Abstract
Patients with low-grade myelodysplastic syndromes (MDS) show high levels of tumor necrosis factor α (TNFα) and up-regulation of apoptosis in the marrow. In contrast, marrow cells in advanced MDS are typically resistant to TNFα-induced apoptosis but are rendered apoptosis-sensitive on coculture with stroma. The present studies show that CD34+ marrow cells in advanced MDS express high levels of TWIST, a basic helix-loop-helix transcription factor that opposes p53 function. TWIST levels correlated with disease stage (advanced > low grade; P = .01). Coculture with HS5 stroma resulted in down-regulation of TWIST and increased apoptosis in response to TNFα in CD34+ cells from advanced MDS; the same effect was achieved by TWIST-specific RNA interference in CD34+ cells. In primary MDS marrow stroma TWIST expression was lower than in healthy controls; suppression of TWIST in stroma interfered with induction of apoptosis sensitivity in cocultured CD34+ cells. Stroma cells so modified expressed reduced levels of intercellular adhesion molecule-1 (ICAM1; CD54); blockade of ICAM1 in unmodified stroma was associated with reduced apoptosis in cocultured CD34+ MDS marrow cells. These data suggest role for dysregulation of TWIST in the pathophysiology of MDS. Conceivably, TWIST or components in the signaling pathway could serve as therapeutic targets for patients with MDS.
Introduction
Myelodysplastic syndromes (MDS) are clonal disorders of hematopoietic stem/precursor cells characterized by ineffective hematopoiesis, generally presenting with single or multilineage cytopenia in peripheral blood.1 Several proinflammatory cytokines and proapoptotic signals such as tumor necrosis factor α (TNFα), interleukin-1β, (IL-1β), Fas-ligand, and TNFα-related apoptosis-inducing ligand (TRAIL) and their respective receptors are up-regulated in MDS marrow and appear to interfere with orderly hematopoiesis.2-4 MDS has been considered a cell-autonomous disorder, and various mutations in hematopoietic cells have been described; however, recent data indicate that, in addition, cellular and noncellular components of the marrow microenvironment are involved in the pathophysiology of MDS and contribute to the broad spectrum of clinical presentations.5-7 We and others have shown that, particularly in early stage/low-grade MDS, apoptosis occurs in both residual normal (polyclonal) and clonal hematopoietic precursors,8,9 albeit at a consistently lower rate in clonal cells.8 This lower apoptotic rate is associated with a survival advantage of the clone. With more advanced MDS, clonal hematopoietic precursors become increasingly resistant to proapoptotic signals8 and in a proportion of patients MDS will evolve to acute myeloid leukemia (AML).8 Moreover, clonal cells generate negative regulatory signals, which contribute to the suppression of residual normal hematopoiesis. A recent study in a murine model suggested in fact that (clonal) leukemia cells create bone marrow niches that interfere with the function of normal hematopoietic precursors.10
Our work has focused on interactions between marrow stroma and hematopoietic precursors in MDS that might differentially affect clonal and nonclonal hematopoiesis.7,11-14 We observed, for example, that treatment of human marrow stroma cell lines with TNFα at concentrations as observed in MDS marrow significantly changed gene expression profiles.11,12 One molecule that was down-regulated in TNFα-treated stroma cells was TWIST,7 a highly conserved basic helix-loop-helix transcription factor that plays a pivotal role in osteoblast differentiation.15 Recent studies show that TWIST is also involved in several pathways that control tumor growth, apoptosis, differentiation, and epithelial–mesenchymal transition.15-17 Further, TWIST directly interacts with and opposes the function of p53.18,19 Although mutations or loss of p53 are the single most common genetic event observed in human cancer in general, p53 mutations are infrequent in patients with MDS.20 It is conceivable, therefore, that other factors modify p53 function. Here, we show that TWIST expression is dysregulated in MDS marrow and is modified in hematopoietic precursors by stroma contact. Furthermore, because TWIST modifies the expression of adhesion molecules that may affect contact-dependent signals, we investigated a potential role of TWIST in stroma/hematopoietic cell interactions.
Methods
Reagents
Recombinant human TNFα was purchased from PeproTech Inc., and TRAIL (Killer TRAIL, soluble [human] recombinant) from Alexis Biochemicals. The soluble TNF receptor etanercept was a gift from Amgen Inc. All reagents were prepared as 1000× stocks and diluted for cultures as appropriate.
Cell lines and cell cultures
KG1a cells (derived from AML) were obtained from ATCC.14 PL-21 cells and HL-60 cells were a gift from Dr Stirewalt (Fred Hutchinson Cancer Research Center [FHCRC]). MDS-L cells were obtained from Prof Tohyama (Hamamatsu University School of Medicine).21 HS5 and HS27a cell lines derived from the marrow of a healthy volunteer and immortalized by transduction with human papilloma virus E6/E7 constructs22-24 were provided by Dr Torok-Storb (FHCRC). Stroma cells were grown, propagated, and used for experiments between passages 8 and 24 as described.25 Some experiments were carried out in serum-free medium, as indicated.12
Primary cells
Primary hematopoietic cells were derived from the marrow aspirates of healthy volunteers and patients with MDS. Patients were 30 to 70 years old, represented all MDS categories, and had international prognostic scoring system scores of 0 to 3.0. Patient and disease characteristics are summarized in Table 1. Healthy donors were 24 to 79 years old. All patients and healthy donors had given informed consent as required by the Institutional Review Board of the FHCRC in accordance with the Declaration of Helsinki. All protocols were reviewed and approved by the Institutional Review Board of the FHCRC. Bone marrow mononuclear cells were separated by Ficoll-Hypaque gradient centrifugation, and CD34+ cells were isolated by magnetic-activated cell sorting (MACS) according to the manufacturer's protocol (Miltenyi Biotec). Cell purity was determined by staining with fluorescently tagged antibody for CD34 and quantified by flow cytometric (fluorescence-activated cell sorting [FACS]) analysis on an LSR2 (BD Biosciences). CD34+ cells were then seeded in 96-well plates with or without HS5 stroma for apoptosis studies.
Patient and disease characteristics and TWIST levels in CD34+ cells
| Diagnosis . | Age, y . | Sex . | Cytogenetics . | IPSS . | BM cellularity . | Marrow blast count, % . | TWIST expression . | P53 expression . | TWIST/p53 ratio . |
|---|---|---|---|---|---|---|---|---|---|
| RA | 59 | F | Normal | 0 | Normal | 3 | 1.1739385 | 0.9980367 | 1.176248 |
| RA | 58 | F | Normal | 0 | Normal | 5 | 1.1937184 | 0.9743392 | 1.225157 |
| RA | 51 | M | Normal | 0.5 | Hypo | 3 | 1.2863391 | 0.9798507 | 1.312791 |
| RA | 61 | F | Normal | NA | Normal | 3 | 1.1456094 | 0.9826767 | 1.165805 |
| RA | 78 | M | Normal | 0.5 | Normal | 2 | 1.1686 | 0.9732649 | 1.200706 |
| RA | 79 | M | Normal | 0.5 | Hypo | 5 | 0.8357093 | 0.9982278 | 0.837193 |
| RA | 58 | M | Normal | 0 | Normal | 4 | 1.2723986 | 1.0561759 | 1.204722 |
| RA | 46 | F | Normal | 0 | Hyper | 3 | 1.0282371 | 1.1760294 | 0.874329 |
| RA | 45 | F | Normal | NA | Hyper | 5 | 1.1807 | 1.1183639 | 1.055771 |
| RA | 58 | M | Del 7 | NA | Hyper | 4 | 1.3967883 | 0.9036251 | 1.545761 |
| RA | 58 | F | Del 5 | 0 | Normal | 5 | 0.9442959 | 0.7455998 | 1.266492 |
| RA | 59 | F | Normal | 0 | Hyper | 5 | 1.0647 | 0.8619087 | 1.235282 |
| RA | 56 | F | Normal | NA | Hyper | 3 | 1.0298 | 0.7042588 | 1.462247 |
| RA | 57 | M | Tri 9, tri 11 | 1 | Normal | 3 | 1.1586889 | 0.9124743 | 1.269832 |
| RA | 76 | M | Tri 5, tri 9, del 6 | 0.5 | Hyper | 1.5 | 1.2326 | 1.079136 | 1.142203 |
| RA | 56 | F | Normal | NA | Normal | 5 | 1.2120478 | 0.9923114 | 1.221439 |
| RA | 65 | F | Normal | NA | Hyper | 5 | 1.1739385 | 0.9980367 | 1.176248 |
| RA | 82 | M | Normal | 0 | Hyper | 4 | 1.5250296 | 1.0450261 | 1.459322 |
| RA | 65 | M | Complex | 0 | Hyper | 5 | 1.0842018 | 0.7963254 | 1.361506 |
| RA | 69 | F | Normal | 1 | Hyper | 2 | 0.7038678 | 0.852639 | 0.825517 |
| RA | 82 | M | Normal | 0 | Hyper | 5 | 0.7189514 | 0.623189 | 1.153665 |
| RA | 78 | M | Normal | 0 | Hyper | 3 | 1.7513135 | 0.9201176 | 1.903358 |
| RA | 43 | F | Tri 1 | 1.0 | Hyper | 5 | 1.1859731 | 0.8560692 | 1.385371 |
| RA | 60 | F | Tri 15 11q− | NA | Hyper | 2 | 1.1355641 | 0.84988 | 1.336146 |
| RARS | 48 | F | t(3,21) | 0 | Hypo | 5 | 1.3050591 | 0.9460929 | 1.37942 |
| RARS | 76 | F | Normal | 0 | Hyper | 5 | 1.2224846 | 0.8011956 | 1.525826 |
| RARS | 40 | F | Del(5q) tri8 | NA | Hypo | 5 | 1.1226 | 1.1519293 | 0.974581 |
| RARS | 48 | M | Tri 8 | 0.5 | Normal | 5 | 0.4877078 | 0.4587859 | 1.06304 |
| RCMD | 50 | F | Mono X | 0.5 | Hyper | 5 | 1.1723329 | 0.8742435 | 1.340968 |
| RCMD | 49 | F | Complex | 1.5 | Hyper | 5 | 1.1680894 | 1.533244 | 0.761842 |
| RCMD | 65 | F | Complex | 1.5 | Normal | 3 | 1.1355641 | 0.84988 | 1.336146 |
| RCMD | 60 | M | Normal | 0.5 | Normal | 5 | 1.3050591 | 0.9460929 | 1.37942 |
| RCMD | 54 | M | Del 7 | 1.5 | Hyper | 5 | 1.565455 | 0.9456547 | 1.656546 |
| RAEB-1 | 59 | M | Normal | 0.5 | Normal | 8 | 1.2224846 | 0.8011956 | 1.525826 |
| RAEB-1 | 71 | M | Complex, del(7) | 1.5 | Normal | 7 | 1.1226 | 1.1519293 | 0.974581 |
| RAEB-1 | 78 | M | Tri8 | 2.5 | Hyper | 6 | 1.0843432 | 0.881764 | 1.229743 |
| RAEB-1 | 71 | F | Mono 7 | 1.5 | Hypo | 6 | 1.2456 | 1.0451892 | 1.19171 |
| RAEB-1 | 52 | F | 11q− | 1.5 | Hyper | 7 | 1.8943619 | 1.1587836 | 1.634785 |
| RAEB-1 | 52 | M | +8, +21 | 0.5 | Hyper | 8 | 1.1292417 | 0.7932933 | 1.423486 |
| RAEB-2 | 50 | M | Normal | 2 | Hyper | 12 | 1.5372 | 1.0765506 | 1.427912 |
| RAEB-2 | 58 | M | Normal | 0.0 | Normal | 15 | 1.1358 | 0.9012022 | 1.260262 |
| RAEB-2 | 61 | M | y− | 2.5 | Hyper | 16 | 1.4958 | 0.686075 | 2.180228 |
| RAEB-2 | 48 | F | Complex | 2.0 | Normal | 19 | 1.1358 | 0.9012022 | 1.260262 |
| RAEB-2 | 56 | M | Del 7 | 2 | Normal | 12 | 1.0385472 | 1.0036802 | 1.034739 |
| RAEB-2 | 68 | M | Complex | 1.5 | Normal | 10 | 1.2003644 | 0.6438533 | 1.864345 |
| RAEB-2 | 76 | F | Complex | 2 | Hyper | 13 | 1.1573 | 0.9615516 | 1.203537 |
| RAEB-2 | 57 | M | Normal | 1.5 | Normal | 15 | 1.2841701 | 0.9245436 | 1.388977 |
| RAEB-2 | 63 | F | Complex | 1.0 | Hyper | 11 | 1.0836589 | 0.8363269 | 1.295736 |
| MDS sec | 65 | M | Normal | Hyper | 2 | 1.0765132 | 0.7689531 | 1.399972 | |
| MDS sec | 58 | M | Normal | Hyper | 5 | 1.2723986 | 1.0561759 | 1.204722 | |
| MDS sec | 57 | M | Complex | Hyper | 10 | 1.670892 | 1.0202264 | 1.637766 | |
| MDS sec | 55 | F | Normal | Normal | 7 | 1.1392 | 0.8290421 | 1.146339 | |
| MDS sec | 48 | M | Complex | Hyper | 11 | 1.390892 | 1.8638578 | 0.746244 | |
| MDS sec | 63 | F | Complex | Hyper | 5 | 1.1392 | 0.8290421 | 1.146339 | |
| MDS sec | 58 | F | Der (5)t(5;17)(p10;q11), −17/idem, t(1;10) | Hyper | 7 | 1.9175455 | 0.8482369 | 2.260625 | |
| MDS sec | 57 | M | Complex | Normal | 12 | 1.3744509 | 0.8745623 | 1.571587 |
| Diagnosis . | Age, y . | Sex . | Cytogenetics . | IPSS . | BM cellularity . | Marrow blast count, % . | TWIST expression . | P53 expression . | TWIST/p53 ratio . |
|---|---|---|---|---|---|---|---|---|---|
| RA | 59 | F | Normal | 0 | Normal | 3 | 1.1739385 | 0.9980367 | 1.176248 |
| RA | 58 | F | Normal | 0 | Normal | 5 | 1.1937184 | 0.9743392 | 1.225157 |
| RA | 51 | M | Normal | 0.5 | Hypo | 3 | 1.2863391 | 0.9798507 | 1.312791 |
| RA | 61 | F | Normal | NA | Normal | 3 | 1.1456094 | 0.9826767 | 1.165805 |
| RA | 78 | M | Normal | 0.5 | Normal | 2 | 1.1686 | 0.9732649 | 1.200706 |
| RA | 79 | M | Normal | 0.5 | Hypo | 5 | 0.8357093 | 0.9982278 | 0.837193 |
| RA | 58 | M | Normal | 0 | Normal | 4 | 1.2723986 | 1.0561759 | 1.204722 |
| RA | 46 | F | Normal | 0 | Hyper | 3 | 1.0282371 | 1.1760294 | 0.874329 |
| RA | 45 | F | Normal | NA | Hyper | 5 | 1.1807 | 1.1183639 | 1.055771 |
| RA | 58 | M | Del 7 | NA | Hyper | 4 | 1.3967883 | 0.9036251 | 1.545761 |
| RA | 58 | F | Del 5 | 0 | Normal | 5 | 0.9442959 | 0.7455998 | 1.266492 |
| RA | 59 | F | Normal | 0 | Hyper | 5 | 1.0647 | 0.8619087 | 1.235282 |
| RA | 56 | F | Normal | NA | Hyper | 3 | 1.0298 | 0.7042588 | 1.462247 |
| RA | 57 | M | Tri 9, tri 11 | 1 | Normal | 3 | 1.1586889 | 0.9124743 | 1.269832 |
| RA | 76 | M | Tri 5, tri 9, del 6 | 0.5 | Hyper | 1.5 | 1.2326 | 1.079136 | 1.142203 |
| RA | 56 | F | Normal | NA | Normal | 5 | 1.2120478 | 0.9923114 | 1.221439 |
| RA | 65 | F | Normal | NA | Hyper | 5 | 1.1739385 | 0.9980367 | 1.176248 |
| RA | 82 | M | Normal | 0 | Hyper | 4 | 1.5250296 | 1.0450261 | 1.459322 |
| RA | 65 | M | Complex | 0 | Hyper | 5 | 1.0842018 | 0.7963254 | 1.361506 |
| RA | 69 | F | Normal | 1 | Hyper | 2 | 0.7038678 | 0.852639 | 0.825517 |
| RA | 82 | M | Normal | 0 | Hyper | 5 | 0.7189514 | 0.623189 | 1.153665 |
| RA | 78 | M | Normal | 0 | Hyper | 3 | 1.7513135 | 0.9201176 | 1.903358 |
| RA | 43 | F | Tri 1 | 1.0 | Hyper | 5 | 1.1859731 | 0.8560692 | 1.385371 |
| RA | 60 | F | Tri 15 11q− | NA | Hyper | 2 | 1.1355641 | 0.84988 | 1.336146 |
| RARS | 48 | F | t(3,21) | 0 | Hypo | 5 | 1.3050591 | 0.9460929 | 1.37942 |
| RARS | 76 | F | Normal | 0 | Hyper | 5 | 1.2224846 | 0.8011956 | 1.525826 |
| RARS | 40 | F | Del(5q) tri8 | NA | Hypo | 5 | 1.1226 | 1.1519293 | 0.974581 |
| RARS | 48 | M | Tri 8 | 0.5 | Normal | 5 | 0.4877078 | 0.4587859 | 1.06304 |
| RCMD | 50 | F | Mono X | 0.5 | Hyper | 5 | 1.1723329 | 0.8742435 | 1.340968 |
| RCMD | 49 | F | Complex | 1.5 | Hyper | 5 | 1.1680894 | 1.533244 | 0.761842 |
| RCMD | 65 | F | Complex | 1.5 | Normal | 3 | 1.1355641 | 0.84988 | 1.336146 |
| RCMD | 60 | M | Normal | 0.5 | Normal | 5 | 1.3050591 | 0.9460929 | 1.37942 |
| RCMD | 54 | M | Del 7 | 1.5 | Hyper | 5 | 1.565455 | 0.9456547 | 1.656546 |
| RAEB-1 | 59 | M | Normal | 0.5 | Normal | 8 | 1.2224846 | 0.8011956 | 1.525826 |
| RAEB-1 | 71 | M | Complex, del(7) | 1.5 | Normal | 7 | 1.1226 | 1.1519293 | 0.974581 |
| RAEB-1 | 78 | M | Tri8 | 2.5 | Hyper | 6 | 1.0843432 | 0.881764 | 1.229743 |
| RAEB-1 | 71 | F | Mono 7 | 1.5 | Hypo | 6 | 1.2456 | 1.0451892 | 1.19171 |
| RAEB-1 | 52 | F | 11q− | 1.5 | Hyper | 7 | 1.8943619 | 1.1587836 | 1.634785 |
| RAEB-1 | 52 | M | +8, +21 | 0.5 | Hyper | 8 | 1.1292417 | 0.7932933 | 1.423486 |
| RAEB-2 | 50 | M | Normal | 2 | Hyper | 12 | 1.5372 | 1.0765506 | 1.427912 |
| RAEB-2 | 58 | M | Normal | 0.0 | Normal | 15 | 1.1358 | 0.9012022 | 1.260262 |
| RAEB-2 | 61 | M | y− | 2.5 | Hyper | 16 | 1.4958 | 0.686075 | 2.180228 |
| RAEB-2 | 48 | F | Complex | 2.0 | Normal | 19 | 1.1358 | 0.9012022 | 1.260262 |
| RAEB-2 | 56 | M | Del 7 | 2 | Normal | 12 | 1.0385472 | 1.0036802 | 1.034739 |
| RAEB-2 | 68 | M | Complex | 1.5 | Normal | 10 | 1.2003644 | 0.6438533 | 1.864345 |
| RAEB-2 | 76 | F | Complex | 2 | Hyper | 13 | 1.1573 | 0.9615516 | 1.203537 |
| RAEB-2 | 57 | M | Normal | 1.5 | Normal | 15 | 1.2841701 | 0.9245436 | 1.388977 |
| RAEB-2 | 63 | F | Complex | 1.0 | Hyper | 11 | 1.0836589 | 0.8363269 | 1.295736 |
| MDS sec | 65 | M | Normal | Hyper | 2 | 1.0765132 | 0.7689531 | 1.399972 | |
| MDS sec | 58 | M | Normal | Hyper | 5 | 1.2723986 | 1.0561759 | 1.204722 | |
| MDS sec | 57 | M | Complex | Hyper | 10 | 1.670892 | 1.0202264 | 1.637766 | |
| MDS sec | 55 | F | Normal | Normal | 7 | 1.1392 | 0.8290421 | 1.146339 | |
| MDS sec | 48 | M | Complex | Hyper | 11 | 1.390892 | 1.8638578 | 0.746244 | |
| MDS sec | 63 | F | Complex | Hyper | 5 | 1.1392 | 0.8290421 | 1.146339 | |
| MDS sec | 58 | F | Der (5)t(5;17)(p10;q11), −17/idem, t(1;10) | Hyper | 7 | 1.9175455 | 0.8482369 | 2.260625 | |
| MDS sec | 57 | M | Complex | Normal | 12 | 1.3744509 | 0.8745623 | 1.571587 |
IPSS indicates International Prognostic Scoring System; RA, refractory anemia; normal, normal karyotype; hypo, cellularity lower than expected for age; hyper, cellularity greater than expected for age; NA, not available because of incomplete information on peripheral blood cell counts or no scores were assigned to secondary MDS; RARS, RA with ring sideroblasts; RCMD, refractory cytopenia with multilineage dysplasia; RAEB, RA with excess blasts (1 or 2); and MDS sec, secondary (treatment-related) myelodysplastic syndrome.
Primary bone marrow stroma cells were isolated from Ficoll-Hypaque–separated bone marrow mononuclear cells by culturing 2.5 to 3 × 107 cells per T75 flask in nonhematopoietic expansion medium (Miltenyi Biotec) at 37°C in a humidified 5% CO2/air atmosphere. Culture medium was replaced weekly. When cultures reached 80% to 90% confluence, cells were detached, analyzed for possible myeloid/lymphoid cell contamination, and plated for coculture experiments. Specifically, adherent cells were washed with phosphate-buffered saline, detached with TrypLE (Invitrogen) for 10 minutes, and analyzed by FACS for markers of myeloid/lymphoid cell populations, including CD45, CD11b, CD14, and CD34. In addition to excluding cells with hematopoietic markers, the adherent (stroma) cells were characterized for expression of CD54, CD73, and CD90. The fluorescently tagged antibodies for these markers were obtained from BD Biosciences and R&D Systems.
Reverse transcriptase polymerase chain reaction and quantitative real-time polymerase chain reaction of pimary cells from patients with MDS and healthy donors
Quantitative reverse transcriptase polymerase chain reaction (RT-PCR) was carried out with the use of the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). cDNA was generated with the μMACS One-Step cDNA kit (Miltenyi Biotec), using a starting population of 5 × 104 CD34+ hematopoietic or stroma cells. Quantitative RT-PCR was performed with SYBR Green Master Mix and 3-step standard cycling conditions with sequence-specific primers. The melting curves were examined to ascertain that a single product was amplified. For quantitative analysis, the observed copy numbers of TWIST and p53 were normalized to the expression of β-actin.
The primer sequences for the amplification of TWIST in RT-PCR experiments were as follows: forward 5′-GGA GTC CGC AGT CTT ACG AG-3′ and reverse 5′-TCT GGA CCT GGT AGA GG-3′. PCR was carried out in a volume of 20 μL containing 1 U of Taq enzyme in the buffer supplied by the manufacturer (Invitrogen), 5 pM of each primer and 0.1mM of each deoxyribonucleotide triphosphate. Program conditions were as follows: 95°C for 5 minutes, followed by 35 cycles (95°C, 30 seconds; 53°C, 30 seconds; 72°C, 30 seconds). Amplification of β2-microglobulin as a housekeeping gene was performed to check the integrity of RNA and the success of the reverse transcription reaction. The PCR products were subjected to 2% agarose gel electrophoresis, stained with ethidium bromide, and visualized under UV illumination.
Western blot analysis
Total cell lysates were prepared by sonicating cells for 4 minutes. Fractions were cleared by centrifugation at 20 000g for 10 minutes. Protein concentrations were quantified by bicinchoninic acid assay (Pierce Biotechnology Inc), and equal amounts of protein (20 μg) from each lysate were diluted in Laemmli sodium dodecyl sulfate sample buffer and resolved by electrophoresis on 4% to 12% Bis-Tris precast NuPage gels (Invitrogen) in running buffer (50mM 2-(N-morpholino) ethane sulfonic acid, 50mM Tris [tris(hydroxymethyl)aminomethane]) base, 0.1% sodium dodecylsulfate, and 1mM EDTA [ethylenediaminetetraacetic acid]) as per the manufacturer's instructions. The proteins were then transferred to polyvinylidene diflouride membranes for immunoblotting. The membranes were blocked in 5% nonfat dry milk diluted in Tris-buffered saline containing 0.1% Tween-20 (TBS-T) for 1 hour at room temperature and then incubated overnight at 4°C in 5% nonfat dry milk /TBS-T containing either rabbit anti-P53 antibody (1:1000; Cell Signaling Technology Inc), rabbit anti-TWIST antibody (1:1000; Cell Signaling Inc), rabbit anti– intercellular adhesion molecule-1 (ICAM1) antibody (1:1000; Cell Signaling Inc) or rabbit anti-PYCARD antibody (1:1000; Sigma). Secondary goat anti–rabbit antibodies (1:2000; Santa Cruz Biotechnology Inc) conjugated to horseradish peroxidase were used for enhanced chemoluminescence (Pierce Biotechnology Inc), and membranes were exposed to film.
Assessment of apoptosis and expression of TWIST and p53 mRNA in primary cells
The response of hematopoietic cells to proapoptotic ligands was assessed with the use of annexin V staining. CD34+ cells from patients with MDS or healthy donors were cultured with stroma contact in the presence of TNFα at 25 ng/mL for 24 hours, based on previous dose response studies.12 Cell plates were centrifuged at 1200 rpm (300g), the supernatant was discarded, and the cells were washed with cold phosphate-buffered saline containing 2% bovine serum albumin. Cells in each well were then labeled with CD45-allophycocyanin or CD34-phycoerythrin (PE) antibodies (BD Biosciences) and assayed with the annexin V–fluorescein isothiocyanate (FITC) apoptosis kit; propidium iodide was used as a counter stain.
Primers for amplifying TWIST mRNA are described already. Primers for p53 amplification were forward 5′-TGAAGACCC AGGTCCAGAT-3′ and reverse 5′-TGTAGTGGATGGTGGTACAGTC-3′; they produced an amplicon of 530 bp (base pairs). cDNA samples were generated with the use of μMACS One-Step cDNA kit (Miltenyi Biotec). DNA was extracted from 5 × 104 primary cells, and 1/50 of each reaction was added to appropriate wells of the PCR plates. The mixtures were denatured at 96°C for 5 minutes, and then cycled for 40 cycles at 96°C for 15 seconds, at 53°C for 30 seconds, followed by 1 minute at 72°C. β2-Microglobulin was used as a housekeeping control. The PCR products were subjected to 2% agarose gel electrophoresis, stained with ethidium bromide, and visualized under UV illumination.
Immunoprecipitation
Immunoprecipitation assays of KG1a, MDS-L, HL-60, and PL-21 cell lysates were performed as described.18 Briefly, 5 × 106 cells were centrifuged and resuspended in 1 mL of lysis buffer, placed on ice for 1 hour, and shaken gently during the incubation. Cells were then sedimented at 15 000g for 10 minutes at 4°C. The supernatants were transferred into clean tubes, and 100 μL of Protein G was added for 1 hour at 4°C. Protein G beads were discarded after centrifugation, and the primary TWIST antibody was added overnight at 4°C; an isotype-matched irrelevant antibody served as negative control. Protein G, 100 μL, was added, and after 1 to 2 hours at 4°C the supernatant was discarded. The pellet was washed 5 times with lysis buffer, and the protein was loaded with 2× loading buffer and dichlorodiphenyltrichloroethane. Thirty microliters of supernatant was then analyzed with the use of 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis under reducing conditions and transferred to nitrocellulose membranes. Membranes were incubated with the appropriate antibody for 1 hour at room temperature, washed, and incubated with horseradish peroxidase–conjugated secondary antibody for another hour and developed by enhanced chemoluminescence.
Assessment of apoptosis
All unmodified myeloid cells and cells modified by short interference RNA (siRNA; see “siRNA knockdown of TWIST”) were cultured in the presence or absence of TNFα (10 ng/mL and 25 ng/mL) or TRAIL (100 ng/mL), and apoptosis was determined.12 Alternatively, myeloid cells were cocultured with stroma in the presence or absence of TNFα at 25 ng/mL for 24 hours. Apoptosis was determined separately in detached cells and in myeloid cells remaining attached to stroma, which were removed with the stroma after trypsin treatment. Cells were then labeled with FITC-conjugated anti-CD45 antibody to distinguish hematopoietic cells (myeloid cells, CD45+) from stroma (CD45−) and analyzed by FACS for apoptosis using annexin V–phycoerythrin and propidium iodide.
siRNA knockdown of TWIST
Stealth siRNA oligonucleotides (oligos) were obtained, along with Lipofectamine 2000 from Invitrogen. The sequences of the siRNA oligos for TWIST were 5′-UGGCGGCAAGGUACAUCGACUUCCU-3′ and 3′-AGGAAGUCGAUGUACCUGGCCGCCA-5′. The scrambled siRNA sequences were: 5′-CGAAUCCUAAUGCUGCUCCCUACUU-3′ and 3′-AAGUAGGGAGGAGCAUUACCAUUCG-5′. Myeloid cells were electroporated with siRNA with the use of the Nucleofector Kit L (Amaxa Biosystems Inc). HS5 and HS27a stroma cells, as well as primary stroma cells, were plated in 12-well plates at 0.2 to 0.5 × 105 cells/well in 1 mL of growth medium without antibiotics, grown to 90% to 95% confluence, and transfected with siRNA oligos and Lipofectamine 2000 (Invitrogen). siRNAs were suspended in growth medium for 5 to 10 minutes. Lipofectamine diluted in Dulbecco modified essential medium was added, and complexation continued for 30 minutes. The Lipofectamine/RNA ratio was 2 μL to 1 ng. After 6 hours, Dulbecco modified essential medium plus 10% fetal bovine serum was added to minimize toxicity.
Assessment of ICAM1 expression and function in HS5 cells in coculture
Stroma cells with TWIST knockdown were stained with anti-ICAM1 antibody and analyzed by FACS and Western blotting. Next, HS5 or HS27a cells (5 × 104 cells/well) were pretreated with 100 μL of a 10 μg/mL solution of neutralizing anti–human ICAM1 (CD54) antibody for 1 hour in preparation for coculture with KG1a or PL-21 cells. After washing off the excess antibody from stroma cells, KG1a or PL-21 cells were added to the wells for overnight coculture. Myeloid cells from coculture were removed by trypsin treatment and stained with anti–human CD54 (FITC conjugated) for 30 minutes and tested by FACS. To determine a potential role of CD11b in the coculture model, myeloid cells were pretreated with anti-CD11b antibody (P4H9), 1 μg/mL, for 1 hour before coculture with stroma.
Blocking of phospholipase C γ 1 by herbimycin in coculture
KG1a cells were treated with herbimycin (1μM) for 4 hours to block the phosphorylation of phospholipase C γ 1 (PLCG1), and then cocultured with HS5 cells overnight. After 24-hour incubation, KG1a cells were removed from coculture, and apoptosis was determined by FACS. Phospho-PLCG1 expression in KG1a cells under this condition was assessed by Western blotting.
Statistical analysis
The relationship between gene expression levels in different disease categories was assessed with the use of the 2-sample t test, and the correlation between TWIST levels and TWIST/p53 ratios and marrow parameters was examined with the use of linear regression.
Results
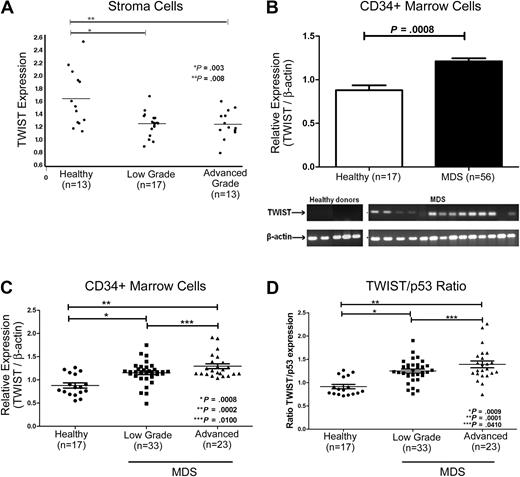
TWIST expression in MDS marrow stroma and hematopoietic precursors
Adherent cells from the marrows of 30 untreated patients with MDS expressed significantly lower levels of steady-state TWIST mRNA than did adherent cells from healthy donors (n = 13; Figure 1A). Levels were lowest in advanced stage MDS (refractory anemia with excess of blasts [RAEB]), and higher in patients with low-grade MDS (refractory anemia [RA]/refractory cytopenia with multilineage dysplasia), although the difference was not statistically significant.
Expression of TWIST in patients with MDS and in healthy donors. (A) Stroma cells. mRNA levels were determined by quantitative real-time PCR. TWIST levels were lower in MDS than in healthy controls (see text). (B) CD34+ cells. Samples from 56 patients with MDS and from 17 healthy donors were analyzed. TWIST mRNA levels were significantly higher in patients with advanced MDS (P < .001). Shown are the means and SEM. The gel shows actual mRNA levels in 14 patients with MDS and 5 healthy controls. (C) TWIST levels in patients with MDS and healthy donors. Relative mRNA levels of TWIST were significantly higher in patients with low-grade (P < .001) and advanced MDS (P < .001) than in healthy donors. (D) Ratio of relative TWIST and p53 levels in patients with MDS and healthy donors. The ratio of TWIST to p53 mRNA differed significantly between cells from healthy donors and cells from patients with low-grade (P < .001) and advanced MDS (P < .001), respectively. Horizontal lines indicate median values.
Expression of TWIST in patients with MDS and in healthy donors. (A) Stroma cells. mRNA levels were determined by quantitative real-time PCR. TWIST levels were lower in MDS than in healthy controls (see text). (B) CD34+ cells. Samples from 56 patients with MDS and from 17 healthy donors were analyzed. TWIST mRNA levels were significantly higher in patients with advanced MDS (P < .001). Shown are the means and SEM. The gel shows actual mRNA levels in 14 patients with MDS and 5 healthy controls. (C) TWIST levels in patients with MDS and healthy donors. Relative mRNA levels of TWIST were significantly higher in patients with low-grade (P < .001) and advanced MDS (P < .001) than in healthy donors. (D) Ratio of relative TWIST and p53 levels in patients with MDS and healthy donors. The ratio of TWIST to p53 mRNA differed significantly between cells from healthy donors and cells from patients with low-grade (P < .001) and advanced MDS (P < .001), respectively. Horizontal lines indicate median values.
In contrast to stroma, TWIST mRNA levels in CD34+ cells from untreated patients with MDS (n = 56) were consistently higher than in healthy controls (n = 17; Figure 1B; Table 1) for both patients with low-grade MDS (RA/refractory anemia with ringed sideroblasts/refractory cytopenia with multilineage dysplasia; n = 33; P < .001) and advanced-stage disease (RAEB-1/RAEB-2; n = 23; P < .001). The difference between low-grade and advanced MDS was significant at P = .01, suggesting that TWIST expression increased with disease progression (Figure 1C). There was a trend for positive correlations of TWIST levels in CD34+ MDS cells with marrow cellularity (R = 0.23; P = .09), marrow blast proportion (R = 0.21; P = .11), and log (blast count; R = 0.26; P = .06). Because TWIST does interact directly with p53,18 we also determined p53 levels in the same samples. Levels of p53 in CD34+ MDS cells did not differ significantly from levels in healthy donors. However, as the levels of TWIST mRNA were higher in advanced MDS (Figure 1D), the TWIST/p53 ratio in advanced MDS (RAEB; ratio, 1.4) was significantly higher than in low-grade MDS (RA; ratio, 1.2; P = .041). Similarly, the TWIST/p53 ratio showed a trend for correlation with marrow cellularity (R = 0.22; P = .10), marrow blast proportion (R = 0.23; P = .09), and log (blast count; R = 0.25; P = .07). There was no significant correlation of TWIST levels and international prognostic scoring system scores (not shown).
Effect of stroma contact on TWIST expression in hematopoietic precursors
p53 levels in myeloid cell lines and primary MDS marrow cells are up-regulated in stroma contact culture, and up-regulation is accompanied by enhanced sensitivity to TNFα-induced apoptosis.12 We were, therefore interested in determining whether TWIST might play a role in those responses.
Myeloid cell lines.
When placed in stroma contact, expression of TWIST in KG1a, MDS-L, and PL-21 cells declined, whereas p53 levels increased, and apoptosis in response to TNFα was enhanced (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Decreased expression of TWIST and increased expression of p53 after stroma contact persisted over an extended period of time (supplemental Figure 2).
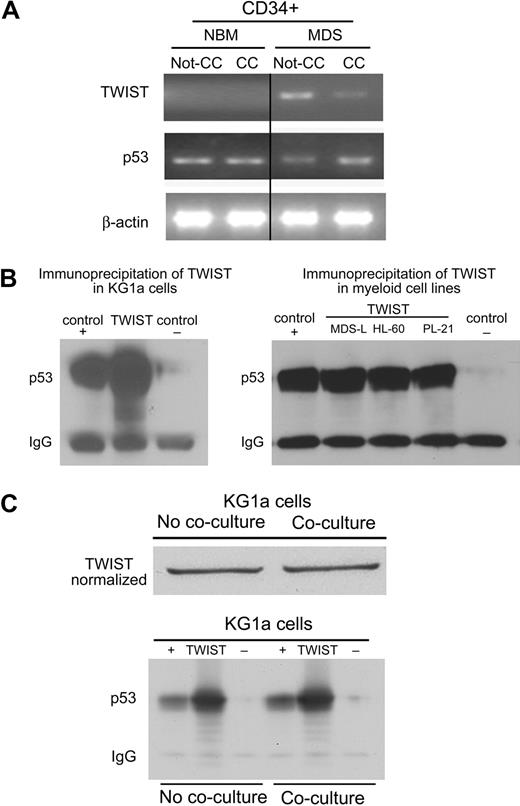
Primary marrow cells.
Next, we determined the mRNA levels of TWIST and p53 in primary bone marrow CD34+ cells before and after contact with HS5 stroma cells. As shown in Figure 2A, in CD34+ cells from healthy donors TWIST mRNA was not detectable, and p53 levels did not change with stroma contact. In contrast, in CD34+ cells from patients with advanced MDS, TWIST levels were down-regulated after contact with HS5 stroma and showed a negative correlation with expression of p53. Thus, as in clonal myeloid cell lines, changes in TWIST expression occurred in (presumably clonal) MDS cells but not in CD34+ cells from healthy donors. These data correlate with the susceptibility of clonal, but not healthy marrow cells, to TNFα-induced apoptosis in the previously described stroma coculture model.12
TWIST expression and interactions with p53 in primary CD34+ marrow cells and myeloid cell lines. (A) Primary CD34+ marrow cells from healthy donors and patients with MDS. Healthy CD34+cells (control) cultured without stroma (Not-CC) or with stroma contact (CC) showed no change in the expression of p53 or TWIST. In cells from patients with MDS, p53 showed prominent up-regulation on stroma contact (CC), whereas levels of TWIST declined. (B) Immunoprecipitation (IP) of TWIST from KG1a, MDS-L, HL-60, and PL-21 cell lysates. p53 was contained in the complex precipitated from all myeloid cell lines by anti-TWIST antibody; DJ-1 served as positive (+) and Stro-1 as negative (−) control. (C) IP as in panel B, but using KG1a cells without and with coculture, normalizing for TWIST content in the cell lysate. IgG indicates immunoglobulin G.
TWIST expression and interactions with p53 in primary CD34+ marrow cells and myeloid cell lines. (A) Primary CD34+ marrow cells from healthy donors and patients with MDS. Healthy CD34+cells (control) cultured without stroma (Not-CC) or with stroma contact (CC) showed no change in the expression of p53 or TWIST. In cells from patients with MDS, p53 showed prominent up-regulation on stroma contact (CC), whereas levels of TWIST declined. (B) Immunoprecipitation (IP) of TWIST from KG1a, MDS-L, HL-60, and PL-21 cell lysates. p53 was contained in the complex precipitated from all myeloid cell lines by anti-TWIST antibody; DJ-1 served as positive (+) and Stro-1 as negative (−) control. (C) IP as in panel B, but using KG1a cells without and with coculture, normalizing for TWIST content in the cell lysate. IgG indicates immunoglobulin G.
TWIST interacts directly with p53 and exerts antiapoptotic function in myeloid cell lines
Direct interactions of TWIST and p53 had been shown by Shiota et al18 in the human prostate cancer cell line PC3 and in MCF7 human breast cancer cells in which TWIST interfered with p53-dependent gene expression. To investigate such a possibility in our model, protein extracts from KG1a, MDS-L, HL-60, and PL-21 cell lysates were precipitated with anti-TWIST antibody and subjected to Western blot analysis with the use of an anti-p53 antibody. As shown in Figure 2B, the protein complex precipitated by anti-TWIST antibody was recognized by anti-p53 antibody, indicative of a physical association of endogenous TWIST and p53. As shown in Figure 2C, when protein content from KG1a cells after stroma coculture was normalized to the same level of TWIST as observed in KG1a cells cultured alone, the amounts of TWIST/p53 precipitated before and after stroma contact did not differ, suggesting that the TWIST/p53 binding affinity was not altered by stroma contact. Rather, taken together these results suggest that reduced levels of TWIST in cocultured KG1a cells resulted in increased levels of free active p53, which led to enhanced apoptosis sensitivity. As shown in supplemental Figure 3, we sequenced p53 in several cell lines and primary cells and we observed either mutated or nonmutated sequences in the cell lines studied. The fact that functional results were similar in myeloid cells with nonmutated p53 (eg, PL-21) and with mutated p53 (eg, KG1a) strongly suggests that factors in addition to p53 (eg, PYCARD, as shown previously12 ) are involved in apoptosis responses after stroma contact.
Inhibition of TWIST by siRNA in hematopoietic cells results in increased expression of p53 and enhanced TNFα-triggered apoptosis
To further characterize a role of TWIST in the modification of apoptosis in myeloid cell lines and primary MDS cells, we determined the effect of inhibition of TWIST by siRNA on TNFα-induced apoptosis. As shown in Figure 3A and B, suppression of TWIST rendered KG1a and PL-21 cells sensitive to apoptosis induced by TNFα or by TRAIL. Apoptosis in the absence of exogenous TNFα was, presumably, related to the release of endogenous TNFα from myeloid cells in culture. In fact, the addition of the soluble TNF receptor etanercept, as shown in Figure 3C, resulted in at least a partial inhibition of apoptosis in KG1a and PL-21 cells in which TWIST had been silenced.
Inhibition of TWIST in KG1a cells and its effect on apoptosis. Early stage apoptosis (annexin V+, propidium iodide−) in KG1a and PL-21 cell lines transfected with either a scrambled siRNA sequence (SCR) or siRNA specific for TWIST (siTWIST), and exposed overnight to normal medium (veh) versus exogenous TNFα (25 ng/mL, A) or TRAIL (100 ng/mL, B), respectively. Apoptosis was determined by flow cytometry. (C) Cell preparation as in panels A and B, but cultured in normal medium (veh) or in the presence of the soluble TNF receptor etanercept (10 μg/mL for 24 hours [concentration based on ancillary studies]). Error bars indicate SEM. (D) Protein lysates from the same cell preparations were separated on 4% to 12% Bis-Tris gels and immunoblotted with antibodies against TWIST, p53-ser46, Bax, and caspase 9, respectively. β-Actin served as loading control. (E) TWIST and p53 levels in primary CD34+ MDS cells in which TWIST was silenced by specific siRNA (KD TWIST) compared with levels in unmodified cells from the same primary CD34+ MDS sample.
Inhibition of TWIST in KG1a cells and its effect on apoptosis. Early stage apoptosis (annexin V+, propidium iodide−) in KG1a and PL-21 cell lines transfected with either a scrambled siRNA sequence (SCR) or siRNA specific for TWIST (siTWIST), and exposed overnight to normal medium (veh) versus exogenous TNFα (25 ng/mL, A) or TRAIL (100 ng/mL, B), respectively. Apoptosis was determined by flow cytometry. (C) Cell preparation as in panels A and B, but cultured in normal medium (veh) or in the presence of the soluble TNF receptor etanercept (10 μg/mL for 24 hours [concentration based on ancillary studies]). Error bars indicate SEM. (D) Protein lysates from the same cell preparations were separated on 4% to 12% Bis-Tris gels and immunoblotted with antibodies against TWIST, p53-ser46, Bax, and caspase 9, respectively. β-Actin served as loading control. (E) TWIST and p53 levels in primary CD34+ MDS cells in which TWIST was silenced by specific siRNA (KD TWIST) compared with levels in unmodified cells from the same primary CD34+ MDS sample.
As shown in Figure 3D, a significant correlation was observed between the decrease in TWIST protein and increased levels of phosphorylated p53 at serine 46. Concurrently, Bax expression was increased, as might be expected because of the effect of p53 on Bax transcription.26 As in myeloid cell lines, inhibition of TWIST also resulted in increased p53 expression in primary CD34+ MDS cells (Figure 3E). These data support a role of TWIST in the control of p53-mediated apoptosis, possibly by Bax, in this model.
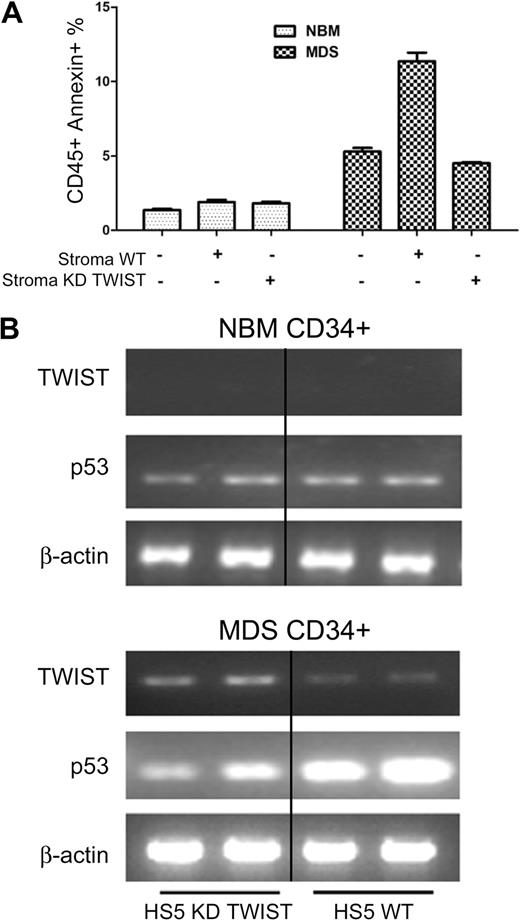
Inhibition of TWIST in HS5 stroma interferes with TWIST and p53 expression as well as apoptosis in cocultured CD34+ MDS cells
As shown in Figure 4A and B, CD34+ cells derived from patients with MDS, when cocultured with HS5 stroma cells in which TWIST was suppressed by siRNA, showed no increase in apoptosis and p53 levels (as had been observed with unmodified HS5 cells). However, TWIST suppression in HS5 stroma did not alter results in cocultured CD34+ cells from healthy donors (Figure 4A-B), consistent with previous results,12 which showed that stroma contact only affected clonal cells but not CD34+ cells from healthy donors.
TWIST and p53 expression and apoptosis in primary CD34+ cells. (A) Apoptosis in CD34+ marrow cells from healthy normal bone marrow donors (NBM) and patients with MDS cocultured with either unmodified HS5 stroma or HS5 with suppression of TWIST by siRNA (KD TWIST). Apoptosis was determined by flow cytometry. Error bars indicate SEM. (B) Levels of TWIST and p53 mRNA in CD34+ cells from healthy donors (NBM) and patients with MDS cocultured with either unmodified HS5 stroma (WT) or HS5 stroma with inhibition of TWIST (KD TWIST).
TWIST and p53 expression and apoptosis in primary CD34+ cells. (A) Apoptosis in CD34+ marrow cells from healthy normal bone marrow donors (NBM) and patients with MDS cocultured with either unmodified HS5 stroma or HS5 with suppression of TWIST by siRNA (KD TWIST). Apoptosis was determined by flow cytometry. Error bars indicate SEM. (B) Levels of TWIST and p53 mRNA in CD34+ cells from healthy donors (NBM) and patients with MDS cocultured with either unmodified HS5 stroma (WT) or HS5 stroma with inhibition of TWIST (KD TWIST).
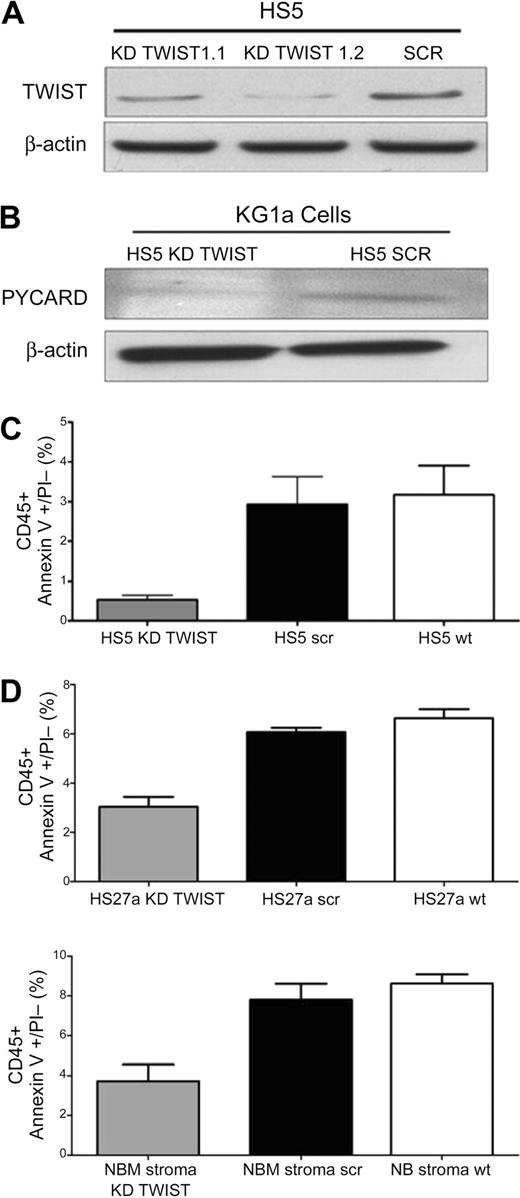
As illustrated in Figure 5A and B, suppression of TWIST in HS5 cells by specific siRNA was also associated with reduced levels of PYCARD, which we had shown previously to be up-regulated after stroma contact.12 In parallel, the rate of apoptosis declined (Figure 5C).
TWIST expression in HS5 stroma and effect on apoptosis in cocultured KG1a cells. (A) Expression of TWIST in HS5 cells transfected with scrambled siRNA (SCR) or TWIST-specific siRNA (KD TWIST 1.1 and KD TWIST 1.2). TWIST expression was determined by Western blot. β-Actin served as control. (B) Western blot of PYCARD protein in KG1a cells in contact cultures with HS5 cells, transfected with either a scrambled sequence (SCR) or with a TWIST-specific siRNA (KD TWIST), respectively. β-Actin served as loading control. PYCARD served as additional protein control for the stroma effect because we had shown previously up-regulation of PYACRD in KG1a cells after stroma contact.12 (C) Earl-stage apoptosis induced by TNFα (25 ng/mL) in KG1a cells that remained stroma-attached in control cultures containing unmodified HS5 cells (HS5 wt) and in cultures with HS5 cells transfected with either a scrambled siRNA sequence (HS5 scr) or siRNA specific for TWIST (HS5 KD TWIST). Apoptosis was determined by flow cytometry. Only CD45+ (KG1a) cells were considered. KG1a cells that had detached from stroma are not included. (D) Early-stage apoptosis induced by TNFα (25 ng/mL) in KG1a cells in contact with HS27a or normal bone marrow stroma (NBM), either unmodified (wt) or modified by transfection with a scrambled siRNA sequence (scr) or siRNA specific for TWIST (KD TWIST). Apoptosis was determined by flow cytometry. Only CD45+ (KG1a) cells were considered. KG1a cells that had detached from stroma are not included. Error bars indicate SEM.
TWIST expression in HS5 stroma and effect on apoptosis in cocultured KG1a cells. (A) Expression of TWIST in HS5 cells transfected with scrambled siRNA (SCR) or TWIST-specific siRNA (KD TWIST 1.1 and KD TWIST 1.2). TWIST expression was determined by Western blot. β-Actin served as control. (B) Western blot of PYCARD protein in KG1a cells in contact cultures with HS5 cells, transfected with either a scrambled sequence (SCR) or with a TWIST-specific siRNA (KD TWIST), respectively. β-Actin served as loading control. PYCARD served as additional protein control for the stroma effect because we had shown previously up-regulation of PYACRD in KG1a cells after stroma contact.12 (C) Earl-stage apoptosis induced by TNFα (25 ng/mL) in KG1a cells that remained stroma-attached in control cultures containing unmodified HS5 cells (HS5 wt) and in cultures with HS5 cells transfected with either a scrambled siRNA sequence (HS5 scr) or siRNA specific for TWIST (HS5 KD TWIST). Apoptosis was determined by flow cytometry. Only CD45+ (KG1a) cells were considered. KG1a cells that had detached from stroma are not included. (D) Early-stage apoptosis induced by TNFα (25 ng/mL) in KG1a cells in contact with HS27a or normal bone marrow stroma (NBM), either unmodified (wt) or modified by transfection with a scrambled siRNA sequence (scr) or siRNA specific for TWIST (KD TWIST). Apoptosis was determined by flow cytometry. Only CD45+ (KG1a) cells were considered. KG1a cells that had detached from stroma are not included. Error bars indicate SEM.
Silencing of TWIST in a second stroma cell line, HS27a, achieved effects similar to those observed with HS5 stroma (Figure 5D). Thus, inhibition of TWIST in stroma interfered with the signal required to convey apoptosis sensitivity to myeloid cells.
TWIST effect on CD54 expression in stroma and crosstalk between stroma and hematopoietic cells
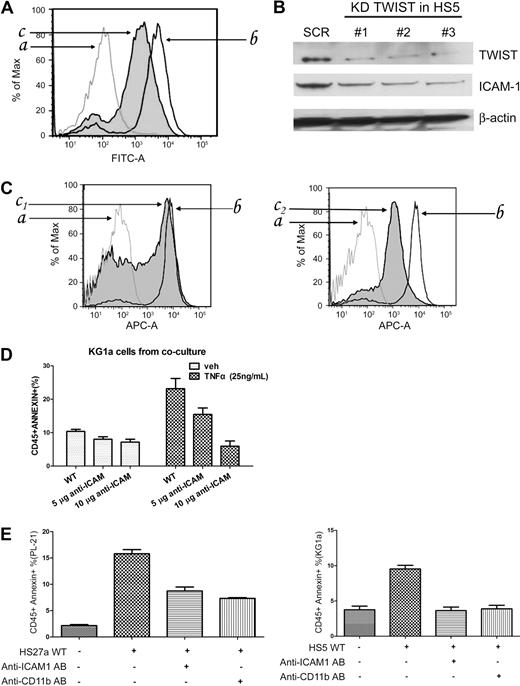
The mechanisms of stroma-mediated cell protection are multiple and involve a complex interplay of stroma-produced cytokines, chemokines, and adhesion molecules. Because TWIST has been shown to be involved in epithelial–mesenchymal transition,27 we speculated that TWIST expression might also affect adhesion molecules, such as CD54 (ICAM1). As determined by flow cytometric analysis of HS5 cells and Western blotting of HS5 lysates, CD54 expression was decreased in HS5 cells modified by TWIST-specific siRNA compared with unmodified cells (Figure 6A-B). The same pattern was observed in HS27a cells (supplemental Figure 4). Consistent with those findings, primary stroma from a patient with RA (low-grade MDS, high rate of apoptosis) expressed high levels of CD54, whereas levels were decreased in stroma from a patient with RAEB (advanced MDS, low rate of apoptosis; Figure 6C).
Inhibition of TWIST and ICAM1 (CD54) expression in HS5 cells. (A) FACS analysis of ICAM1 expression in HS5 cells transfected with scrambled siRNA (SCR) or TWIST-specific siRNA (KD TWIST). The cells were stained with FITC conjugated anti–human CD54 antibody (AB) for 30 minutes (a indicates isotype control; b, HS5 transfected with scrambled siRNA sequences; c, HS5 transfected with TWIST-specific siRNA.) (B) Western blot of TWIST and ICAM1 proteins in HS5 cells transfected with scrambled siRNA (SCR) or with 1 of 3 TWIST-specific siRNAs (KD TWIST in HS5). β-Actin served as loading control. (C) IACM1 expression in primary MDS stroma cells; a indicates normal bone marrow stroma labeled with isotype control antibody; b, normal bone marrow stroma labeled with CD54+-allophycocyanin flow antibody; c1, patients with RA MDS; c2, patients with RAEB-2 MDS. (D) Early stage apoptosis (annexin V+/propidium iodide−) in KG1a cells in control cultures containing unmodified HS5 cells (WT) or HS5 cells pretreated with either 5 or 10 μg of anti-ICAM1 antibody. Cells were cultured in the absence or presence of TNFα (25 ng/mL). Apoptosis was determined by flow cytometry. Only CD45+ (KG1a) cells were considered. (E) Early-stage apoptosis in PL-21 and KG1a cells cultured with either unmodified stroma or stroma pretreated with anti-IACM1 antibody, and apoptosis in PL-21 or KG1a cells treated with anti-CD11b antibody. Error bars indicate SEM.
Inhibition of TWIST and ICAM1 (CD54) expression in HS5 cells. (A) FACS analysis of ICAM1 expression in HS5 cells transfected with scrambled siRNA (SCR) or TWIST-specific siRNA (KD TWIST). The cells were stained with FITC conjugated anti–human CD54 antibody (AB) for 30 minutes (a indicates isotype control; b, HS5 transfected with scrambled siRNA sequences; c, HS5 transfected with TWIST-specific siRNA.) (B) Western blot of TWIST and ICAM1 proteins in HS5 cells transfected with scrambled siRNA (SCR) or with 1 of 3 TWIST-specific siRNAs (KD TWIST in HS5). β-Actin served as loading control. (C) IACM1 expression in primary MDS stroma cells; a indicates normal bone marrow stroma labeled with isotype control antibody; b, normal bone marrow stroma labeled with CD54+-allophycocyanin flow antibody; c1, patients with RA MDS; c2, patients with RAEB-2 MDS. (D) Early stage apoptosis (annexin V+/propidium iodide−) in KG1a cells in control cultures containing unmodified HS5 cells (WT) or HS5 cells pretreated with either 5 or 10 μg of anti-ICAM1 antibody. Cells were cultured in the absence or presence of TNFα (25 ng/mL). Apoptosis was determined by flow cytometry. Only CD45+ (KG1a) cells were considered. (E) Early-stage apoptosis in PL-21 and KG1a cells cultured with either unmodified stroma or stroma pretreated with anti-IACM1 antibody, and apoptosis in PL-21 or KG1a cells treated with anti-CD11b antibody. Error bars indicate SEM.
In a second, functional approach, we used a neutralizing antibody to block CD54. As shown in Figure 6D, in coculture with stroma pretreated with CD54 neutralizing antibody, KG1a cells showed a lower rate of apoptosis than in cocultures with unmanipulated HS5 stroma. These data suggested that CD54 was involved in conveying an apoptosis-facilitating signal from stroma to hematopoietic cells. Identical results were obtained with PL-21 cells cocultured with HS27a stroma (Figure 6E). Because one counter receptor of CD54 is CD11b/CD18, which is involved in apoptosis,28 we used the anti-CD11b antibody P4H9 to block CD11b on myeloid cell lines. Figure 6E illustrates that pretreatment with P4H9 resulted in decreased apoptosis sensitivity compared with unmanipulated myeloid cells.
PLCG1 in hematopoietic cells is involved in stroma-facilitated apoptosis
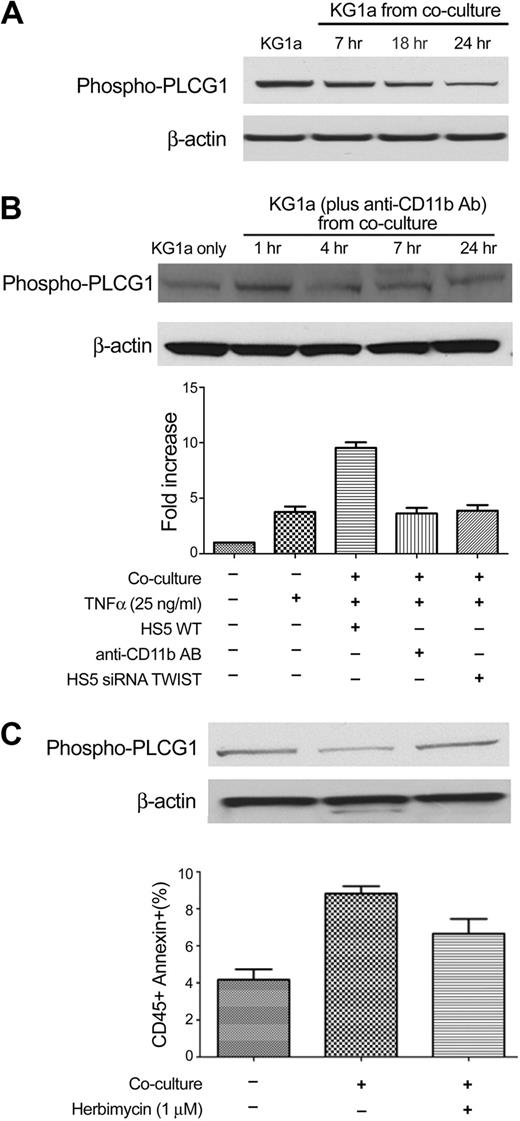
An analysis that used Ingenuity software (Ingenuity Systems Inc) showed that one factor downstream of CD11b/CD18 signaling was PLCG1, which is involved in cellular proliferation and differentiation.29,30 Furthermore, previous studies in our laboratory had shown time-dependent changes in the degree of phosphorylation of PLCG1 in KG1a cells when cocultured with stroma.31 As shown in Figure 7A, KG1a cells in contact with stroma exhibited decreased phosphorylation of PLCG1 over time relative to KG1a cells cultured without stroma. Blockade of CD11b by antibody P4H9 prevented the decrease in phosphorylation of PLCG1 in myeloid cells and reduced the rate of apoptosis (Figure 7B). These data, together with results shown in Figure 6, support the hypothesis that down-regulation of CD54 on stroma after inhibition of TWIST (in stroma) resulted in blockade of the signal, which, by interaction with CD11b/CD18, led to decreased phosphorylation of PLCG1 in cocultured KG1a cells.
PLCG1 is involved in the coculture model. (A) PLCG1 expression in KG1a cells without stroma contact (KG1a) and after coculture with HS5 cells for 7 to 24 hours. (B) Western blotting of phospho-PLCG1 in KG1a cells before and after coculture with HS5 stroma treated with CD11b antibody for various time intervals (top). (Bottom) The fold-increase in apoptosis in KG1a cells treated under different conditions is shown. The anti-CD11b antibody (AB) used was P4H9. (C) Early apoptosis in KG1a cells cultured alone (KG1a only), unmodified KG1a cocultured with HS5 (KG1a from coculture), and KG1a pretreated with herbimycin (see “Methods”) before HS5 coculture. Error bars indicate SEM.
PLCG1 is involved in the coculture model. (A) PLCG1 expression in KG1a cells without stroma contact (KG1a) and after coculture with HS5 cells for 7 to 24 hours. (B) Western blotting of phospho-PLCG1 in KG1a cells before and after coculture with HS5 stroma treated with CD11b antibody for various time intervals (top). (Bottom) The fold-increase in apoptosis in KG1a cells treated under different conditions is shown. The anti-CD11b antibody (AB) used was P4H9. (C) Early apoptosis in KG1a cells cultured alone (KG1a only), unmodified KG1a cocultured with HS5 (KG1a from coculture), and KG1a pretreated with herbimycin (see “Methods”) before HS5 coculture. Error bars indicate SEM.
To further substantiate the relevance of phosphorylation of PLCG1, we used herbimycin to block phosphorylation. Although herbimycin affects multiple molecules, Figure 7C shows that addition to coculture resulted in higher levels of phosphorylated PLCG1 and lower rates of apoptosis than that observed in control cocultured KG1a cells.
Discussion
Ineffective hematopoiesis in patients with MDS has been attributed to altered differentiation and increased apoptosis in parallel with the expansion of clonal hematopoietic stem or precursor cells. Although long considered a cell autonomous disorder, recent data suggest that the microenvironment contributes to the pathophysiology of MDS.32-36 Stroma cells are one of several key components of the microenvironment. We have shown, for example, that the co-injection of human marrow stroma cells with hematopoietic MDS cells into mice enabled propagation of the MDS clone, suggesting that stroma cells may contribute to the survival of MDS precursors.37 However, in vitro studies indicated that stroma contact conveyed signals to clonal hematopoietic precursors that rendered them sensitive to TNFα-mediated apoptosis.12 Here, we show that the transcription factor TWIST is dysregulated in MDS and is involved in apoptosis. The results obtained in a coculture model that used myeloid and stroma cell lines were confirmed with primary CD34+ marrow cells and primary stroma. The data suggest that expression of TWIST and its interactions with p53 in clonal hematopoietic cells control apoptosis. A major effect of stroma contact consisted in changes in the expression and interaction of TWIST and p53 in cocultured hematopoietic cells. Strikingly, among primary cells those changes occurred only in cells derived from MDS marrows, but not in CD34+ cells from healthy persons (Mhyre et al12 and present study). Our results show, furthermore, that TWIST expression in stroma contributed to the signaling events that reduced apoptosis resistance in cocultured hematopoietic cells. The data suggest that the stroma-derived signals were “translated” into apoptosis-facilitating events in hematopoietic cells by CD54 (ICAM1)/CD11b interactions.
TWIST was originally described in Drosophila in which mutation of the gene caused the twisted phenotype in embryos.38 In humans, a germline mutation and reduced expression of TWIST are responsible for Saethre-Chotzen syndrome.39 TWIST has been invoked to play a role in the development and progression of human cancers. For example, up-regulation of TWIST was reported in human rhabdomyosarcoma,40 gastric carcinoma,41,42 melanoma,43 breast cancer,27 prostate cancer,17 and glioma,44,45 suggesting that TWIST functions as an oncogene. TWIST was also shown to mediate taxol resistance by suppression of BAX, supporting its role as an antiapoptotic factor in mediating drug resistance.15
The present study showed significant dysregulation of TWIST expression in hematopoietic and marrow stroma cells from patients with MDS. Previous work suggests that low levels in stroma are related to high concentrations of TNF α in the marrow,7 and experiments presented here indicate that subnormal expression of TWIST in stroma is associated with up-regulation of TWIST in cocultured myeloid cells. The up-regulation of TWIST in primary CD34+ marrow cells from patients with advanced MDS suggests that TWIST is an important factor in the regulation of apoptosis, apparently involving p53, but probably also other factors. One might argue that the noted increase in TWIST was because of an expansion of the myeloblast pool, and a statistical analysis suggested, in fact, a correlation of TWIST levels and marrow myeloblasts. However, because myeloblasts in most patients with MDS express CD34, and we restricted our analysis to CD34+ cells and corrected for cell numbers, results should reflect a true increase in TWIST protein; we cannot exclude the possibility that nonclonal CD34+ cells were included in the population analyzed.
Previous studies showed that TWIST interfered with cell differentiation38 and had antiapoptotic effects.15 The present study did not examine differentiation, but the data support an antiapoptotic role of TWIST as suppression of TWIST by siRNA enhanced the rate of TNFα-induced apoptosis. We showed that p53 protein was contained in the complex precipitated by anti-TWIST antibody, indicating protein/protein interactions of the 2 molecules. Inhibition of TWIST resulted in higher expression of p53 in cell lines and in primary CD34+ MDS marrow cells and led to increased levels of Bax in KG1a cells.26 However, the fact that inhibition of TWIST enhanced apoptosis in cell lines with either wild type (eg, PL-21) or mutated p53 (eg, KG1a), suggests that factors in addition to p53, for example, PYCARD, as shown previously,12 contributed to apoptosis up-regulation, although residual function of mutated p53 has been described in other models.46
That additional pathways of apoptosis facilitation may be involved is also suggested by our observations on CD54/CD11b interactions. Reports by others had shown that TWIST regulates the expression of adhesion molecules in mesothelial cells47,48 and that adhesion molecules are involved in MDS pathophysiology.49,50 We speculated, therefore, that down-regulation of TWIST (in stroma) may modify the expression of adhesion molecules such as CD54. In keeping with reported data,51,52 our results indicate that CD54 via its counter receptor CD11b/CD18, conveyed signals to clonal hematopoietic cells, which modified PLCG1 activation and thereby facilitated apoptosis.
The present results are not necessarily in conflict with the concept proposed by others that stroma contact protects leukemic cells.53,54 Clonal myeloid cells became apoptosis sensitive only in the presence of TNFα (which is up-regulated in MDS) while in stroma contact. Furthermore, previously reported data were obtained from patients with AML, rather than MDS, although up-regulation of TWIST has also been observed in AML.55
In summary, the current studies present evidence for a role of TWIST in the pathophysiology of clonal myeloid diseases and provide data in patients with MDS, which offer new insights into stroma and hematopoietic cell interactions. The data also add to the complex picture of the pathophysiology of MDS. Although the in vivo role of TWIST remains to be proven, TWIST (or components of TWIST-dependent signaling pathways) could serve as therapeutic targets for patients with MDS.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Emily Spaulding for technical assistance. We also thank Helen Crawford and Bonnie Larson for help with manuscript preparation.
This work was supported by the National Institutes of Health (grants HL036444 and HL082941) and by a J. P McCarthy Development Grant from the Community Foundation for Southeast Michigan (A.M.M. and L.X.).
National Institutes of Health
Authorship
Contribution: X.L. proposed the investigation of TWIST in the model, carried out the experiments, and wrote the manuscript; A.M.M. contributed to some of the experiments and offered critique on the manuscript; T.A.G. carried out the statistical analyses and critiqued the manuscript; and H.J.D. co-designed the study, offered critical input, and co-wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: H. Joachim Deeg, Fred Hutchinson Cancer Research Center, D1-100, 1100 Fairview Ave North, Seattle, WA 98109; e-mail: jdeeg@fhcrc.org.

![Figure 3. Inhibition of TWIST in KG1a cells and its effect on apoptosis. Early stage apoptosis (annexin V+, propidium iodide−) in KG1a and PL-21 cell lines transfected with either a scrambled siRNA sequence (SCR) or siRNA specific for TWIST (siTWIST), and exposed overnight to normal medium (veh) versus exogenous TNFα (25 ng/mL, A) or TRAIL (100 ng/mL, B), respectively. Apoptosis was determined by flow cytometry. (C) Cell preparation as in panels A and B, but cultured in normal medium (veh) or in the presence of the soluble TNF receptor etanercept (10 μg/mL for 24 hours [concentration based on ancillary studies]). Error bars indicate SEM. (D) Protein lysates from the same cell preparations were separated on 4% to 12% Bis-Tris gels and immunoblotted with antibodies against TWIST, p53-ser46, Bax, and caspase 9, respectively. β-Actin served as loading control. (E) TWIST and p53 levels in primary CD34+ MDS cells in which TWIST was silenced by specific siRNA (KD TWIST) compared with levels in unmodified cells from the same primary CD34+ MDS sample.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/13/10.1182_blood-2009-09-242313/4/m_zh89991057670003.jpeg?Expires=1769117740&Signature=Evg25BFQI8pT1DigvEQkZX1u3jrmZ7n4fSPoEHBg~rW3NqIGmH3YYffmLFvGA9Oc4gKTIYWQ6kuQ8jl3yZZ1w59iZE82KzzlpGHGImvXRZvhvGp33Q01cjLvzlJR1yQClXyNaJETuk47rsi6t1rJgw0ehFc4paRYbTIW8Hf7VV0ydDaircvKX9MhGO-w7P~zHHKpQ15hBhEEgIGdl4MhRS5PMmph-ueArTXFVUC7Q9nWTTmltmoUVOg9w~8WkXOLecpKGV5TIS-g~4jWeo2vrEmjk4chRxQjZ0uAb7FZ~XO9c2qUrT9H8UvHlcv4uYrXsgyRCajEfhLevpXhcvIrRA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)