKII (Table 3) could suggest that PKB/Akt and CAMKII represent weaker signaling nodes. Alternatively, these sites may be falsely predicted substrates for PKB/Akt and CAMKII and could also be novel PKA substrates. The analysis of signaling differences between distinct lymphocyte cell subtypes showed that there is a clear difference in the PGE2 signaling profile of CD3+ and CD3− lymphocytes. In the latter, some phospho-epitopes (VASP Ser239, heat shock protein 27 Ser78, and Histone H3 Ser10) were more sensitive to PGE2-induced phosphorylation, whereas the signaling response is dampened at phospho-epitopes such as S6-ribosomal protein-Ser235/236 (Figure 4B).
On the basis of the reported immunomodulating role of PGE2, it is interesting to note the phosphorylation of proteins important in T-cell activation (reviewed in Torgersen et al40 ). These include direct phosphorylation of CARMA1, FYN binding protein/adhesion and degranulation promoting adapter protein, VASP, Nck, phospholipase C γ, Wiskott-Aldrich syndrome protein–interacting protein, GADS, and nuclear factor of activated T cells cytoplasmic 2 (supplemental Table 2; supplemental data). These are all proteins that either directly or indirectly regulate enzymatic activity and or protein complex formation downstream of TCR, and phosphorylation of any of these proteins could potentially affect protein–protein interactions, enzyme activity, or subcellular location, depending on the protein domain affected. Although some of the identified phosphorylation sites are uncharacterized, it clearly points to signal crosstalk between the 2 receptor systems and possible novel sites of action for the inhibitory effect of PGE2 on antigen receptor signaling.
The detailed analysis of signaling profiles in CD4+ and CD8+ effector/memory T cells versus signaling profiles in naive T cells identifies fundamental differences in basal states between these cell types that directly affect the signal amplitude of potential antigen stimulation through TCR. We describe that effector/memory cells have a blunted TCR-mediated signaling in response to TCR stimulation, consistent with earlier observations of reduced tyrosine phosphorylation in effector/memory cells.43 The regulatory role of PGE2 and PKA in CD8+CD45RO+ cells could provide a link to better understand immune dysfunction in different diseases, such as cancer and autoimmunity in which immune control is important.
Overall, we show that phospho flow cytometry not only allows the validation of phosphoproteomic mass spectrometry data but also provides crucial additional information about signaling kinetics, the effect of kinase inhibitors, and differences in cell subtype signaling. Moreover, if antibodies detecting both unphosphorylated and phosphorylated forms of each protein of interest were included in the setup, differences in signaling responses of single cells within a subset population to a uniform stimulus could be assessed in a similar manner as recently described by Spencer et al.44 Taken together, the data resulting from the combination of all 3 approaches provide a detailed signaling map of PGE2 in lymphocytes (Figure 5A). Importantly, this present study showed an absolute requirement for studying individual cell subpopulations from complicated cell mixtures. This approach can easily be adapted to study signaling networks in a large variety of cell types and because of the comparatively small cell number required for the method, this approach is optimal for the study of limited patient material such as from patients with cancer. Furthermore, improvements in the fields of proteomics as well as the development of a larger variety of specific antibodies against phospho-epitopes and other posttranslational epitopes will increase the potential of the combinational approach described here.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Torkel Vang for his support and useful discussions.
This work was supported by grants from the Norwegian Functional Genomics Program, the Research Council of Norway, the Norwegian Cancer Society, Novo Nordic Foundation, and European Union (grant LSHB-CT-2006-037189, thera-cAMP; K.T.) and by the Netherlands Proteomics Center, a program embedded in the Netherlands Genomics Initiative (A.J.R.H.). N.G.O. is a postdoctoral fellow of the Norwegian Cancer Society.
Authorship
Contribution: N.G.O., S.L., and M.E.K. did experiments and analyzed data; A.J.R.H. and K.T. supervised the project; K.M.T. provided essential advice and analyzed data; and N.G.O., S.L., K.M.T., and K.T. wrote the paper. All authors read and commented on the draft versions of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for S.L. is Lehrstuhl für Bioanalytik, Technische Universität München, Freising, Germany.
Correspondence: Albert J. R. Heck, The Netherlands Proteomics Centre, Utrecht University, Sorbonnelaan 16, 3584 CA Utrecht, The Netherlands; e-mail: a.j.r.heck@uu.nl; or Kjetil Taskén, The Biotechnology Centre of Oslo, University of Oslo, PO Box 1125, Blindern, N-0317 Oslo, Norway; e-mail: kjetil.tasken@biotek.uio.no.
References
Author notes
N.G.O. and S.L. contributed equally to this study.

![Figure 1. PGE2 signaling kinetics. (A) Immunoblot of CD3+ T lymphocytes stimulated with PGE2 (10μM) over a 60-minute time course with the use of a phospho-PKA substrate antibody (RRXpS/pT). (B) Relative density of 4 independent immunoblots plus SEM as depicted in panel A. (C) Mass spectrometry work flow. Three samples from 1 blood donor (CT, control; 10 and 60 minutes of PGE2 [10μM] stimulated) were lysed separately and in-solution trypsin digested. The resulting peptides were labeled with stable isotope labeling by reductive amination. Subsequently, the 3 samples of the 3 different time points were mixed at a ratio of 1:1:1. After SCX fractionation, phosphopeptides were subjected to online TiO2-LC enrichment, coupled to a LTQ Orbitrap mass spectrometer. Data analysis was conducted as described in the “Bioinformatic handling of mass spectrometry data.” LC-MS/MS indicates liquid chromatography tandem mass spectrometry.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/13/10.1182_blood-2010-01-266650/4/m_zh89991057590001.jpeg?Expires=1767730086&Signature=VBp9dFRoJspjUYK~VnQHj4eZJgutfbY0EsUyAriYHXLE9XebEEp838MqeZb-ZPKAQSZh5SL-lwUpW2ExrIoABaqP9FaR0Okv7sw3v105OVUohXlhvWxdRXv~Agk3BaeafFdgzpe0WELsUkthTOfVw88uls9UcwXB6m8JjRJnIJCygrgzknUSpdT3HyNuExCQ573ttZ5sVMIyyaAhmLdmGtmGiFQhDV~P1vWSIll8RxqDMJBrD32~23hbQzR8DLZ2rfyfPfevmhBfnydUX5H99Eil9FraUvb~Ni0aZGsgClj-NS1jyPc0CS2Zns1hZxQqM4VnTzA-qgaXlrJ71mB~jw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. Identification and relative quantitation of detected phosphopeptides. (Top) Extracted ion chromatograms (XICs) of monoisotopic peaks were used to calculate peptide ratios (blue indicates control sample; purple, 10 minutes of PGE2 [10μM]; green, 60 minutes of PGE2 [10μM]). (Bottom) Tandem mass spectrometry spectra of detected phosphopeptides. (A) Phosphopeptide from AP-3 complex subunit δ-1 protein with phosphorylation at Ser567. (B) Phosphopeptide from Histone H1.4 protein with phosphorylation at Ser35. For complete mass spectrometry results, please refer to supplemental Table 4 and supplemental data.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/13/10.1182_blood-2010-01-266650/4/m_zh89991057590002.jpeg?Expires=1767730086&Signature=MgbDr0n8tW3bg8bN8~liZpHad1bkZPFYM~~eTxTuOymi9RBl1fAtnfBMrunzHMESdSvn3OGj3X-8iHs27Dry51gTet7nAeRIg-RRgO0UVAyJ3PiWefW~SHMvDjl00ibCpn0vdkBu6fKZQJKs6Y-7shBQlPBFCVtjdao2IsrW4X0k56WUZ2GtiAuoXRKB9gVw9OvzztaD4TNvQkRJdVMbWrej-Lk9wAkdFpCbfKuO~7r~EOePLvBIp4DiDdb-iNQxg~Rs36d-~n6fJYHgoze69IELiArK1i6ttZ3Zi17Rz03O2gG7qLZjJQ-hbrVIEZvj6p8gDRJh41s2idL9G1YsVQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. A combinational approach provides high-resolution signaling data. (A) Schematic overview of the combined phosphoproteomic and FCB-phospho flow methods that provide a high-resolution map of intracellular signaling and stimulus-specific signaling networks. (B) Summary depicting results obtained from kinase prediction, phosphoproteomics, and FCB-phospho flow (from left to right). Kinase prediction shows a probable connection between PKA and its substrates. All 3 depicted substrates (VASP, GSK3α, and heat shock protein 27 [Hsp27]) have been detected by mass spectrometry, and the relative changes in phospho-status at the specified sites have been determined. In addition, the detected phosphosites were studied with the use of FCB-phospho flow to show high-resolution kinetic information over extensive time courses in 5 lymphocyte cell populations, both in the absence and presence of a PKA inhibitor (H89). Color coding of histograms is according to a scale of 2 for VASP and GSK3α, and to a scale of 1 for Hsp27.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/13/10.1182_blood-2010-01-266650/4/m_zh89991057590005.jpeg?Expires=1767730086&Signature=GwQbHJBvt7BwUJFIuWDhSdAi0FZF3aRDP4X6vwX61hShhK6DtEyq1P5-8yJjhg2-4PrDxaTd0MBZ6NcAAjwOulPJD1pbFIpam0Y4BRAw1eX9gkZq-h8K6JhMPsxPyr7l22jnPwYgzxI0Si16HSL9YBKuUVgvRaBRiONr7C9wB~4dQPqEZw51mDS7bKJxw9me79WYglKX66vSicjHdYbFL3dbXtH~BLJeSEEXizw-UbqcXOLbKN-4Cd~7bWcJ-lrSZDoVtrD3~lJ3M5fUleuDChpzcsXnX~J-l5eiZGhtC4aTcGzFoxk~H-a4obe~LSHP47UOhyN-epc-MlNte~7M4w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 6. CD3+ T-cell subset analysis shows a highly active PKA signaling node in CD8+CD45RO+ T cells. (A) Heatmap depicting differences in Arcsinh median of the antiphospho PKA substrate (RRXpS/pT) antibody in CD3+ T-cell subsets after 10 minutes of PGE2 (10μM) alone or with IBMX (3-isobutyl-1-methylxanthine) or rolipram (5mM and 20μM, respectively; 30 minutes) preincubation, or 8-CPT-cAMP (300μM; 5 minutes). Heatmap depicts net signaling profiles from 1 representative blood donor with the first column in each row set as reference. (B) Statistical analysis of PKA signaling in selected T-cell subsets from 3 individual blood donors analyzed independently as in panel A (mean ± SD; n = 3). (C) Heatmap depicting total signaling dynamics of the same samples as in panel A with the control sample of the CD3+ T-cell population set as reference. (D) Statistical analysis from 3 individual blood donors analyzed independently, using the difference in basal PKA signaling of C (mean ± SD; n = 3). (E) TCR (anti-CD3 [OKT3] 1 μg/mL; anti-CD28 [CD28.2] 5 μg/mL) stimulation for the indicated time period with and without PGE2 (10μM; 10 minutes preincubation) measured by Arcsinh median differences in CD3ζ-chain phosphorylation at Tyr142. Heatmap depicts the relative level of signaling with the control sample of the CD3+ T-cell population set as reference from 1 representative blood donor. (F) Statistical analysis from 5 individual blood donors analyzed independently (mean ± SD; n = 5) of samples as in panel E. To account for large differences in TCR responses between different donors, data are normalized to percentages with TCR stimulation (1 minute) representing 100%. (G-H) Heatmaps depicting total signaling dynamics for the indicated phospho-epitope–specific antibodies in CD8+ T cells from 1 representative blood donor with the control sample of the CD8+ T-cell population as reference. One minute of TCR (anti-CD3 [OKT3] 1 μg/mL; anti-CD28 [CD28.2] 5 μg/mL) stimulation and PGE2 (10μM) preincubation for 10 minutes before TCR stimulation. In all heatmaps, warmer colors (yellow) indicate increases and cooler colors (blue) indicate decreases in Arcsinh median differences displayed on a scale of 1.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/13/10.1182_blood-2010-01-266650/4/m_zh89991057590006.jpeg?Expires=1767730086&Signature=3EmVICA2F4K8srVV1R7Sx8PVT7Rr7EAs7oPdpvAEEX9xCNgGUyLl2JQpgQOm70hXxtps0cAKqKhEafIXIBYC~qt3uQ7KcmkYNHqTWy5OETAuSLn~1MMwMQNIjXLd4CHmF86AN1ws~hZQBZrgnYxVuXeeuTnidchc5OMk1jaaeCFujre6nuEgi9mUfjl0GwGUYnnOEUC4ju0MAh3ZQL8htlkpRWm9cSecUmZTwqVRsiHXT2AUSTFYdD8WVRF35IanNMm-mhtVXdrT7VT0m9~ZQaFEr2R5GO7BdmBILN3tICrbfHm72RyXs9JuoIgZJBDqhwvFYtxnuwN9iiKOwZ2stw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)