Abstract
Degradation of BCR-ABL oncoproteins by heat shock protein 90 (Hsp90) inhibitors in chronic myelogenous leukemia is expected to overcome resistance to ABL tyrosine kinase inhibitors. However, the precise mechanisms still remain to be uncovered. We found that while c-Cbl E3 ligase induced ubiquitin-dependent degradation of mature and phosphorylated BCR-ABL proteins, another E3 ligase CHIP (carboxyl terminus of the Hsc70-interacting protein) degraded immature BCR-ABL proteins and efficiently suppressed BCR-ABL–dependent leukemic growth. Interestingly, Bag1 (Bcl-2-associated athanogene-1), a nucleotide exchange factor for Hsc70, directly bound BCR-ABL with a high affinity, which was enhanced by CHIP and Hsp90 inhibitors, inhibited by imatinib and competed with Hsc70. Bag1 knockdown abrogated Hsp90 inhibitor-induced BCR-ABL degradation. Bag1 induced binding of immature BCR-ABL to proteasome. Expression of Bag1 induced BCR-ABL degradation and growth suppression in Ba/F3 cells when Hsc70 was knocked down with or without CHIP induction. CHIP appears to sort newly synthesized Hsp90-unchaperoned BCR-ABL to the proteasome not only by inhibiting Hsc70 and thereby promoting Bag1 to bind BCR-ABL, but also by ubiquitinating BCR-ABL. Bag1 may direct CHIP/Hsc70-regulated protein triage decisions on BCR-ABL immediately after translation to the degradation pathway.
Introduction
Chronic myelogenous leukemia (CML) is a hematopoietic stem cell malignancy caused by the BCR-ABL tyrosine kinase chimeric oncoprotein.1 Specific inhibitors of ABL tyrosine kinase have provided a remarkable success in the treatment of CML. However, resistance to the inhibitors mainly by acquisition of mutations of specific amino acids within the ABL kinase or by overexpression of BCR-ABL protein is still an obstacle.2-5 Although mechanistic insights into the relapse could be provided by sequence analysis of the oncogene, the mechanisms of overexpression of BCR-ABL are poorly understood.2,6
Heat shock protein 90 (Hsp90) inhibitors would be expected to overcome the resistance of ABL-specific inhibitors, because BCR-ABL proteins with or without mutations (eg, T315I, E255K) are ubiquitinated and degraded by the proteasome machinery.7-10 Indeed, 17-allylamino-17-demethoxygeldanamycin (17-AAG), a less toxic analog of Hsp90 inhibitor geldanamycin (GA), is currently in clinical trials not only for CML but also other malignant disorders. Hsp90 inhibitors have been shown to induce dissociation of Hsp90 from client proteins for degradation. Some of them subsequently bind to and allow ubiquitination by CHIP (carboxyl terminus of the heat shock cognate protein 70 [Hsc70]–interacting protein).9,11,12 However, E3 ligases for BCR-ABL are not known. CHIP was identified as a negative regulator for the chaperone ATPase activity of Hsc70 and also serves as an ubiquitin ligase for the quality control for unfolded or misfolded proteins.13-15 However, CHIP-null cells are recently found to be still capable of degrading client proteins as efficiently as wild-type (wt) cells when treated with Hsp90 inhibitors.16 Hsp90 inhibitor-induced apoptosis in tumor cells is promoted by silencing both Hsc70 and Hsp72 that contribute to tumorigenesis by their antiapoptotic activity.17 This suggests as yet unidentified mechanisms in Hsp90 inhibitor-induced protein degradation that may depend on individual nature of client proteins.
Another E3 ubiquitin ligase c-Cbl, which ubiquitinates and down-regulates many receptor tyrosine kinases, acts as a multifunctional adaptor protein regulating the phosphatidylinositol (PI) 3-kinase pathways in BCR-ABL–transformed hematopoietic cells.18,19 The different biochemical effects between CHIP and c-Cbl on BCR-ABL and the unexpected finding that Bag1 (Bcl-2-associated athanogene-1), a nucleotide exchange factor for Hsc70,20,21 directly binds BCR-ABL prompted us to show for the first time how immature proteins immediately after translation are processed through Hsc70 to either CHIP-dependent degradation or Hsp90-mediated maturation.
Methods
Cell culture
COS7 cells were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum. K562 and Ba/F3 cells were cultured in RPMI with 10% fetal calf serum and 10% WEHI-conditioned medium as a source of interleukin-3 (IL-3). p185 BCR-ABL-expressing Ba/F3 cells with the tet regulatory expression system for CHIP and c-Cbl were deprived of tet for the indicated time.22 Viable cell numbers were determined by trypan blue staining, flow cytometry and colorimetric assay using Almar Blue (Biosource International).
Plasmid, antibody, and reagents
Expression plasmids for p185 BCR-ABL, BCR and ABL, and BCR 1-413 have been described previously.23,24 KD (ABL K290R), Δkina (ABL Δ240-500), and kina (ABL 240-500) were subcloned in-frame into pFlag-CMV2 vector (Sigma-Aldrich). The cDNA encoding BCR-ABL and its mutants were also subcloned into pTNT (Promega) or in-frame into pFlag-TNT vector that was constructed by insertion of a DNA fragment encoding Flag peptide. Human CHIP, c-Cbl, cdc37, Bag1M, Bag1S, p23, Ubc4, UbcH5c, and UbcH7 were subcloned in-frame into pGEX2T (Amersham Pharmacia Biotech), pFlag-CMV2 or pEF1/His vector (Invitrogen). Flag-tagged or GST-tagged-Hsp90, -Hsc70, and -Hsc54 expression plasmids have been described previously.25 Hsc70, CHIP, and Bag1 were subcloned in-frame into pFASTBAC HT (Invitrogen). Bag1M Δubiquitin-like domain mutant (ΔUb, Bag1M Δ89-153) was generated by polymerase chain reaction and subcloned in-frame into pFASTBAC HT. Flag-tagged mouse c-Cbl cDNA was a kind gift from T. Tezuka and T. Yamamoto at the University of Tokyo. Mouse c-Cbl ΔRing mutant and human c-Cbl C381A mutant were subcloned in-frame into pGEX2T. CHIP and c-Cbl cDNAs were also subcloned into pUHD10-3.22 Anti-Bag1, anti-Hsc70, anti-CHIP, and anti–c-Cbl siRNAs, and anti-ABL, anti–c-Cbl, anti-BCR, and anti-GST antibodies were purchased from Santa Cruz Biotechnology, anti-CHIP from CALBIOCHEM, anti-actin from CHEMICON, anti-Xpress from Invitrogen, anti-Flag from Sigma, anti-Hsc70 and anti-Hsp70 from Stressgen, anti-Hsp90 from NeoMarkers, anti–green fluorescent protein (GFP) from Wako and Clontech, anti-phosphotyrosine (PY-20) from ICN Biomedicals, anti-histidine from QIAGEN, anti-20S from BIOMOL. Imatinib mesylate was purchased from Novartis, and 17-AAG from AG. Scientific, MG-132 from CALBIOCHEM, GA, radicicol (Rad) lactacystin, bafilomycin, methylamine, E64, ALLN, and zVAD-FMK from Sigma.
Immunoblotting and immunoprecipitation
BCR-ABL–expressing Ba/F3 cells with the tet system for CHIP or c-Cbl were transiently transfected with or without Flag-tagged Hsp90, Hsc70, Hsc54, or control vector, and then were deprived of tet for 24 hours. K562 cells were transiently transfected with CHIP, c-Cbl, or control vector (−) under the control of the tet system with or without tet for 48 hours, or with Xpress-tagged CHIP, Flag-tagged c-Cbl, or control vector. K562 cells were daily transfected with anti-CHIP, anti–c-Cbl siRNAs, or control siRNA (20 pmol/2 × 106 cells) for 5 days. Twenty hours after the last transfection, the cells were incubated with or without GA (3μM) for 8 hours. The cells were also transiently transfected with anti-Bag1 or control siRNA (20 pmol/2 × 106 cells) for 24 hours, and then GA was applied to cell cultures for 8 hours. Ba/F3 cells expressing p185 BCR-ABL-wt were preincubated with imatinib (10μM) for 30 minutes and with GA (3μM), Rad (3μM), and 17-AAG (3μM) for additional 8 hours. COS7 cells were transiently transfected with the wt p185 BCR-ABL, BCR, ABL (type 1b), or BCR-ABL mutants together with CHIP, c-Cbl, Bag1, or control vector under the tet system. The cells were also transiently transfected with Flag-tagged p185 BCR-ABL and Flag-tagged Hsc70, Flag-tagged Hsp90, Xpress-tagged Bag1M, or S together with CHIP or c-Cbl under the control of the tet system with or without tet for 24 hours.
For immunoprecipitation, COS7 cells were transiently transfected with Flag-tagged BCR-ABL wt, KD, or Δkina together with Xpress-tagged Bag1. Empty vector was added to balance the total DNA amount. After transfection, cells were lysed in NP40 buffer (50mM Tris-HCl, pH 7.5, 100mM NaCl, 1% NP40, 10% glycerol, 5mM sodium orthovanadate, 5mM NaF, 1mM phenylmethylsulfonyl fluoride, 20μg/mL leupeptin, 20μg/mL aprotinin), immunoprecipitated with anti-Flag antibody. Flag-tagged BCR-ABL proteins were in vitro–transcribed/translated in rabbit reticulocyte lysates (RRLs) with (+) or without (−) GA (10μM) and then incubated with or without His-tagged Bag1 or His-tagged Bag1ΔUb. Anti-Flag immunoprecipitates were subjected to immunoblotting with the indicated antibodies. K562 and Ba/F3 cells were transfected using the Amaxa Nucleofector kit according to the instructions of the manufacturer (Lonza). The transfection efficiency of GFP constructs was more than 80% as judged by GFP signals. COS7 cells were transiently transfected using Superfect transfection reagent (QIAGEN) under the conditions recommended by the supplier.
GST-pull down
Bag1M, Bag1S, CHIP, Hsc70, Hsp90, cdc37, p23 proteins were produced as glutathione-S-transferase (GST) fusion proteins in Escherichia coli and purified. GST alone or GST-fused proteins bound with glutathione-Sepharose 4B beads (Amersham Pharmacia Biotech) were incubated with p185 BCR-ABL, ABL, BCR, or BCR-ABL mutants that were in vitro–transcribed/translated in RRLs in the presence or absence of GA (10μM) or imatinib (10μM). The proteins associated with GST alone or GST-fused proteins in NP40 buffer at 4°C for 90 minutes were analyzed by immunoblotting. To examine the effects of Hsc70 and CHIP on the binding, histidine (His)–tagged Hsc70 and His-CHIP were produced in E coli, purified, and added in the buffer. For immunodepletion of Hsc70, RRLs were incubated with anti-Hsc70 antibody prebound with protein G-sepharose beads, and the supernatant was used for in vitro translation of BCR-ABL.
ELISA
Microtiter plates (NunclonTM; NUNC NS) were coated with a His-tagged ABL kinase domain fragment or His-tagged Hsc70 in phosphate-buffered saline (PBS). After treatment with blocking buffers (4% bovine serum albumin in PBS or protein-free blocking buffer; Thermo Scientific), the plates were incubated with various concentrations of His-tagged Bag1 diluted with PBS. The plates were then incubated with anti-Bag1 antibody and with horseradish peroxide (HRP)–conjugated second antibody. After being washed in PBS, the plates were allowed to react with 3,3′,5,5′-tetramethylbenzidine substrate reagent (Becton Dickinson) and analyzed at 450 nm using a microplate reader.
In vitro ubiquitination assay
Anti-ABL or anti-Flag immunoprecipitates from aliquot of Ba/F3 cell lysates expressing p185 BCR-ABL (wt and Δkina), in vitro–transcribed/translated Flag-tagged BCR-ABL proteins in RRLs with or without GA (10μM) and GST-Bag1 were collected using protein A/G-sepharose or glutathione-Sepharose 4B beads, washed with NP40 buffer and/or 50mM Tris-HCl, pH7.5, and then subjected to in vitro ubiquitination assay. The proteins bound with beads were incubated with GST alone, GST/His-CHIP, or GST-c-Cbl in 50 μL reaction buffers (50mM Tris-HCl, pH 7.5, 20nM E1 [Calbiochem], 200nM E2 [GST-UbcH5c/Ubc4 or UbcH1-3, 6-10, 12, 13, 5a-c; BIOMOL], biotin-labeled ubiquitin [2 μg/mL; AFFINITI Research Products], 5mM ATP, 10mM phosphocreatine [Sigma], 3.5 U creatine kinase [Sigma], 10mM dithiothreitol, and 5mM magnesium chloride, 1mM phenylmethylsulfonyl fluoride, 20 μg/mL leupeptin, 20 μg/mL aprotinin) for 90 minutes at 30°C. The beads were washed 5 times, and the ubiquitinylated proteins were detected by immunoblotting with HRP-conjugated streptavidin.
Flow cytometry
BCR-ABL–expressing Ba/F3 cells with the tet system for CHIP and K562 cells were transiently transfected with Xpress-tagged Bag1 together with or without anti-Hsc70 siRNA (20 pmol/2 × 106 cells) using the Amaxa Nucleofector kit according to the instructions of the manufacturer (Lonza). Empty vector or control siRNA was added to balance the total DNA or siRNA amount, respectively. After 2 days, the viable cells were analyzed based on volume (Coulter volume or electronic impedance) and side-scattering gating and GFP-positive (CHIP-expressing) cells based on fluorescent intensity by flow cytometry on a Beckman-Coulter Cell Lab Quanta TM SC MPL system.
Statistical analysis
Data are expressed as mean ± SE. Statistical significance of differences was evaluated by analysis of variance followed by Newman-Keuls multiple range test. Significant discrimination into 2 groups was evaluated by Akaike Information Criterion and t test.
Results
CHIP or c-Cbl promotes degradation of BCR-ABL leading to attenuation of BCR-ABL–dependent cell growth
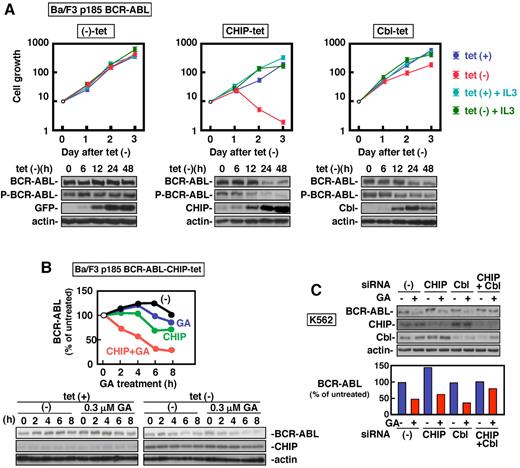
To determine whether CHIP or c-Cbl induces degradation of BCR-ABL and inhibits BCR-ABL–dependent cell growth, we established the stable tet regulatory system for expression of CHIP and c-Cbl in hematopoietic Ba/F3 cells. As shown in Figure 1A, after removal of tet, the protein levels of CHIP were time-dependently increased with concomitant decrease of total and phosphorylated protein amounts of BCR-ABL. The effect was prominent at 24 hours. This was clearly associated with the inhibition of BCR-ABL–dependent (IL-3–independent), but not IL-3–dependent cell growth. Induction of c-Cbl expression by deprivation of tet was also achieved, and it reduced BCR-ABL protein amounts and slightly, but appreciably, attenuated BCR-ABL–dependent, but not IL-3–dependent cell growth (Figure 1A). Transient induction of CHIP or c-Cbl also induced down-regulation of p210 BCR-ABL in K562 cells (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). These data indicated that both CHIP and c-Cbl could decrease the stability of BCR-ABL protein. However, the ability of CHIP for suppression of IL-3–independent cell growth was more potent than that of c-Cbl in hematopoietic Ba/F3 cells expressing BCR-ABL (Figure 1A). The CHIP induction failed to up-regulate unfolded protein response-related proteins such as ATF6 and phosphorylated PERK (data not shown). Studies using a set of inhibitors for protein degradation demonstrated that CHIP degrades BCR-ABL mainly via proteasome, while c-Cbl via lysosome (supplemental Figure 2).
CHIP and c-Cbl induce degradation of BCR-ABL leading to attenuation of BCR-ABL–dependent cell growth. (A) BCR-ABL–expressing Ba/F3 cells with the tet regulatory expression system for CHIP and c-Cbl were deprived of tet for the indicated time. Viable cell numbers were determined by trypan blue staining (n = 3). Whole cell lysates were analyzed by immunoblotting with antibodies against ABL (BCR-ABL), phosphotyrosine (P-BCR-ABL), CHIP, c-Cbl, GFP, and actin. (B) p185 BCR-ABL–expressing Ba/F3 cells with the tet regulatory expression system for CHIP were deprived of tet for 14 hours. The cells were further incubated with cycloheximide (200μg/mL) together with or without GA (0.3μM) for indicated times. Whole cell lysates at each time point were analyzed by immunoblotting with antibodies against ABL, CHIP, and actin. (C) K562 cells were daily transfected with anti-CHIP and anti–c-Cbl siRNAs for 5 days. Twenty hours after the last transfection, the cells were incubated with or without GA (3μM) for 8 hours and analyzed by immunoblotting with antibodies against ABL, CHIP, and actin. The data were expressed as percentage of the signals obtained with each control. The bottom graph shows quantification of relative amounts of BCR-ABL proteins.
CHIP and c-Cbl induce degradation of BCR-ABL leading to attenuation of BCR-ABL–dependent cell growth. (A) BCR-ABL–expressing Ba/F3 cells with the tet regulatory expression system for CHIP and c-Cbl were deprived of tet for the indicated time. Viable cell numbers were determined by trypan blue staining (n = 3). Whole cell lysates were analyzed by immunoblotting with antibodies against ABL (BCR-ABL), phosphotyrosine (P-BCR-ABL), CHIP, c-Cbl, GFP, and actin. (B) p185 BCR-ABL–expressing Ba/F3 cells with the tet regulatory expression system for CHIP were deprived of tet for 14 hours. The cells were further incubated with cycloheximide (200μg/mL) together with or without GA (0.3μM) for indicated times. Whole cell lysates at each time point were analyzed by immunoblotting with antibodies against ABL, CHIP, and actin. (C) K562 cells were daily transfected with anti-CHIP and anti–c-Cbl siRNAs for 5 days. Twenty hours after the last transfection, the cells were incubated with or without GA (3μM) for 8 hours and analyzed by immunoblotting with antibodies against ABL, CHIP, and actin. The data were expressed as percentage of the signals obtained with each control. The bottom graph shows quantification of relative amounts of BCR-ABL proteins.
We next examined whether Hsp90 inhibitor-induced degradation of BCR-ABL is dependent on CHIP and/or c-Cbl. Although a lower concentration of GA at 0.3μM or overexpression of CHIP individually degraded BCR-ABL protein roughly by 25% by 8 hours, their combination dramatically induced BCR-ABL degradation by more than 70% in Ba/F3-p185 BCR-ABL cells (Figure 1B). When both anti-CHIP and c-Cbl siRNAs were transfected into K562 cells, Hsp90 inhibitor-induced degradation of BCR-ABL was almost completely inhibited (Figure 1C). These results suggest that both CHIP and c-Cbl E3 ligases are involved in BCR-ABL degradation when Hsp90 is inhibited.
c-Cbl induces ubiquitin-dependent degradation of mature and phosphorylated BCR-ABL proteins, while CHIP degrades immature BCR-ABL proteins
To characterize the necessary domain of BCR-ABL for CHIP and c-Cbl–induced degradation, we examined a series of BCR-ABL mutants using COS7 cells under the tet-regulated expression system for CHIP and c-Cbl. CHIP clearly down-regulated the wt and all types of BCR-ABL mutants even including BCR 1-413 devoid of the whole ABL sequence, except for intact ABL and BCR (Figure 2A-B). In contrast, c-Cbl could efficiently degrade only the wt BCR-ABL, ΔSH3, Δ40, and imatinib-resistant mutants, T315I and E255K. BCR, ABL, kinase-negative mutant KD, kinase-deficient Δkina, ΔSH2, ΔSH2 bind, and ΔSH2biΔSH3 were not significantly affected by c-Cbl. These results indicate that CHIP and c-Cbl have a different substrate-recognizing property toward BCR-ABL. c-Cbl–mediated down-regulation of BCR-ABL requires the kinase activity. Because BCR-ABL as a whole molecule has never been crystallized, contribution of the BCR sequences to the structural maturation of BCR-ABL still lacks physical evidence. It can safely be mentioned that mature BCR-ABL should achieve autophosphorylation, because it is a constitutively active enzyme. Given the possible intramolecular interaction between the ABL SH2 domain and the BCR SH2-binding domain in BCR-ABL,23 deletion of either of the domains could artificially create immature forms or unfolded conformation even after posttranslational time enough for maturation. This prompted us to hypothesize that CHIP may target unfolded proteins.
c-Cbl induces ubiquitin-dependent degradation of mature and phosphorylated BCR-ABL proteins, while CHIP degrades immature BCR-ABL proteins. (A) Structure of BCR-ABL, BCR, ABL, and BCR-ABL mutants used in this study. Deletions of functional domains of p185 BCR-ABL are shown, including the coiled-coil oligomerization domain (Δ40), SH2-binding domain (ΔSH2 bind), SH3 domain (ΔSH3), ΔSH2 domain (ΔSH2), kinase domain (Δkina), SH2-binding domain and SH3 domain (ΔSH2biΔSH3), kinase-negative mutant (ABL K290R, KD), imatinib-resistant mutants (ABL T315I, E255K), ABL, BCR, and BCR 1-413. (B) Effects of CHIP and c-Cbl on the stabilities of BCR-ABL, BCR, ABL, and BCR-ABL mutants. COS7 cells were transiently transfected with the wt p185 BCR-ABL or its mutants [KD, T315I, E255K, Δ40, Δkina, ΔSH2, ΔSH2 bind, ΔSH3, ΔSH2biΔSH3, BCR 1-413], BCR, and ABL (type 1b) together with CHIP, c-Cbl, or control vector (−) under the tet regulatory expression system. The cells were incubated with or without tet for 24 hours and analyzed by immunoblotting with antibodies against ABL, BCR, CHIP, c-Cbl, and actin. The top graphs indicate the intensities of the signals of tet (−) cells relative to those of tet (+) cells that were normalized against those of the control vector. The data were expressed as percentage of the signals obtained with each control. The red lines, at 53.2% and 73.3% in the CHIP and c-Cbl panels, respectively, are drawn to make a statistically significant discrimination into 2 groups (Akaike Information Criterion and t test, P < .001). (C) Immunoprecipitated Flag-tagged BCR-ABL proteins that were in vitro–transcribed/translated with (+) or without (−) GA (10μM) were incubated with biotin-labeled ubiquitin, GST-CHIP, GST-c-Cbl, E1, mixtures of E2s as indicated. The numbers indicate the signals of ubiquitinated BCR-ABL. (D) Immunoprecipitated BCR-ABL proteins from p185 wt- and Δkina-expressing Ba/F3 cell lysates were incubated with GST-CHIP, GST-c-Cbl, E1, E2s (Ubc4 and UbcH5c), and biotin-labeled ubiquitin. Ubiquitinated proteins were detected by streptavidin-HRP (C-D).
c-Cbl induces ubiquitin-dependent degradation of mature and phosphorylated BCR-ABL proteins, while CHIP degrades immature BCR-ABL proteins. (A) Structure of BCR-ABL, BCR, ABL, and BCR-ABL mutants used in this study. Deletions of functional domains of p185 BCR-ABL are shown, including the coiled-coil oligomerization domain (Δ40), SH2-binding domain (ΔSH2 bind), SH3 domain (ΔSH3), ΔSH2 domain (ΔSH2), kinase domain (Δkina), SH2-binding domain and SH3 domain (ΔSH2biΔSH3), kinase-negative mutant (ABL K290R, KD), imatinib-resistant mutants (ABL T315I, E255K), ABL, BCR, and BCR 1-413. (B) Effects of CHIP and c-Cbl on the stabilities of BCR-ABL, BCR, ABL, and BCR-ABL mutants. COS7 cells were transiently transfected with the wt p185 BCR-ABL or its mutants [KD, T315I, E255K, Δ40, Δkina, ΔSH2, ΔSH2 bind, ΔSH3, ΔSH2biΔSH3, BCR 1-413], BCR, and ABL (type 1b) together with CHIP, c-Cbl, or control vector (−) under the tet regulatory expression system. The cells were incubated with or without tet for 24 hours and analyzed by immunoblotting with antibodies against ABL, BCR, CHIP, c-Cbl, and actin. The top graphs indicate the intensities of the signals of tet (−) cells relative to those of tet (+) cells that were normalized against those of the control vector. The data were expressed as percentage of the signals obtained with each control. The red lines, at 53.2% and 73.3% in the CHIP and c-Cbl panels, respectively, are drawn to make a statistically significant discrimination into 2 groups (Akaike Information Criterion and t test, P < .001). (C) Immunoprecipitated Flag-tagged BCR-ABL proteins that were in vitro–transcribed/translated with (+) or without (−) GA (10μM) were incubated with biotin-labeled ubiquitin, GST-CHIP, GST-c-Cbl, E1, mixtures of E2s as indicated. The numbers indicate the signals of ubiquitinated BCR-ABL. (D) Immunoprecipitated BCR-ABL proteins from p185 wt- and Δkina-expressing Ba/F3 cell lysates were incubated with GST-CHIP, GST-c-Cbl, E1, E2s (Ubc4 and UbcH5c), and biotin-labeled ubiquitin. Ubiquitinated proteins were detected by streptavidin-HRP (C-D).
The in vitro transcription/translation system with Hsp90 inhibitors has been reported to produce Hsp90-unchaperoned immature checkpoint kinase.26 As expected, autophosphorylation, but not synthesis, of BCR-ABL protein was clearly inhibited by addition of GA into in vitro–transcribed/translated system of RRLs (supplemental Figure 3A). MG132, a proteasome inhibitor, induced accumulation of BCR-ABL proteins without autophosphorylation in Ba/F3-p185 BCR-ABL cells treated with GA in a dose-dependent manner (supplemental Figure 3B). An in vitro ubiquitination assay demonstrated that CHIP preferred immature to mature BCR-ABL for ubiquitination and used UbcH5a/b/c, but not other E2s (Figure 2C). In contrast, c-Cbl–induced ubiquitination of mature BCR-ABL required the ABL kinase domain (Figure 2C-D), which correlates with the result of the protein degradation.
Direct recognition of BCR-ABL protein by Bag1
Because the ability of CHIP to suppress BCR-ABL–dependent cell growth was more prominent than that of c-Cbl, we further examined molecular details in CHIP-induced BCR-ABL degradation. To investigate how unfolded or misfolded BCR-ABL proteins are recognized by CHIP-mediated protein degradation pathway, a series of molecular chaperones were screened for binding to BCR-ABL translated in vitro by GST-pull down assay. To our surprise, stronger binding of BCR-ABL with Bag1M or Bag1S was observed than with other molecules including Hsc70, CHIP, Hsp90, cdc37, and p23 (Figure 3A). Bag1 (termed Bag1M throughout this study unless otherwise indicated) bound in vitro–translated ABL, BCR-ABL, BCR, BCR 1-413, and the Flag-tagged kinase domain of ABL (Flag-kina; Figure 3B-D). Although Raf-1 binds BCR 240-413 via 14-3-3 protein,27 BCR 1-242 also binds Bag1 (data not shown), excluding the possibility that 14-3-3/Raf-1 bridges the binding between Bag1 and BCR 1-413. Cotransfection experiments using COS7 cells demonstrated that the wt, KD, and Δkina mutants of BCR-ABL also bound Bag1, but the binding ability estimated by intensities of the signal ratio of Xpress/Flag in immunoprecipitation of Δkina, but not KD, was lower (approximately 47%) than that of the wt (Figure 3E). BCR-ABL bound to Bag1 also in K562 cells (Figure 3F). Enzyme-linked immunosorbent assay (ELISA) revealed that a dissociation constant (Kd) of Bag1 with the ABL kinase domain was 8.34nM and that with Hsc70 was 10.11nM (Figure 3G), which was comparable to that of the previous report.28 Bag1S behaved in a similar fashion to Bag1M (supplemental Figure 4A-C), but binding with BCR-ABL was stronger in Bag1M than Bag1S in both ELISA and in cells (supplemental Figure 4D-F).
Correlation between direct binding of Bag1 and BCR-ABL degradation. (A) GST-(−), -CHIP, -Hsc70, -Hsp90, -cdc37, -Bag1M, -Bag1S, or -p23 were incubated with in vitro–transcribed/translated BCR-ABL with (+) or without (−) GA (10μM) or imatinib (10μM). The proteins associated with those GST-fusion proteins were analyzed by immunoblotting with the indicated antibodies. Vertical lines are inserted to indicate a repositioned gel lane. Because of a large number of samples, Western blotting was performed using 2 separate gels with completely identical experimental conditions. (B-D) GST-Bag1 was incubated with BCR-ABL, BCR, ABL (B), Flag-tagged ABL kinase domain fragments (wt and KD; C), and BCR 1-413 (D) that were in vitro–transcribed/translated in the presence (+) or absence (−) of GA (10μM) in RRLs. GST-Bag1–bound proteins were analyzed by immunoblotting with the indicated antibodies. (E) COS7 cells were transiently transfected with Flag-tagged BCR-ABL wt, KD, or Δkina together with Xpress (Xp)–tagged Bag1. Twenty-four hours after transfection, anti-Flag immunoprecipitates were immunoblotted with antibodies against Xpress and Flag. (F) K562 cells were treated with GA (1μM) for 4 hours. Anti-Bag1 or mouse control immunoglobulin G (IgG) immunoprecipitates were immunoblotted with antibodies against ABL and Bag1. (G) His-tagged ABL kinase domain fragments or His-tagged Hsc70 proteins coated on microtiter plates were incubated with various concentrations of His-tagged Bag1 and bound Bag1 was detected by anti-Bag1 antibody.
Correlation between direct binding of Bag1 and BCR-ABL degradation. (A) GST-(−), -CHIP, -Hsc70, -Hsp90, -cdc37, -Bag1M, -Bag1S, or -p23 were incubated with in vitro–transcribed/translated BCR-ABL with (+) or without (−) GA (10μM) or imatinib (10μM). The proteins associated with those GST-fusion proteins were analyzed by immunoblotting with the indicated antibodies. Vertical lines are inserted to indicate a repositioned gel lane. Because of a large number of samples, Western blotting was performed using 2 separate gels with completely identical experimental conditions. (B-D) GST-Bag1 was incubated with BCR-ABL, BCR, ABL (B), Flag-tagged ABL kinase domain fragments (wt and KD; C), and BCR 1-413 (D) that were in vitro–transcribed/translated in the presence (+) or absence (−) of GA (10μM) in RRLs. GST-Bag1–bound proteins were analyzed by immunoblotting with the indicated antibodies. (E) COS7 cells were transiently transfected with Flag-tagged BCR-ABL wt, KD, or Δkina together with Xpress (Xp)–tagged Bag1. Twenty-four hours after transfection, anti-Flag immunoprecipitates were immunoblotted with antibodies against Xpress and Flag. (F) K562 cells were treated with GA (1μM) for 4 hours. Anti-Bag1 or mouse control immunoglobulin G (IgG) immunoprecipitates were immunoblotted with antibodies against ABL and Bag1. (G) His-tagged ABL kinase domain fragments or His-tagged Hsc70 proteins coated on microtiter plates were incubated with various concentrations of His-tagged Bag1 and bound Bag1 was detected by anti-Bag1 antibody.
Bag1 directs triage decisions on Hsp90 inhibitor-induced BCR-ABL degradation
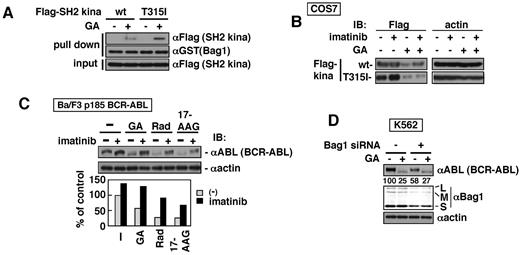
We further examined a possible relationship between Bag1 binding to BCR-ABL and Hsp90 inhibitor-induced BCR-ABL degradation. As shown in Figure 3A through D and F and supplemental Figure 4A through C, binding of Bag1M and Bag1S with ABL, BCR-ABL, Flag-kina, or Flag-kina KD was drastically enhanced by GA. Such enhancement was not observed in BCR and BCR 1-413. The ABL kinase domain is required for the enhancement, implying that the structure by itself rather than kinase activity is necessary for BCR-ABL to bind Bag1 (supplemental Figure 5). The imatinib binding site mutant T315I mutant degrades more readily with and CML mice expressing the mutant respond better to Hsp90 inhibitors than the wt BCR-ABL.8,29 T315I was found to bind Bag1 more strongly than the wt (Figure 4A). The imatinib-bound ABL kinase domain has been shown to display a specific inactive conformation.30,31 We found that Hsp90 inhibitor-induced degradation of the wt BCR-ABL, but not T315I, was inhibited by imatinib (Figure 4B-C). Imatinib or ATP attenuated the binding of Bag1M but not Bag1S to BCR-ABL (Figure 3A and supplemental Figure 6A-C), suggesting that mature BCR-ABL may not be able to bind Bag1M. To assess Bag1 dependence in BCR-ABL degradation, endogenous Bag1 was knocked down by anti-Bag1 siRNA by roughly 50%. Basal BCR-ABL levels were reduced by 40%-60% in K562 cells. Bag1 has been shown to act as a nucleotide exchange factor for Hsc70.20,21 Therefore it is possibly due to reduced protein-folding activity of Hsc70 regulated by Bag1. However, the efficiency of GA-induced degradation of BCR-ABL was significantly attenuated (75% versus 53%; Figure 4D). These results suggest that Bag1 functions like a double-edged sword and preferentially recognizes immature BCR-ABL and directs triage decisions on Hsp90 inhibitor-induced protein degradation.
Bag1 mediates Hsp90 inhibitor-induced BCR-ABL degradation. (A) GST-Bag1 was incubated with SH2 kinase domain fragments (wt and T315I) that were in vitro–transcribed/translated in the presence (+) or absence (−) of GA (10μM) in RRLs. GST-Bag1–bound proteins were analyzed by immunoblotting with the indicated antibodies. (B) COS7 cells were transiently transfected with Flag-tagged kina-wt and -T315I. Twenty-four hours after transfection, cells were treated with imatinib and GA for 8 hours. (C) Ba/F3 cells expressing p185 BCR-ABL-wt were preincubated with imatinib (10μM) for 30 minutes and with GA (3μM), Rad (3μM), and 17-AAG (3μM) for additional 8 hours. The culture was performed with IL-3 to guarantee the cell survival after BCR-ABL inhibition. The bottom graph shows quantification of relative amounts of BCR-ABL proteins. (D) Twenty-four hours after transfection of anti-Bag1 siRNA to K562 cells, GA was added for 8 hours. Numbers indicate the intensities of the bands of BCR-ABL. The cell lysates were analyzed by immunoblotting with the indicated antibodies (B-D).
Bag1 mediates Hsp90 inhibitor-induced BCR-ABL degradation. (A) GST-Bag1 was incubated with SH2 kinase domain fragments (wt and T315I) that were in vitro–transcribed/translated in the presence (+) or absence (−) of GA (10μM) in RRLs. GST-Bag1–bound proteins were analyzed by immunoblotting with the indicated antibodies. (B) COS7 cells were transiently transfected with Flag-tagged kina-wt and -T315I. Twenty-four hours after transfection, cells were treated with imatinib and GA for 8 hours. (C) Ba/F3 cells expressing p185 BCR-ABL-wt were preincubated with imatinib (10μM) for 30 minutes and with GA (3μM), Rad (3μM), and 17-AAG (3μM) for additional 8 hours. The culture was performed with IL-3 to guarantee the cell survival after BCR-ABL inhibition. The bottom graph shows quantification of relative amounts of BCR-ABL proteins. (D) Twenty-four hours after transfection of anti-Bag1 siRNA to K562 cells, GA was added for 8 hours. Numbers indicate the intensities of the bands of BCR-ABL. The cell lysates were analyzed by immunoblotting with the indicated antibodies (B-D).
Hsc70 attenuates and CHIP stimulates Bag1 binding BCR-ABL
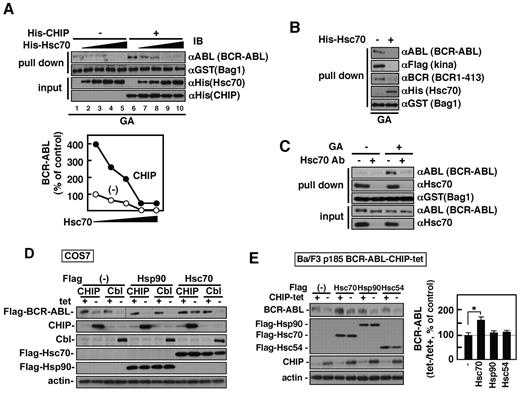
Bag1 cooperates with CHIP to degrade unfolded or misfolded proteins in the proteasome.20,21 Bag1 is also implicated in tumor cell survival partly by binding Raf with which Hsc70 competes.28,32-34 Given the structural similarity between ABL and Raf as a protein kinase, we expected that Hsc70 and BCR-ABL also have competitive binding to Bag1. The binding of GST-Bag1 to in vitro–translated immature BCR-ABL was dose-dependently inhibited by His-tagged Hsc70 (Figure 5A, lanes 1-5). Hsc70-mediated inhibition of Bag1 binding to the ABL kinase domain and BCR1-413 was also demonstrated (Figure 5B). A deletion mutant of the Bag domain bound neither Hsc70 nor BCR-ABL (data not shown). Interestingly, addition of CHIP alone after in vitro translation increased the binding of full-length BCR-ABL to Bag1 by 4-fold (Figure 5A, compare lanes 1 and 6) without affecting the half maximal (50%) inhibitory concentration (IC50) of Hsc70 (Figure 5A, lanes 6-10). In vitro–translated Flag-tagged kina switched its binding partner from Hsp90 to Hsc70 when GA was added (supplemental Figure 7). We suppose that in vitro–translated immature BCR-ABL binds either Bag1 or Hsc70 of RRLs origin and CHIP sequesters and inhibits Hsc70 allowing BCR-ABL to bind exogenous GST-Bag1. This idea was supported by poor binding of GST-Bag1 to newly synthesized immature BCR-ABL in RRLs immunodepleted of Hsc70 (Figure 5C). Binding of immature BCR-ABL may be saturated with Hsc70-unbound Bag1 preexisting in RRLs. Taken together, Bag1binds to BCR-ABL separately through BCR1-413 and the ABL kinase domain. Both of the binding are inhibited by Hsc70 through competition and promoted by CHIP via sequestration of Hsc70. However, GA-induced enhancement was observed only in the latter.
Hsc70 attenuates Bag1 binding BCR-ABL and inhibits CHIP-induced BCR-ABL degradation. (A) GST-Bag1 and an increasing amount of His-tagged Hsc70 were incubated with in vitro–transcribed/translated BCR-ABL in the presence (+) or absence (−) of His-tagged CHIP. The bottom graph shows quantification of relative amounts of BCR-ABL proteins. (B) BCR-ABL, a Flag-tagged ABL kinase domain fragment, or Flag-tagged BCR 1-413 was in vitro transcribed/translated in RRLs with GA and incubated with GST-Bag1 with (+) or without (−) His-tagged Hsc70. Bag1 and Hsc70 panels represent the 3 experiments with BCR-ABL, kina, and BCR 1-413 for each. (C) GST-Bag1 was incubated with in vitro–transcribed/translated BCR-ABL with (+) or without (−) GA (10μM) in RRLs immunodepleted of Hsc70. The proteins bound to GST-Bag1 were analyzed by immunoblotting with the indicated antibodies (A-C). (D) COS7 cells were transiently transfected with Flag-tagged BCR-ABL and Flag-tagged Hsp90, Hsc70, or control vector together with the tet system of CHIP, c-Cbl, or control vector. A vertical line indicates where lanes of the identical experiment were rearranged for consistency. (E) BCR-ABL–expressing Ba/F3 cells with the tet system for CHIP were transiently transfected with Flag-tagged Hsp90, Hsc70, Hsc54, or control vector. The cells were incubated with or without tet for 24 hours and were analyzed by immunoblotting with antibodies as indicated (D-E). The graph shows the ratios of BCR-ABL protein amount without tet (−) relative to that with tet (+) in E that were normalized against those of the control vector. The values are mean ± SEs. *P < .05.
Hsc70 attenuates Bag1 binding BCR-ABL and inhibits CHIP-induced BCR-ABL degradation. (A) GST-Bag1 and an increasing amount of His-tagged Hsc70 were incubated with in vitro–transcribed/translated BCR-ABL in the presence (+) or absence (−) of His-tagged CHIP. The bottom graph shows quantification of relative amounts of BCR-ABL proteins. (B) BCR-ABL, a Flag-tagged ABL kinase domain fragment, or Flag-tagged BCR 1-413 was in vitro transcribed/translated in RRLs with GA and incubated with GST-Bag1 with (+) or without (−) His-tagged Hsc70. Bag1 and Hsc70 panels represent the 3 experiments with BCR-ABL, kina, and BCR 1-413 for each. (C) GST-Bag1 was incubated with in vitro–transcribed/translated BCR-ABL with (+) or without (−) GA (10μM) in RRLs immunodepleted of Hsc70. The proteins bound to GST-Bag1 were analyzed by immunoblotting with the indicated antibodies (A-C). (D) COS7 cells were transiently transfected with Flag-tagged BCR-ABL and Flag-tagged Hsp90, Hsc70, or control vector together with the tet system of CHIP, c-Cbl, or control vector. A vertical line indicates where lanes of the identical experiment were rearranged for consistency. (E) BCR-ABL–expressing Ba/F3 cells with the tet system for CHIP were transiently transfected with Flag-tagged Hsp90, Hsc70, Hsc54, or control vector. The cells were incubated with or without tet for 24 hours and were analyzed by immunoblotting with antibodies as indicated (D-E). The graph shows the ratios of BCR-ABL protein amount without tet (−) relative to that with tet (+) in E that were normalized against those of the control vector. The values are mean ± SEs. *P < .05.
Hsc70 inhibits CHIP-induced BCR-ABL degradation
The effects of Hsc70 on the CHIP and c-Cbl–induced BCR-ABL degradation were examined. Using the tet expression system for CHIP and c-Cbl, BCR-ABL, Hsp90, and Hsc70 were transiently coexpressed in COS7 cells (Figure 5D and supplemental Figure 8). Overexpression of Hsc70 inhibited CHIP- but not c-Cbl–induced BCR-ABL degradation in COS7 cells, whereas that of Hsp90 failed to do so. Hsc54 that lacks a portion in the protein-binding and variable domain of Hsc70,25 failed to protect BCR-ABL against CHIP-induced degradation in Ba/F3-185 BCR-ABL cells (Figure 5E). Interestingly, Hsc70 overexpression alone augmented BCR-ABL protein levels by 1.9-fold, suggesting that Hsc70 prevents its misfolding and aggregation.35,36 We assume that Hsc70 binding to Bag1 not only stimulates chaperoning activity itself but also inhibits Bag1 binding to BCR-ABL for protein degradation. Folding of BCR-ABL by Hsc70 may precede chaperoning by Hsp90 for activation.9,35 Hsc70 associates with BCR-ABL proteins released from Hsp90 by Hsp90 inhibitors (supplemental Figure 7).12,37 Therefore, Hsc70 may protect newly synthesized Hsp90-unchaperoned immature BCR-ABL against CHIP-induced protein degradation in vivo. This is also supported by the finding that CHIP augmented GA-induced BCR-ABL degradation (Figure 1B). In contrast to GA-induced BCR-ABL degradation, anti-Bag1 siRNA failed to rescue CHIP-induced BCR-ABL degradation (Figure 4D and supplemental Figure 9). We assume that Hsc70 in the absence of Bag1 may not be competent enough to block CHIP.
Bag1 sorts BCR-ABL to proteasome and stimulates CHIP-induced BCR-ABL degradation
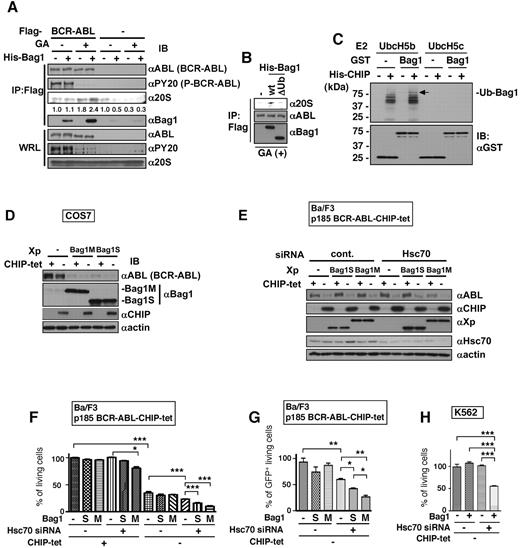
Bag1 associates with the proteasome through its ubiquitin-like domain.20,21 We found that Bag1 stimulated binding of immature BCR-ABL with 20S proteasome (Figure 6A), which is attenuated by the deletion of ubiquitin-like domain of Bag1 (Figure 6B). Significant CHIP-induced ubiquitination of Bag1M and Bag1S was detected by in vitro ubiquitination assay, which may also increase Bag1 affinity to the proteasome (Figure 6C and supplemental Figure 10A). The effects of Bag1 on the CHIP-induced BCR-ABL degradation were examined in COS7 cells. Overexpression of Bag1 under the tet system in combination with CHIP (Bag1/tet plus CHIP) revealed that Bag1 degraded BCR-ABL, which was enhanced by CHIP (supplemental Figure 10B). The opposite pattern of expression (Bag1 plus CHIP/tet) also gave a similar result (Figure 6D). These results suggest that Bag1 sorts BCR-ABL to proteasome and thus, stimulates CHIP-induced BCR-ABL degradation.
Bag1 stimulates CHIP-induced BCR-ABL degradation and combination of Bag1 overexpression and Hsc70 knockdown promotes CHIP-induced suppression of BCR-ABL–dependent cell growth. (A) Flag-tagged BCR-ABL proteins were in vitro transcribed/translated with (+) or without (−) GA and then incubated with 20S proteasome with (+) or without (−) His-tagged Bag1. Numbers indicate the intensities of the bands of 20S proteasome. (B) Flag-tagged BCR-ABL proteins were in vitro–transcribed/translated with GA and then incubated with 20S proteasome with His-tagged Bag1 (wt) or His-tagged ubiquitin like domain-deletion mutant of Bag1 (ΔUb). Anti-Flag immunoprecipitates and whole reticulocyte lysates (WRL) were subjected to immunoblotting with the indicated antibodies (A-B). (C) Glutathione sepharose-bound GST-tagged Bag1 proteins were incubated with His-tagged CHIP, E1, E2 (UbcH5b or UbcH5c), and biotin-labeled ubiquitin. Ubiquitinated proteins were detected by streptavidin-HRP. An arrow indicates ubiquitinated Bag1. (D) COS7 cells were transiently transfected with the BCR-ABL together with or without Xpress-tagged Bag1M, Bag1S, or control vector, and the tet system of CHIP or control vector. (E) BCR-ABL–expressing Ba/F3 cells with the tet system for CHIP were transiently transfected with Xpress-tagged Bag1S or Bag1M together with or without Hsc70 siRNA. The cells were incubated with or without tet for 24 hours (D) or 48 hours (E) and were analyzed by immunoblotting with the indicated antibodies (D-E). (F-G) BCR-ABL–expressing Ba/F3 cells with the tet system for CHIP were transiently transfected with Xpress-tagged Bag1S or Bag1M together with or without Hsc70 siRNA. After 2 days, the viable cell numbers were analyzed based on volume and side-scattering gating (F) and then GFP-positive (CHIP-expressing) cell numbers were analyzed by fluorescent intensity (G) with flow cytometry. (H) K562 cells were transiently transfected with the tet system for CHIP together with or without Xpress-tagger Bag1 and Hsc70 siRNA. After 2 days, the viable cell numbers were analyzed based on volume and side-scattering gating with flow cytometry. The values are mean ± SEs determined by 4 independent experiments. *P < .05, **P < .01, ***P < .001.
Bag1 stimulates CHIP-induced BCR-ABL degradation and combination of Bag1 overexpression and Hsc70 knockdown promotes CHIP-induced suppression of BCR-ABL–dependent cell growth. (A) Flag-tagged BCR-ABL proteins were in vitro transcribed/translated with (+) or without (−) GA and then incubated with 20S proteasome with (+) or without (−) His-tagged Bag1. Numbers indicate the intensities of the bands of 20S proteasome. (B) Flag-tagged BCR-ABL proteins were in vitro–transcribed/translated with GA and then incubated with 20S proteasome with His-tagged Bag1 (wt) or His-tagged ubiquitin like domain-deletion mutant of Bag1 (ΔUb). Anti-Flag immunoprecipitates and whole reticulocyte lysates (WRL) were subjected to immunoblotting with the indicated antibodies (A-B). (C) Glutathione sepharose-bound GST-tagged Bag1 proteins were incubated with His-tagged CHIP, E1, E2 (UbcH5b or UbcH5c), and biotin-labeled ubiquitin. Ubiquitinated proteins were detected by streptavidin-HRP. An arrow indicates ubiquitinated Bag1. (D) COS7 cells were transiently transfected with the BCR-ABL together with or without Xpress-tagged Bag1M, Bag1S, or control vector, and the tet system of CHIP or control vector. (E) BCR-ABL–expressing Ba/F3 cells with the tet system for CHIP were transiently transfected with Xpress-tagged Bag1S or Bag1M together with or without Hsc70 siRNA. The cells were incubated with or without tet for 24 hours (D) or 48 hours (E) and were analyzed by immunoblotting with the indicated antibodies (D-E). (F-G) BCR-ABL–expressing Ba/F3 cells with the tet system for CHIP were transiently transfected with Xpress-tagged Bag1S or Bag1M together with or without Hsc70 siRNA. After 2 days, the viable cell numbers were analyzed based on volume and side-scattering gating (F) and then GFP-positive (CHIP-expressing) cell numbers were analyzed by fluorescent intensity (G) with flow cytometry. (H) K562 cells were transiently transfected with the tet system for CHIP together with or without Xpress-tagger Bag1 and Hsc70 siRNA. After 2 days, the viable cell numbers were analyzed based on volume and side-scattering gating with flow cytometry. The values are mean ± SEs determined by 4 independent experiments. *P < .05, **P < .01, ***P < .001.
Combination of Hsc70 knockdown and Bag1 overexpression promotes CHIP-induced suppression of BCR-ABL–dependent cell growth
To assess the biologic significance of our findings, we examined cell growth of Ba/F3-p185 BCR-ABL cells with the tet-regulated CHIP expression system (shown in Figure 1A) or K562 cells by manipulating the expression levels of Bag1 and/or Hsc70. In contrast to the results in COS cells as stated in the previous paragraph, Bag1 expression alone in those 2 leukemic cells failed to induce significant degradation of BCR-ABL (Figure 6E and supplemental Figure 10C) and had almost no effect in cell growth. However, anti-Hsc70 siRNAs treatments caused Bag1M-dependent BCR-ABL degradation and growth suppression in Ba/F3-p185 BCR-ABL-CHIP-tet cells and K562 cells (Figure 6E-F and supplemental Figure 10C-D). An additive negative influence to CHIP induction on protein degradation and cell growth was observed by the combinatory expression of Bag1 and anti-Hsc70 siRNAs in those 2 leukemic cells and the effect was more prominent in Bag1M than Bag1S (Figure 6F-H). In both indicator cell lines, the numbers of live cells decreased in 48 hours without affecting the trypan blue-stainable cell numbers. However, in 72 hours, we observed increased dead cell numbers. Therefore we suppose the biologic effect in this condition is biphasic, initial growth inhibition followed by apoptosis.
Discussion
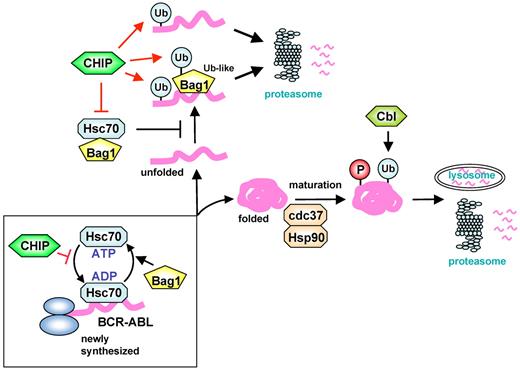
Down-regulation of BCR-ABL is a crucial anticancer strategy for CML. Hsc70 associates with newly synthesized polypeptides to promote their proper folding and also facilitates recognition of misfolded abnormal proteins by CHIP followed by proteasomal elimination.35,36 However, it is not fully understood how unfolded or misfolded proteins are recognized. We showed here that both c-Cbl and CHIP ubiquitin ligases are involved in Hsp90 inhibitor-induced BCR-ABL degradation. Immature BCR-ABL unchaperoned by Hsp90 is directly recognized by Bag1 for proteasomal degradation, which is promoted by CHIP and attenuated by Hsc70 (see Figure 7 for schematic representation).
Schematic representation of regulation of molecular fate of BCR-ABL by CHIP and c-Cbl. Newly synthesized BCR-ABL proteins are initially stabilized by Hsc70 and then passed on to Hsp90 and cdc37 to achieve maturation or kinase activation. c-Cbl induces ubiquitination-mediated degradation of mature BCR-ABL in a phosphorylation-dependent manner. Bag1 facilitates Hsc70-mediated folding of BCR-ABL. A large and multidomain protein such as BCR-ABL is inefficiently amenable to Hsc70-mediated folding. Bag1 recognizes immature BCR-ABL via BCR 1-413 and the ABL kinase domain, which is competed by Hsc70. CHIP acts not only as an E3 ligase for both immature BCR-ABL and Bag1 but also as a negative regulator of Hsc70 to promote the interaction of Bag1 and immature BCR-ABL. Bag1 binds to the proteasome and stimulates CHIP-induced BCR-ABL degradation. The mechanism can also apply to BCR-ABL degradation by Hsp90 inhibitors.
Schematic representation of regulation of molecular fate of BCR-ABL by CHIP and c-Cbl. Newly synthesized BCR-ABL proteins are initially stabilized by Hsc70 and then passed on to Hsp90 and cdc37 to achieve maturation or kinase activation. c-Cbl induces ubiquitination-mediated degradation of mature BCR-ABL in a phosphorylation-dependent manner. Bag1 facilitates Hsc70-mediated folding of BCR-ABL. A large and multidomain protein such as BCR-ABL is inefficiently amenable to Hsc70-mediated folding. Bag1 recognizes immature BCR-ABL via BCR 1-413 and the ABL kinase domain, which is competed by Hsc70. CHIP acts not only as an E3 ligase for both immature BCR-ABL and Bag1 but also as a negative regulator of Hsc70 to promote the interaction of Bag1 and immature BCR-ABL. Bag1 binds to the proteasome and stimulates CHIP-induced BCR-ABL degradation. The mechanism can also apply to BCR-ABL degradation by Hsp90 inhibitors.
Folding of many tyrosine kinases by Hsc70 precedes chaperoning by Hsp90 for activation, disruption of which by Hsp90 inhibitors causes degradation of client proteins with ubiquitination.9,36 Because only double knockout of CHIP and c-Cbl rescued the Hsp90 inhibitor-induced BCR-ABL degradation, both c-Cbl and CHIP may be involved in Hsp90 inhibitor-induced BCR-ABL degradation. We as well as other groups show that Hsp90 inhibitors induce the dissociation of Hsp90 from BCR-ABL, with concomitant formation of the Hsc70-BCR-ABL complex (supplemental Figure 7A,C).12,37 This may seem to be in disagreement with the result that Hsc70 overexpression increased BCR-ABL protein levels as shown in Figure 5E. This can be explained as follows. c-Cbl induces ubiquitin-dependent degradation of mature and phosphorylated BCR-ABL proteins, while CHIP degrades immature BCR-ABL proteins. We assume that activated BCR-ABL proteins released from the Hsp90-BCR-ABL complex by Hsp90 inhibitors may be targeted by c-Cbl for protein degradation. In contrast, Hsp90 inhibitors also increase Hsp90-unchaperoned immature BCR-ABL immediately after translation. Hsc70 overexpression partially counteracts against GA-induced BCR-ABL degradation (supplemental Figure 7B). During the Hsc70-mediated refolding of immature BCR-ABL, cochaperone Bag1 not only stimulates Hsc70 chaperoning activity, but also directly binds and sorts the Hsp90 unchaperoned immature BCR-ABL to the proteasome for degradation. GA promoted binding of purified ABL kinase and Bag1 in vitro (supplemental Figure 7A). Increased BCR-ABL binding to Hsc70, Hsp70 (supplemental Figure 7C), and Bag1 (Figure 3F) is also shown in K562 cells. CHIP stimulates Bag1-induced BCR-ABL degradation not only by ubiquitination of BCR-ABL and Bag1, but also by sequestration and inhibition of Hsc70. Hsc70 may protect newly synthesized Hsp90-unchaperoned immature BCR-ABL against CHIP-induced protein degradation in vivo by inhibition of Bag1 binding to BCR-ABL for protein degradation.
The cocrystal structure of imatinib and the ABL kinase domain revealed that the drug specifically trapped by the kinase leads to specific inactive conformation.30,31 Binding of BCR-ABL with imatinib structurally “closes” the kinase domain. Our results showed that treatment with imatinib attenuated Bag1 binding to BCR-ABL and also Hsp90 inhibitor-induced degradation of BCR-ABL. Previous reports from others showed that imatinib enhanced the cytotoxic effect of Hsp90 inhibitors.29,38 We have also found a similar combinatory effect (see the effects at a fixed dose of GA in supplemental Figure 11A). However, the higher the imatinib concentration is, the less prominent the dose-dependent synergistic effect of GA is, and GA-induced down-regulation of BCR-ABL is partially abrogated in the presence of imatinib (supplemental Figure 11A-B). T315I was found to bind Bag1 more strongly than the wt, which correlates with the sensitivity of Hsp90 inhibitors for BCR-ABL degradation and CML mice with the mutant.8,29 These informations suggest that imatinib binding induces structural maturation of BCR-ABL to which Bag1 can no longer bind, and the T315I mutant is presumed to be less tightly bound to Hsp90 than the wt. The ABL kinase domain bound to imatinib may escape its degradation for the time being. To overcome imatinib resistance, combination of imatinib with other reagents has been investigated. Radujkovic et al38 reported that the combination of imatinib and Hsp90 inhibitor, 17-AAG, may be useful to overcome imatinib resistance in a clinical setting. However, in our experiment, imatinib attenuated the Hsp90 inhibitor-induced degradation of BCR-ABL. Therefore, the combination of imatinib and Hsp90 inhibitor should be used carefully in cancer treatment, which may depend upon the site of mutation in the kinase domain.
Significant degradation of BCR-ABL by CHIP is associated with complete inhibition of BCR-ABL–dependent cell growth. c-Cbl–induced BCR-ABL degradation and inhibition of the IL-3–independent cell growth was less prominent than those of CHIP. Tyrosine-phosphorylated c-Cbl by BCR-ABL is directly bound to the SH2 domain of BCR-ABL, which also recruits Grb-2 and the p85 subunit of the PI 3-kinase. Activation of the PI 3-kinase pathway by c-Cbl may contradict with its inhibitory effect on the BCR-ABL–dependent cell growth.19 Bag1 has been reported to stimulate survival of cytokine-dependent and BCR-ABL-untransformed cells.39 This may partly explain our findings that Bag1 alone could efficiently degrade BCR-ABL proteins in COS7 cells but failed to inhibit BCR-ABL–dependent growth in Ba/F3 cells. However, Hsc70 knockdown with or without CHIP induction gave the Bag1-dependent biologic effects. Because Hsc70 knockdown without Bag1 expression slightly inhibited cell growth under CHIP induction without reducing the BCR-ABL protein amount in Ba/F3 cells, we cannot completely exclude a possibility that CHIP could target other growth-promoting client proteins than BCR-ABL. Our findings may benefit not only understanding the molecular triage decision of protein degradation but also future strategy in the treatment of imatinib-resistant CML.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr O. N. Witte at University of California Los Angeles for critical reading of the manuscript and supplying tet constructs and Drs T. Yamamoto and T. Tezuka at Tokyo University for the c-Cbl plasmid.
This work was supported in part by a Grant-in-Aid for Scientific Research from Ministry of Education, Science, Sports, and Culture of Japan nos. 19590259 to F.T. and 20013041 to Y.M.
Authorship
Contribution: F.T. and Y.M. designed and performed the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yoshiro Maru, Department of Pharmacology, Tokyo Women's Medical University School of Medicine, 8-1, Kawada-cho, Shinjuku-ku, Tokyo 162-8666, Japan; e-mail: ymaru@research.twmu.ac.jp.

![Figure 2. c-Cbl induces ubiquitin-dependent degradation of mature and phosphorylated BCR-ABL proteins, while CHIP degrades immature BCR-ABL proteins. (A) Structure of BCR-ABL, BCR, ABL, and BCR-ABL mutants used in this study. Deletions of functional domains of p185 BCR-ABL are shown, including the coiled-coil oligomerization domain (Δ40), SH2-binding domain (ΔSH2 bind), SH3 domain (ΔSH3), ΔSH2 domain (ΔSH2), kinase domain (Δkina), SH2-binding domain and SH3 domain (ΔSH2biΔSH3), kinase-negative mutant (ABL K290R, KD), imatinib-resistant mutants (ABL T315I, E255K), ABL, BCR, and BCR 1-413. (B) Effects of CHIP and c-Cbl on the stabilities of BCR-ABL, BCR, ABL, and BCR-ABL mutants. COS7 cells were transiently transfected with the wt p185 BCR-ABL or its mutants [KD, T315I, E255K, Δ40, Δkina, ΔSH2, ΔSH2 bind, ΔSH3, ΔSH2biΔSH3, BCR 1-413], BCR, and ABL (type 1b) together with CHIP, c-Cbl, or control vector (−) under the tet regulatory expression system. The cells were incubated with or without tet for 24 hours and analyzed by immunoblotting with antibodies against ABL, BCR, CHIP, c-Cbl, and actin. The top graphs indicate the intensities of the signals of tet (−) cells relative to those of tet (+) cells that were normalized against those of the control vector. The data were expressed as percentage of the signals obtained with each control. The red lines, at 53.2% and 73.3% in the CHIP and c-Cbl panels, respectively, are drawn to make a statistically significant discrimination into 2 groups (Akaike Information Criterion and t test, P < .001). (C) Immunoprecipitated Flag-tagged BCR-ABL proteins that were in vitro–transcribed/translated with (+) or without (−) GA (10μM) were incubated with biotin-labeled ubiquitin, GST-CHIP, GST-c-Cbl, E1, mixtures of E2s as indicated. The numbers indicate the signals of ubiquitinated BCR-ABL. (D) Immunoprecipitated BCR-ABL proteins from p185 wt- and Δkina-expressing Ba/F3 cell lysates were incubated with GST-CHIP, GST-c-Cbl, E1, E2s (Ubc4 and UbcH5c), and biotin-labeled ubiquitin. Ubiquitinated proteins were detected by streptavidin-HRP (C-D).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/18/10.1182_blood-2009-10-249623/4/m_zh89991059850002.jpeg?Expires=1765901279&Signature=hdCnUDjXFAD2ZIIgZFRk0mBlhhP67bwUJdXuytP5fF3tX6lRZppnS~wI~6dftl8pU6oGaPesetza1T3-HZQ9urz5iU~EMNYHomXj52ePDqaNWnKsPebSm39izz5cYgdIEVW0Q~vXrG2P4583n5~khO08Q5k1DBwFho78yRDBCpSCK80-1uBklxB7TRAsNMycpW~JGAwMEOscEtUmcfA5-PCu0rRkVS1pFmu3zxjmXUlQQBUJ0PtGyU2U2MnLAlGrDPFLi0TGgPtUhUv4ny98rfELWDfP2y-C7ZzjsEi2HRgNe7Os~Y41PxzsL1q258OR~Fpvky9JY6bh108tK8dwJg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal