Abstract
Trib1 is a myeloid oncogene that cooperates with Hoxa9 and Meis1. Although the MAPK pathway and C/EBP transcription factors are known to interact with Trib proteins, the mechanisms by which Trib1 contributes to myeloid leukemogenesis remains to be clarified. Here we report that interaction between Trib1 and MEK1 is required for Trib1-induced leukemogenesis. The C-terminal ILLHPWF motif that is well conserved among Trib family proteins is required for MEK1 binding, enhancement of ERK phosphorylation, enhanced self-renewal activity of bone marrow cells and leukemogenic activity by Trib1. The motif is also important for Trib1-induced C/EBPα degradation though interaction between Trib1 and C/EBPα is not necessary. Inhibition of ERK phosphorylation suppressed Trib1-induced C/EBPα degradation, indicating an important role for Trib1/MEK1 interaction. These results suggest that Trib1 may be a key mediator between the RTK-MAPK pathway and the C/EBP transcription factor in myeloid leukemogenesis.
Introduction
Leukemic cells are characterized by molecular abnormalities that dysregulate proliferation, survival, and differentiation. Signal transduction is important for regulating such cellular processes and it is frequently involved in malignant transformation of myeloid progenitors. A number of gain-of-function mutations within the RTK-RAS-MAPK pathway have been identified in human myeloid leukemias, and these mutations enhance proliferation and survival of myeloid cells.1,2 Most mutations are found in RTK such as ABL1, PDGFR, FLT3 and c-KIT.1 Mutant downstream messengers or regulators including RAS, JAK2, SHP2 and NF1 are found less frequently.2 As a result of the mutations downstream molecules such as ERK, ELK, jun/fos and CDKs are activated,2,3 resulting in increased proliferative activity of hematopoietic cells.
Tribbles regulates String/CDC25 activity and affects cell proliferation, migration and morphogenesis.4,5 In addition, tribbles regulates both MEK/ERK phosphorylation and C/EBP degradation.6-8 Recently, 2 of 3 mammalian homologues of tribbles, Trib1 and Trib2, have been found causally activated in myeloid leukemia.9,10 The Trib1 gene cooperates with Hoxa9/Meis1 and is a transforming gene by itself in myeloid leukemogenesis,9 and over-expression of Trib1 enhances ERK phosphorylation, reducing apoptosis of leukemic cells upon interleukin-3 (IL-3) depletion. In addition, over-expression of Trib2 induces proteasomal-dependent degradation of C/EBPα by physical interaction.10 These results raised the possibility that Trib1 might be important for myeloid cell transformation, functioning as a key mediator between the MAPKK/MAPK pathway and C/EBP transcription factors.
Here, we show that Trib1 interacts with MEK1 through its C-terminal ILLHPWF motif, resulting in enhanced ERK phosphorylation. The MEK1 interaction motif is required for self-renewal activity of bone marrow cells as well as leukemogenesis. Moreover, the motif is important for C/EBPα degradation by Trib1. These results demonstrate a novel functional role of Trib1 as well as linkage between MEK/ERK and C/EBPα in myeloid leukemogenesis.
Methods
Plasmid constructs
Retroviral constructs bearing Trib1 and Hoxa9/Meis1 were described previously.9 Mek1 and Trib1 deletion mutants were generated in pCDNA3.1 (Invitrogen) or p3xFLAG CMV (Sigma-Aldrich), respectively, by PCR using the full-length murine Mek1 cDNA (a gift from Dr Katsuji Yoshioka, Kanazawa University) or Trib1 cDNA as templates. The Trib1 point mutant W337A was generated using the KOD-Plus Mutagenesis kit (Toyobo) according to the manufacturer's protocol. These mutants were also transferred into the pMYs-IRES-GFP retrovirus vector. The retrovirus stock was generated as described previously.9
Cell culture and transfection
HeLa, U2OS and 293 cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum (FBS). NH112 acute myeloid leukemia (AML) cells that were derived from the NUP98-HOXA9 transgenic mouse,11 and BaF3 cells were maintained in RPMI1640 medium supplemented with 10% FBS and IL-3 (10 ng/mL; R&D Systems). Cytokine-starvation was performed in RPMI1640 medium containing 0.5% FBS, after which IL-3 (20 ng/mL) was added for stimulation. Transfection of plasmid DNA to HeLa or U2OS cells was carried out using LipofectAmine 2000 reagents (Invitrogen). NH112 and BaF3 cells were infected with retrovirus by spinfection at 1400g for 2 hours. For the C/EBPα degradation study NH112 cells were cultured with 125 μg/mL cycloheximide (Sigma-Aldrich) and/or 10μM U0126 (Cell Signaling Technologies) for 1, 2, or 4 hours.
Immunoprecipitation and immunoblotting
Immunoprecipitation and immunoblotting were performed using cell lysates in RIPA buffer as described.12 The following primary antibodies were used: monoclonal anti-HA (Invivogen), polyclonal anti-HA (MBL), anti-FLAG (monoclonal and polyclonal; Sigma-Aldrich), anti–β-tubulin (Sigma-Aldrich), anti-p44/42 ERK (Cell Signaling Technologies), anti–phospho-p44/42 ERK (Cell Signaling Technologies), anti-MEK1/2 (Cell Signaling Technologies) and anti–p-C/EBPα (Cell Signaling Technologies). Secondary anti–mouse and anti–rabbit horseradish peroxidase-conjugated antibodies were obtained from Amersham.
Immunofluorescence
Cells cultured in 4-well chamber slides (BD Falcon) were transfected with plasmids. Twenty-four hours after transfection, cells were washed with PBS and fixed with 100% methanol. Samples were stained with the indicated primary antibodies followed by FITC- or TRITC-conjugated secondary antibodies. Nuclei were stained with Hoechst 33342 and mounted in ProlongGold (Invitrogen). Images were obtained and analyzed by a Leica AS MDW confocal microscopy (Leica Microsystems) equipped with a 100×/1.40 numeric aperture (NA) oil objective or Leica CTR6500 microscope. Images were acquired with a DFC 350FX CCD camera (Leica) using Leica Confocal software Version 2.61 and were processed using Adobe Photoshop CS2 Version 9.0 (Adobe Systems).
Replating assay
Retroviral transduced bone marrow cells were subjected to replating assays without subsequent selection as described.9 Colonies were counted and analyzed morphologically after 10 days, then harvested and the same number of cells were replated in the same way.
Animals, retroviral infection, and bone marrow transfer
Bone marrow cells were prepared from 8-week-old female C57Bl/6J mice purchased from CLEA Japan Inc 5 days after injection of 150 mg/kg body weight of 5-fluorouracil (5-FU; Kyowa Hakko Kogyo). Retroviral infection of bone marrow cells and bone marrow transfer experiments were performed as described.9 Animals were housed, observed daily, and handled in accordance with the guidelines of the animal care committee at the Japanese Foundation for Cancer Research, which gave ethical approval for these studies. Mice were monitored daily for evidence of disease, and smear samples and GFP-positive populations of peripheral blood were examined every week. The onset of AML was determined by detecting myeloblasts in peripheral blood. All the diseased mice were subjected to autopsy and analyzed morphologically and the blood was examined by flowcytometric techniques. When mice had more than 20% myeloid blasts in the bone marrow, they were diagnosed as positive for AML, as indicated in the classification of the Bethesda proposal.13 The survival rate of each group was evaluated using the Kaplan-Meier test.
Flow cytometry
GFP-positivity of blood cells was examined using a FACSCalibur flow cytometer (Becton Dickinson). Single-cell suspensions of 1 × 106 cells were incubated with fluorescein isothiocyanate-conjugated antibodies, Gr-1, Mac-1, B220, or CD3 (Pharmingen) for 30 minutes, washed 3 times in PBS and analyzed with the FACSCalibur.
Results
Identification of Trib1 interaction domains in MEK1
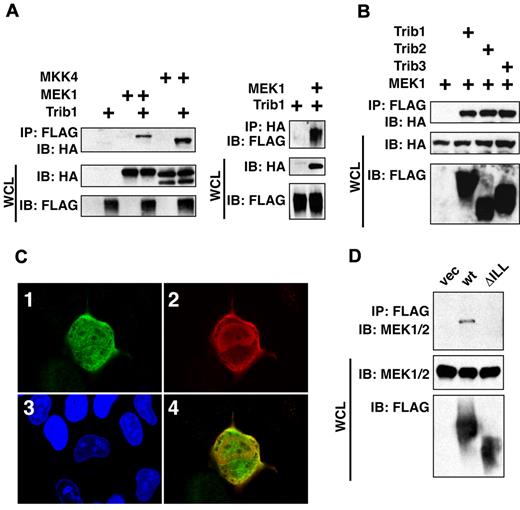
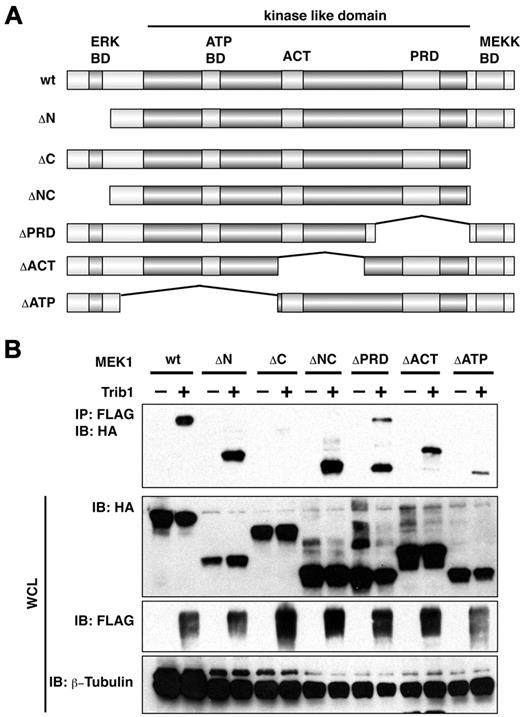
Trib1 was coimmunoprecipitated with MEK1 and MKK4 (Figure 1A), and all 3 Trib family members were shown to interact with MEK1 (Figure 1B). When introduced into U2OS cells Trib1 was predominantly localized in the nucleus, but it was also in the cytoplasm where it colocalized with MEK1 (Figure 1C). Furthermore, coimmunoprecipitation and partial colocalization between Trib1 and endogenous MEK1 was also exhibited (Figure 1D). A series of deletion constructs of MEK1 were then generated and interaction with Trib1 was examined. As shown in Figure 2A and B, Trib1 binding activity was significantly reduced in both ΔC and ΔATP mutants. The reduction was more significant in ΔC, and Trib1 interaction was recovered in ΔNC that lacks both N- and C-terminal domains. Immunofluorescence showed that the nuclear localization of MEK1 ΔC was abrogated, whereas ΔNC remained in both the nucleus and the cytoplasm (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). These findings are consistent with previous observation that the ΔC mutant is translocated to membrane compartments outside of the nucleus,14 suggesting that interaction between Trib1 and MEK1 might occur within portions of the nucleus and that there might be 2 interaction domains, the C-terminus and the ATP binding domain. Trib1 enhances ERK phosphorylation upon cytokine stimulation,6,9 suggesting a possible conformational change that might affect the interaction.
Trib family proteins interact with MAPKKs in vivo. (A) HeLa cells were transiently transfected with FLAG-tagged Trib1 and HA-tagged MEK1 or MKK4. The cell lysates were immunoprecipitated with an anti-FLAG monoclonal antibody and immunoblotted with a rabbit anti-HA antibody (left) or immunoprecipitated with an anti-HA followed by immnunoblotting with anti-FLAG (right). The expression level of each protein was assessed by immunoblotting whole cell lysates (WCL) with anti-HA or anti-FLAG antibodies. (B) HeLa cells were transiently transfected with FLAG-tagged Trib1, Trib2 or Trib3 with HA-tagged MEK1. The cell lysates were immunoblotted with an anti-HA polyclonal antibody. The expression level of each protein was assessed by immunoblotting whole cell lysates with anti-HA or anti-FLAG antibodies. (C) Colocalization of MEK1 and Trib1. Trib1 and MEK1 were detected by immunofluorescence using an anti-FLAG antibody followed by a FITC-labeled secondary antibody or an anti-HA antibody followed by a TRITC-labeled secondary antibody. i, HA; ii, FLAG; iii, DNA; iv, Merge. (D) Coimmunoprecipitation between Trib1 and endogenous MEK1 in HeLa cells. The cell lysates were immunoprecipitated with an anti-FLAG monoclonal antibody and immunoblotted with a rabbit anti-MEK1/2 antibody. The expression level of each protein was assessed by immunoblotting whole cell lysates (WCL) with anti-MEK1/2 or anti-FLAG antibodies.
Trib family proteins interact with MAPKKs in vivo. (A) HeLa cells were transiently transfected with FLAG-tagged Trib1 and HA-tagged MEK1 or MKK4. The cell lysates were immunoprecipitated with an anti-FLAG monoclonal antibody and immunoblotted with a rabbit anti-HA antibody (left) or immunoprecipitated with an anti-HA followed by immnunoblotting with anti-FLAG (right). The expression level of each protein was assessed by immunoblotting whole cell lysates (WCL) with anti-HA or anti-FLAG antibodies. (B) HeLa cells were transiently transfected with FLAG-tagged Trib1, Trib2 or Trib3 with HA-tagged MEK1. The cell lysates were immunoblotted with an anti-HA polyclonal antibody. The expression level of each protein was assessed by immunoblotting whole cell lysates with anti-HA or anti-FLAG antibodies. (C) Colocalization of MEK1 and Trib1. Trib1 and MEK1 were detected by immunofluorescence using an anti-FLAG antibody followed by a FITC-labeled secondary antibody or an anti-HA antibody followed by a TRITC-labeled secondary antibody. i, HA; ii, FLAG; iii, DNA; iv, Merge. (D) Coimmunoprecipitation between Trib1 and endogenous MEK1 in HeLa cells. The cell lysates were immunoprecipitated with an anti-FLAG monoclonal antibody and immunoblotted with a rabbit anti-MEK1/2 antibody. The expression level of each protein was assessed by immunoblotting whole cell lysates (WCL) with anti-MEK1/2 or anti-FLAG antibodies.
Trib1 interaction domains in MEK1. (A) Constructs of MEK1 mutants. BD indicates binding domain. (B) U2OS cells were transiently transfected with FLAG-tagged Trib1 and HA-tagged MEK1. The cell lysates were immunoprecipitated with an anti-FLAG polyclonal antibody and immunoblotted with an anti-HA monoclonal antibody. The expression level of each protein was assessed by immunoblotting whole cell lysates with anti-HA or anti-FLAG antibodies. ACT indicates activation domain; and PRD, proline-rich domain
Trib1 interaction domains in MEK1. (A) Constructs of MEK1 mutants. BD indicates binding domain. (B) U2OS cells were transiently transfected with FLAG-tagged Trib1 and HA-tagged MEK1. The cell lysates were immunoprecipitated with an anti-FLAG polyclonal antibody and immunoblotted with an anti-HA monoclonal antibody. The expression level of each protein was assessed by immunoblotting whole cell lysates with anti-HA or anti-FLAG antibodies. ACT indicates activation domain; and PRD, proline-rich domain
The C-terminus of Trib1 is required for MEK1 binding as well as enhanced self-renewal activity of bone marrow cells
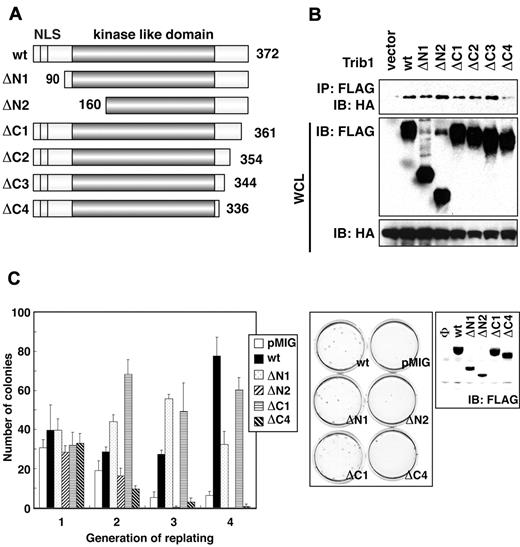
The Trib1 gene cooperates with Hoxa9 and Meis1 in myeloid leukemogenesis, and Trib1 by itself shows transforming activity for murine bone marrow cells.9 Although we have shown enhanced ERK phosphorylation and its role in inhibition of apoptosis,9 the important functional mechanisms of Trib1 in leukemogenesis remained unclear and a possible role of interaction between Trib2 and C/EBPα in leukemogenesis has been reported.10 Trib family proteins retain high sequence homologies to one another and each possesses a highly conserved, single kinase-like domain in the middle of the proteins.15 Short conserved motifs are also scattered both in N- and C-terminal regions.16 To examine the domains responsible for MEK1 binding as well as enhanced self-renewal activity of bone marrow cells, we generated a series of Trib1 deletion mutants (Figure 3A). Complete removal of the N-terminal region did not affect MEK1 binding activity (Figure 3A-B). The ΔC3 mutant that lacked 28 amino acids from the C-terminus still retained binding activity. However, MEK1 binding was abolished by deleting additional 8 amino acids (ΔC4 in Figure 3A). Because Trib1 enhances murine bone marrow cell self-renewal activity,9 a replating assay was performed using retroviral vectors bearing deletion constructs. As expected, the ΔC4 mutant lost enhanced self-renewal activity (Figure 3C). The ΔN2 mutant (in which a long sequence was deleted from the kinase-like domain) also lost enhanced self-renewal activity for bone marrow cells.
The Trib1 regions required for enhanced self-renewal activity of bone marrow cells. (A) Constructs of Trib1 mutants. NLS indicates a nuclear localization signal. (B) HeLa cells were transiently transfected with FLAG-tagged Trib1 and HA-tagged MEK1. The cell lysates were immunoblotted with an anti-HA polyclonal antibody. The expression level of each protein was assessed by immunoblotting whole cell lysates with anti-HA or anti-FLAG antibodies. (C) Replating assay. Primary murine bone marrow cells were infected with retroviruses bearing the indicated mutants. The colony numbers in methylcellulose culture during 4 serial replatings were measured. The means ± SDs from 3 independent experiments are shown (left). Representative results of methylcellulose cultures are shown at center. Expression of FLAG-tagged Trib1 mutants in packaging cells was detected by immunoblotting with anti-FLAG mAb (right).
The Trib1 regions required for enhanced self-renewal activity of bone marrow cells. (A) Constructs of Trib1 mutants. NLS indicates a nuclear localization signal. (B) HeLa cells were transiently transfected with FLAG-tagged Trib1 and HA-tagged MEK1. The cell lysates were immunoblotted with an anti-HA polyclonal antibody. The expression level of each protein was assessed by immunoblotting whole cell lysates with anti-HA or anti-FLAG antibodies. (C) Replating assay. Primary murine bone marrow cells were infected with retroviruses bearing the indicated mutants. The colony numbers in methylcellulose culture during 4 serial replatings were measured. The means ± SDs from 3 independent experiments are shown (left). Representative results of methylcellulose cultures are shown at center. Expression of FLAG-tagged Trib1 mutants in packaging cells was detected by immunoblotting with anti-FLAG mAb (right).
The ILLHPWF motif is important for the enhanced replating activity of Trib1
A homology search identified a septapeptide IL(D/L)HPW(F/L) conserved among the 3 Trib proteins (Figure 4A). The sequence of the motif is also well conserved in drosophila tribbles as IFLTPWL.17 The role of the motif in MEK1 binding was further confirmed by showing that the binding was again abrogated by deletion of the septapeptide (ΔILLHPWF) or replacement of a conserved tryptophan residue with alanin (W337A; Figures 1D, 4B). To examine the role of the septapeptide in Trib1's interaction with MEK1 and the enhancement of ERK phosphorylation, the mutant constructs as well as wild-type were introduced into murine bone marrow cells. As expected ΔC4, ΔILLHPWF and W337A mutants lost their capacity to enhance ERK phosphorylation (Figure 4C), indicating that interaction with MEK1 is required for Trib1's enhancement of ERK phosphorylation.
Identification of the Trib1 motif required for MEK1 binding, enhanced ERK phosphorylation and self-renewal activity. (A) Constructs of Trib1 mutants and a conserved amino acid motif in Trib family proteins. (B) HeLa cells were transiently transfected with FLAG-tagged Trib1 and HA-tagged MEK1. The cell lysates were immunoblotted with an anti-HA polyclonal antibody. The expression level of each protein was assessed by immunoblotting whole cell lysates with anti-HA or anti-FLAG antibodies. (C) Mouse primary bone marrow cells were infected with indicated FLAG-Trib1 retroviruses. The cell lysates were immunoblotted with the indicated antibodies. (D) Replating assay. Primary bone marrow cells were infected with the indicated retroviruses. The colony numbers in methylcellulose culture during 3 replatings were measured. The means ± SDs from 3 independent experiments are shown (left). The representative results of methylcellulose culture are shown (right).
Identification of the Trib1 motif required for MEK1 binding, enhanced ERK phosphorylation and self-renewal activity. (A) Constructs of Trib1 mutants and a conserved amino acid motif in Trib family proteins. (B) HeLa cells were transiently transfected with FLAG-tagged Trib1 and HA-tagged MEK1. The cell lysates were immunoblotted with an anti-HA polyclonal antibody. The expression level of each protein was assessed by immunoblotting whole cell lysates with anti-HA or anti-FLAG antibodies. (C) Mouse primary bone marrow cells were infected with indicated FLAG-Trib1 retroviruses. The cell lysates were immunoblotted with the indicated antibodies. (D) Replating assay. Primary bone marrow cells were infected with the indicated retroviruses. The colony numbers in methylcellulose culture during 3 replatings were measured. The means ± SDs from 3 independent experiments are shown (left). The representative results of methylcellulose culture are shown (right).
The importance of the ILLHPWF motif was further confirmed using a replating assay (Figure 4D). Both ΔILLHPWF and W337A mutants lacking MEK1 interaction lost the enhanced replating activity, similar as the ΔC4 mutant. These results strongly suggest that interaction with MEK1 is essential for Trib1-induced increased self-renewal activity.
The ILLHPWF motif is required for Trib1-induced leukemogenesis as well as cooperation with Hoxa9 and Meis1
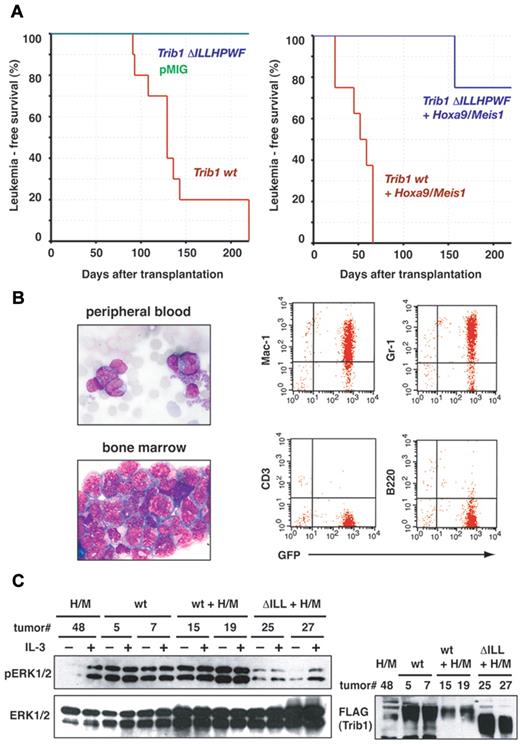
The role of the ILLHPWF motif in leukemogenesis was examined by bone marrow transfer experiments. Wild-type Trib1 and ΔILLHPWF mutant cDNAs were introduced into the pMYs-IRES-GFP retrovirus and 5FU-treated mouse bone marrow cells were infected with the Trib1 retrovirus alone or coinfected with the Hoxa9/Meis1 retrovirus. As shown in our previous study expression of wild-type Trib1 induced AML, and coexpression with Hoxa9/Meis1 induced AML with a very short latency (Figure 5A-B and supplemental Figure 2). On the other hand the ΔILLHPWF mutant did not induce AML nor did it cooperate with Hoxa9/Meis1.
The ILLHPWF sequence is required for Trib1 leukemogenic activity. (A) Leukemia-free survival of animals was compared. Kaplan-Meier survival curves are shown for Trib1 wt (n = 10), Trib1 ΔILLHPWF (n = 10) and pMYs-GFP (n = 8; left), or for Hoxa9/Meis1 + Trib1 wt (n = 8) and Hoxa9/Meis1 + Trib1 ΔILLHPWF (n = 8; right). (B) Representative phenotypes of Trib1-induced AML. Left: leukemic cells in peripheral blood and bone marrow smears (Wright-Giemsa staining). Original magnification, 1000×. Right: flow cytometric analysis. (C) Leukemic cells expressing Hoxa9-Meis1 (H/M), Trib1 wt (wt), Trib1 wild-type/Hoxa9-Meis1 (wt + H/M) and Trib1 ΔILLHPWF/Hoxa9-Meis1 (ΔILL + H/M) were cultured in the absence of IL-3 for 16 hour (−IL-3) and then stimulated with IL-3 for 15 minutes (+IL-3). The cell lysates were immunoblotted with indicated antibodies.
The ILLHPWF sequence is required for Trib1 leukemogenic activity. (A) Leukemia-free survival of animals was compared. Kaplan-Meier survival curves are shown for Trib1 wt (n = 10), Trib1 ΔILLHPWF (n = 10) and pMYs-GFP (n = 8; left), or for Hoxa9/Meis1 + Trib1 wt (n = 8) and Hoxa9/Meis1 + Trib1 ΔILLHPWF (n = 8; right). (B) Representative phenotypes of Trib1-induced AML. Left: leukemic cells in peripheral blood and bone marrow smears (Wright-Giemsa staining). Original magnification, 1000×. Right: flow cytometric analysis. (C) Leukemic cells expressing Hoxa9-Meis1 (H/M), Trib1 wt (wt), Trib1 wild-type/Hoxa9-Meis1 (wt + H/M) and Trib1 ΔILLHPWF/Hoxa9-Meis1 (ΔILL + H/M) were cultured in the absence of IL-3 for 16 hour (−IL-3) and then stimulated with IL-3 for 15 minutes (+IL-3). The cell lysates were immunoblotted with indicated antibodies.
Enhanced phosphorylation of ERK in Trib1-induced AML was then examined. Over-expression of Trib1 frequently induces constitutive phosphorylation of ERK in AML. Figure 5C shows that both Trib1 and Trib1/Hoxa9/Meis1-induced AML enhanced phosphorylation of ERK regardless of the presence of IL-3. On the other hand, AML in which Hoxa9/Meis1 alone or Hoxa9/Meis1 plus the ΔILLHPWF mutant was expressed did not show enhanced ERK phosphorylation in the absence of IL-3 stimulation. These results again indicate the important role of the ILLHPWF motif in leukemogenesis, and suggest that the enhanced MEK1/ERK pathway cooperates with Hoxa9 and Meis1 in the induction of transcriptional dysregulation in myeloid cells.
Trib1/MEK1 interaction induces down-regulation of C/EBPα
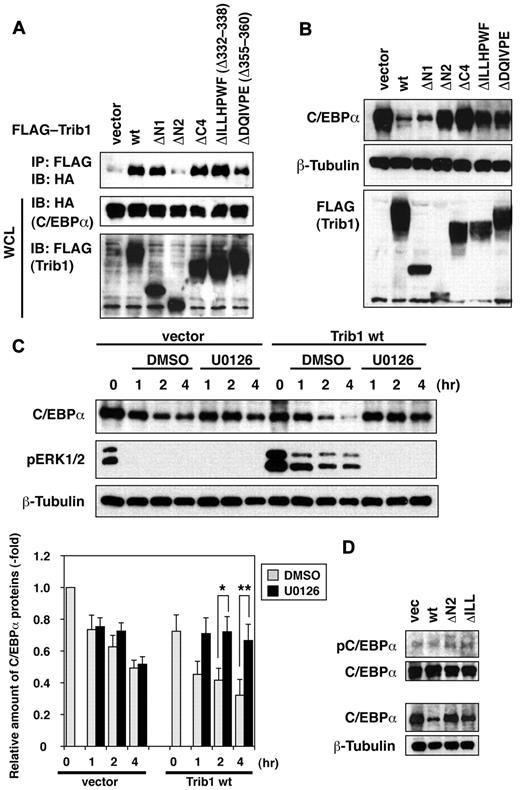
In addition to interacting with MEK1/MAPK, Trib family proteins also interact with C/EBP transcription factors including C/EBPα, C/EBPβ and CHOP.10,18,19 When C/EBPα was coexpressed with Trib1 constructs in HeLa cells, its interaction with wild-type Trib1 was confirmed (Figure 6A). The ΔN1 mutant retained the capacity to interact with C/EBPα whereas the ΔN2 mutant failed to associate with C/EBPα (Figure 6A). This result indicated that Trib1's binding motif for C/EBPα is located at the N-terminal end of the kinase-like domain. The C-terminal deletion mutants, ΔC4 and ΔILLHPWF, retained the binding activity for C/EBPα, indicating that Trib1 uses different mechanisms to interact with MEK1 and C/EBPα. To investigate the effect on C/EBPα down-regulation, Trib1-expressing retroviral vectors were introduced into NH112 AML cells that expressed C/EBPα. As expected, the wild-type Trib1 and ΔN1 that retained C/EBPα interaction significantly down-regulated the expression of C/EBPα p42 but not p30 whereas p42 expression was not affected by ΔN2 (Figure 6B and data not shown). Surprisingly, ΔC4 and ΔILLHPWF both of which retain the interaction domain failed to down-regulate C/EBPα. The ΔDQIVPE mutant, which lacks the binding motif for the E3 ubiquitin ligase COP1,20 also failed to show C/EBPα down-regulation. The Trib1-mediated decrease in C/EBPα protein stability was confirmed by cycloheximide treatment of NH112 cells (Figure 6C). When ERK phosphorylation was inhibited by the MEK1 inhibitor U0126, down-regulation of C/EBPα by Trib1 was abrogated (Figure 6C). The results indicate involvement of the MEK/ERK pathway in Trib1-induced C/EBPα down-regulation. It has been reported that ERK1/2 phosphorylates C/EBPα at serine,21 and we have tested whether Trib1 could modify this phosphorylation. As shown in Figure 6D, residual C/EBPα in Trib1-expressing NH112 cells were phosphorylated at the similar level as in NH112 cells with an empty vector or Trib1 mutants, suggesting that serine 21 phosphorylation might not be responsible for C/EBPα degradation.
Degradation of C/EBPα by Trib1 is dependent on the MAP kinase pathway. (A) Interaction between Trib1 and C/EBPα. 293 cells were transiently transfected with FLAG-tagged Trib1 and HA-tagged C/EBPα. The expression level of each protein was assessed by immunoblotting whole cell lysates with anti-HA or anti-FLAG antibodies. (B) NH112 cells were infected with the indicated FLAG-Trib1 retroviruses. The cell lysates were immunoblotted with indicated antibodies. (C) NH112 cells infected with an empty retrovirus (vector) or the Trib1 retrovirus (Trib1 wt) were treated with cycloheximide with/without U0126 for the indicated periods. The cell lysates were immunoblotted with the indicated antibodies. The relative intensities of C/EBPα bands compared with β-tubulin bands are measured (bottom). The values are means ± SE s from 4 independent experiments. Statistical signicicances were confirmed using the Mann-Whitney test with the Dunn-Sidak post hoc test (*P = .026, **P = .00866), and 1-way analysis of variance for repeated measures with the Dunnett test was also performed. Full description for the statistical analysis is available as supplemental information. (D) Phosphorylation status of C/EBPα serine 21 was assessed in NH112 cells infected with an empty retrovirus (vec), wild-type Trib1 (wt), ΔN2 mutant (ΔN2), or ΔILLHPWF (ΔILL) mutant retroviruses. The expression of total C/EBPα is shown at the bottom with β-tubulin as a loading control, and phosphorylated C/EBPα (pC/EBPα) in equal amounts of total C/EBPα is exhibited at the top panel.
Degradation of C/EBPα by Trib1 is dependent on the MAP kinase pathway. (A) Interaction between Trib1 and C/EBPα. 293 cells were transiently transfected with FLAG-tagged Trib1 and HA-tagged C/EBPα. The expression level of each protein was assessed by immunoblotting whole cell lysates with anti-HA or anti-FLAG antibodies. (B) NH112 cells were infected with the indicated FLAG-Trib1 retroviruses. The cell lysates were immunoblotted with indicated antibodies. (C) NH112 cells infected with an empty retrovirus (vector) or the Trib1 retrovirus (Trib1 wt) were treated with cycloheximide with/without U0126 for the indicated periods. The cell lysates were immunoblotted with the indicated antibodies. The relative intensities of C/EBPα bands compared with β-tubulin bands are measured (bottom). The values are means ± SE s from 4 independent experiments. Statistical signicicances were confirmed using the Mann-Whitney test with the Dunn-Sidak post hoc test (*P = .026, **P = .00866), and 1-way analysis of variance for repeated measures with the Dunnett test was also performed. Full description for the statistical analysis is available as supplemental information. (D) Phosphorylation status of C/EBPα serine 21 was assessed in NH112 cells infected with an empty retrovirus (vec), wild-type Trib1 (wt), ΔN2 mutant (ΔN2), or ΔILLHPWF (ΔILL) mutant retroviruses. The expression of total C/EBPα is shown at the bottom with β-tubulin as a loading control, and phosphorylated C/EBPα (pC/EBPα) in equal amounts of total C/EBPα is exhibited at the top panel.
Discussion
In this study, we determined that Trib1's MEK1 interaction domain resides at the border between the kinase-like domain and the C-terminal region. The IL(L/D)HPW(F/L) motif is well conserved in 3 mammalian Trib proteins.15 Moreover, the conservation is extended to invertebrates and the tryptophan residue is invariably conserved even in drosophila tribbles.16 We found that the motif is required for MEK1 binding, and the mutant that lacks the entire motif as well as the tryptophan mutant lost the binding activity. Replating assays revealed that the motif is critical for increased self-renewal activity of bone marrow cells by Trib1. Moreover, the ΔILLHPWF mutant lost leukemogenic activity and cooperative activity for Hoxa9 and Meis1 in leukemogenesis. In addition, expression of the ΔILLHPWF mutant might suppress the leukemogenic activity of Hoxa9/Meis1. Although additional experiments will be required to confirm the dominant negative effect of the mutant, we confirmed that the copy numbers of the Hoxa9/Meis1 retrovirus in bone marrow cells were similar between leukemic and nonleukemic BMT mice (supplemental Figure 3).
Two Trib1 interaction domains that correspond to ATP binding and C-terminal regions of MEK1 have been identified in the present study. The C-terminal deletion mutant of MEK1 showed less interaction activity for Trib1. The MEK1 C-terminal region contains the Raf1-interaction domain. MEK1 activation is inhibited and is translocated to lysosomal compartments by deletion of this region.14 Altered localization of MEK1 might cause greater inhibition of its Trib1 interaction. Overlapping of RAF and Trib1 interaction domains is also intriguing, given the possible Trib1 modulation of MEK1 activation by RAF. We examined the effect of Trib1 on BRAF-induced MEK1 phosphorylation by an in vitro kinase assay; however, no significant modulation was observed (data not shown).
Trib1 has been identified as a common retroviral integration site in Hoxa9/Meis1-induced AML.9 There is specific genetic interaction between Trib1 and Hoxa9/Meis1 in myeloid leukemogenesis, suggesting that Trib1 and homeodomain proteins are located in functionally independent molecular pathways. Our previous study showed that Trib1 overexpression induces enhanced ERK phosphorylation in hematopoietic cells. Enhanced and prolonged ERK phosphorylation by Trib1 is closely linked to suppression of apoptosis after cytokine depletion. These results indicate the important role of MEK/ERK function for Trib1 leukemogenic activity, and suggest that Trib1 activation may be equivalent to other mutations in the RTK-ERK pathway such as FLT3 and JAK2 mutations. In human AML possible TRIB1 involvement has been reported in cases with 8q24 amplification.22,23 It would therefore be intriguing to know whether TRIB1 activation and FLT3 or JAK2 mutations are mutually exclusive in human AML cases. In Meis1/Hoxa9-induced AML up-regulation of FLT3 expression is induced,24 which may enhance the effect of Trib1 on ERK phosphorylation.
There are important functional linkages between Trib family proteins and C/EBP family transcription factors.8,19,25 C/EBPα plays an important role in myeloid differentiation and its mutations are frequently associated with AML.26 Proteasome-dependent degradation of C/EBPα by Trib2 has been proposed as an important mechanism in murine myeloid leukemogenesis.10 The E3 ubiquitin ligase COP1 mediates C/EBP degradation by Trib3,20 and the COP1 binding motif is conserved in all 3 mammalian Trib proteins in their C-termini. Our present study confirmed that the mutant lacking the COP1 binding motif lost the capacity to down-regulate C/EBPα. Moreover, C/EBPβ was found interacting with Trib1 and up-regulated in Trib1-deficient macrophages.25 The data indicate that Trib1 is an important negative regulator of C/EBP family transcription factors.
It remains to be clarified how MEK1/ERK function modulates C/EBPα activity.27 Activation of MEK1/ERK signaling phosphorylates C/EBPα and as a result of phosphorylation its function is suppressed.28 Our present results suggest that Trib1 promotes C/EBPα down-regulation through enhancement of MEK1/ERK activities. Furthermore, down-regulation of C/EBPα by Trib1 requires interaction between Trib1 and MEK1, whereas Trib1 interaction with C/EBPα is dispensable. The functional significance of physical interaction between Trib1 and C/EBPα is unclear. One possible mechanism is that Trib1 itself protects C/EBPα from degradation by direct interaction. Alternatively, excess amounts of Trib1 that can no longer bind endogenous C/EBPα may bind MEK1 and promote C/EBPα degradation. Further experiments are needed to distinguish between these mechanisms.
Although we and others have proposed that Trib1 and Trib2 are dominant oncogenes for AML,9,10 the role of the genes in tumorigenesis is not yet fully clarified. A recent study showed that over-expression of Trib1 and Trib2 suppressed growth of Me-1 AML cells and knockdown of either genes showed slight enhancement of Me-1 proliferation.29 It is well-known that some oncogenic stimuli result in growth advantage or growth suppression/apoptosis depends on cellular context and the mutation status of other cancer-related genes.30-32 It is therefore intriguing to attempt more comprehensive analysis of signaling network involving Trib family proteins, and to clarify the role of specific interaction between Trib1 and HoxA/Meis1. In addition, a large scale gene expression analysis of human AML patients indicated reduced expression of Trib1 in a group of AML with CEBPα mutation.29,33 However, enhanced expression of Trib2 was observed in this group (cluster 4) suggests that Trib2 might replace Trib1 function in this cluster.10
Leukemogenesis is a complex process requiring multiple genetic and epigenetic alterations that provide the target cell with growth advantages, and combinations of such alterations are important.34 In the present study, we show a novel functional role of Trib1 in myeloid leukemogenesis that mediates RAS-MAPK and C/EBPα pathways. We thus propose 2 important pathways in myeloid leukemogenesis: one is MLL-Meis1/Hox and the other is MEK-Trib1-ERK-C/EBPα. The function of Trib1 as an adaptor is thus important to regulate hematopoietic cell homeostasis as well as the proliferation and survival of malignant myeloid cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Katsuji Yoshioka for providing MEK1 and MKK4 cDNAs, and Manabu Yamashita for technical assistance.
This work was supported by KAKENHI (Grant-in-Aid for Scientific Research) on Priority Areas “Integrative Research Toward the Conquest of Cancer” (T.N.) and for Young Scientists (T.Y.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Authorship
Contribution: T.Y. designed and performed the research and wrote the manuscript; Y.K., Y.Y., T.T., and S.M. performed the research and analyzed the data; and T.N. designed the research and wrote the manuscript.
Conflict-of interest disclosure: The authors declare no competing financial interests.
Correspondence: Takuro Nakamura, Division of Carcinogenesis, The Cancer Institute, Japanese Foundation for Cancer Research, 3-8-31 Ariake, Koto-ku, Tokyo 135-8550, Japan; e-mail: takuro-ind@umin.net.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal