Abstract
Leukemia, with its origin in a specific genetic abnormality, will only arise if the cell properly folds and processes the oncogenic protein encoded by the mutant gene. In this issue of Blood, Tsukahara and Maru describe a set of proteins that control the processing of the nascent BCR-ABL oncoprotein, providing new avenues for potential therapeutic intervention in chronic myelogenous leukemia (CML).
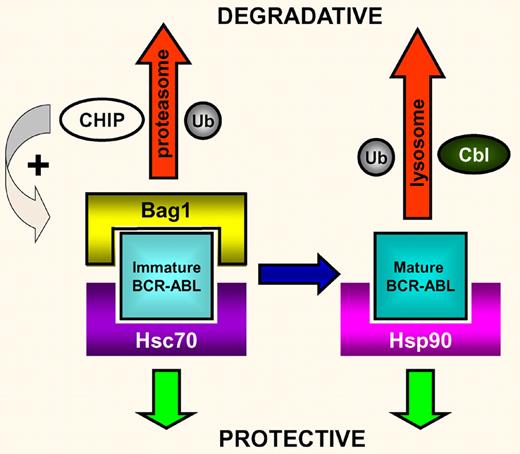
The key proteins regulating the fate of newly synthesized BCR-ABL oncoprotein. In this model, Bag1 and Hsc70 compete for binding to immature BCR-ABL. Bag1 binding will traffic the BCR-ABL for degradation in the proteasome using the CHIP ubiquitin ligase. Conversely, Hsc70 binding will protect BCR-ABL and allow it to fully mature, when it is then bound/protected by Hsp90. Alternatively, any unbound mature BCR-ABL is ubiquitinated by Cbl and degraded in the lysosome (adapted from Figure 7 of the Tsukahara and Maru article).
The key proteins regulating the fate of newly synthesized BCR-ABL oncoprotein. In this model, Bag1 and Hsc70 compete for binding to immature BCR-ABL. Bag1 binding will traffic the BCR-ABL for degradation in the proteasome using the CHIP ubiquitin ligase. Conversely, Hsc70 binding will protect BCR-ABL and allow it to fully mature, when it is then bound/protected by Hsp90. Alternatively, any unbound mature BCR-ABL is ubiquitinated by Cbl and degraded in the lysosome (adapted from Figure 7 of the Tsukahara and Maru article).
CML is caused by the constitutive tyrosine kinase activity of the BCR-ABL fusion protein produced by a balanced t(9;22) translocation in the hematopoietic stem cell.1 Small molecule inhibitors of this tyrosine kinase activity (hence the name TKIs) are potent drugs, with the majority of patients showing rapid and durable eradication of leukemic cells.2 Such therapeutics act by targeting the altered enzymatic activity of the cancer-causing protein. However, despite their considerable success, resistance to TKIs is still a clinical problem in a significant number of patients3 and novel methods of targeting BCR-ABL at the mRNA or protein level are still sought.
The chaperone protein Hsp90 has long been known to protect several proteins, including BCR-ABL, from intracellular proteasomal degradation.4 Hsp90 inhibitors have been developed that prevent the chaperone from binding BCR-ABL (the unprotected BCR-ABL is then degraded), and it is hoped that these will be of clinical benefit in the near future.5,6 Hsp90 is a member of a complex pathway of molecules that regulate the fate of newly synthesized proteins within the cell. Through a painstaking series of experiments, Tsukahara and Maru7 have made great strides in describing the complexities of both newly synthesized and mature BCR-ABL protein processing. Using a combination of techniques and an array of differentially tagged proteins they have made some exciting discoveries (see figure).
First, they identified the proteins CHIP and Cbl as ubiquitin ligases directly involved in BCR-ABL degradation. They further demonstrated that Hsp90 inhibition in combination with CHIP overexpression is dramatically more effective at inducing BCR-ABL degradation than either agent alone. The differential nature of the ubiquitination of BCR-ABL during its degradation by CHIP and Cbl is then described. After investigating a range of candidate molecular chaperones, the authors discovered that BCR-ABL is strongly bound by Bag1 (Bcl-2–associated athanogene-1), through domains derived from both BCR and ABL. Interestingly, Hsc70 (another heat-shock cognate protein) also strongly binds Bag1 and inhibits its binding to BCR-ABL. Conversely, CHIP expression enhances Bag1 binding to BCR-ABL. Hsc70 was shown to inhibit CHIP-induced degradation of BCR-ABL, consistent with Hsc70 playing a protective role. After Hsp90 inhibition, Bag1 sorts BCR-ABL to the proteasome and stimulates its degradation. In some cell lines, a combination of both Hsc70 down-regulation and Bag1 overexpression is needed for BCR-ABL degradation.
While the study has generated a working model of the decision points in BCR-ABL protein processing, several questions remain unanswered. The critical binding sites for Hsc70 and Bag1 could be mapped at a finer level by point mutagenesis of BCR-ABL. A more complete characterization of the evolution from immature to mature forms of BCR-ABL needs to be undertaken. There is also the following curious (and at first glance, contradictory) observation. Inhibition of Hsp90 leads to more Hsc70 being bound to BCR-ABL which should lead to the oncoprotein being stabilized/protected. However, Hsp90 inhibition leads to a reduction in the amount of detectable BCR-ABL protein.
Although most of the experiments concerned the p190 form of BCR-ABL that causes acute lymphoblastic leukemia (ALL), some of the important findings were also shown to be valid for the p210 form of BCR-ABL that is seen in the majority of patients with CML. In the clinical context of this leukemia, some aspects of the data call for pondering. Thus, the authors speculate that the structure of the BCR-ABL protein itself, rather than its kinase activity, is required for BCR-ABL to bind Bag1. If this is the case, it is intriguing that imatinib treatment abolishes Bag1 binding and BCR-ABL degradation completely (as shown in Figure 3A of their article). Similarly, the contention that the binding of the T315I BCR-ABL mutant protein to Bag-1 is due to this mutant being less structurally mature than wild-type BCR-ABL still needs to be demonstrated. Even more important is the apparent contradiction between the presence of mature BCR-ABL in CML stem cells and their lack of imatinib responsiveness. Further investigations in this area will hopefully clarify these questions.
The study by Tsukahara and Maru goes a long way toward understanding the nature of the complex interactions between the various protein-processing factors and their role in determining the fate of the BCR-ABL protein after its synthesis. As such, it provides a wealth of potential new CML targets, from Hsc70 and CHIP antagonists, to Bag1 agonists, or a combination of both.
Conflict-of-interest disclosure: The authors declare no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal