Abstract
Hemokinin-1 (HK-1), encoded by the TAC4 gene, is a tachykinin peptide that is predominantly expressed in non-neuronal cells, such as immune cells. We have disrupted the mouse TAC4 gene to obtain a better understanding of the actions of HK-1 during hematopoiesis. We demonstrate here that TAC4−/− mice exhibit an increase of CD19+CD117+HSA+BP.1− “fraction B” pro-B cells in the bone marrow, whereas pre-B, immature, and mature B cells are within the normal range. We show that in vitro cultures derived from TAC4−/− bone marrow, sorted “fraction B” pro-B cells or purified long-term reconstituting stem cells, contain significantly higher numbers of pro-B cells compared with controls, suggesting an inhibitory role for HK-1 on developing B cells. Supporting this idea, we show that addition of HK-1 to cultures established from long-term reconstituting stem cells and the newly described intermediate-term reconstituting stem cells leads to a significant decrease of de novo generated pro-B cells. Based on our studies, we postulate that HK-1 plays an inhibitory role in hematopoiesis, and we hypothesize that it may be part of the bone marrow microenvironment that supports and regulates the proliferation and differentiation of hematopoietic cells.
Introduction
The tachykinins are a family of structurally related neuropeptides that share the common C-terminal motif FXGLM-NH2, which is essential for their biologic activities (see Figure 1A). In the mouse, TAC1 encodes both substance P (SP) and neurokinin A (NKA) through alternative splicing. TAC2 produces the peptide neurokinin B (NKB), and TAC4 encodes hemokinin-1 (HK-1). Tachykinins mediate their actions through at least 3 transmembrane G-protein coupled receptors, neurokinin-1 (NK-1), NK-2, and NK-3 receptors. Similar to SP, HK-1 acts as a full agonist of all 3 NK receptors with the highest affinity for NK-1, leading to SP-like responses.1-6
Unlike all other tachykinin peptides whose sequence is identical in all mammalian species, the sequence of human HK-1 differs from mouse/rat HK-1.7 Furthermore, human TAC4 was found to be spliced into 4 alternative splice variants (α, β, γ, and δ), giving rise to 4 novel peptides: endokinin A (EKA), EKB, EKC, and EKD.6,8
There is accumulating evidence that HK-1 plays a role during hematopoiesis with important implications for hematology as it impacts immune cells of various lineages. TAC4 was first discovered in mice in studies designed to identify genes that are important for B-cell development. The newly identified tachykinin peptide HK-1 was shown to promote the survival of B lineage cells.9 Recently, HK-1 was reported to act as a costimulatory factor for B-cell activation as it enhanced cell survival, proliferation, and IgM secretion in splenic B cells induced with suboptimal concentrations of lipopolysaccharide (LPS), anti-IgM, and anti-CD40.10 HK-1 has also been shown to enhance the proliferation of T-cell precursors11 and has been reported to rescue dendritic cells from apoptosis induced by deprivation of granulocyte-macrophage colony-stimulating factor (GM-CSF) and interleukin-4 (IL-4).12 These immunoregulatory effects may have important implications in various diseases as a recent study demonstrated that HK-1 suppressed cell proliferation and induced differentiation of the human promyelocytic leukemia cell line HL-60, suggesting a role for HK-1 in cancer development.13
The discovery of a tachykinin gene that encodes for a new and species-divergent group of novel tachykinin peptides raises new questions with respect to function of this family of peptides. Knowledge about TAC4 and its gene products is limited, and the physiologic role of HK-1 remains to be determined. To obtain a better understanding of the functions of HK-1 in vivo, we have generated mice with a targeted deletion of exon 2 of the TAC4 gene, which encodes the entire HK-1 peptide. This study is the first to show that the lack of HK-1 altered the B-cell compartment in TAC4−/− mice leading to increased numbers of pro-B cells in vivo and in vitro, pointing to a novel inhibitory role for HK-1 during the early stages of hematopoiesis and lymphocyte development.
Methods
Generation of TAC4−/− mice
A TAC4 targeting vector was constructed as shown in Figure 1B. We replaced exon 2 of TAC4, which encodes the entire HK-1 peptide, with an inverted PGK-neomycin cassette (NEO). A diphtheria toxin cassette (DT-A) was used as a negative selection marker. TAC4 mutant mice were generated by homologous recombination in E14 embryonic stem cells. G418-resistant embryonic stem cell clones were picked and screened for the presence of the expected recombination event. Two independent TAC4 mutant clones were used to derive chimeras by blastocyst injection, resulting in the generation of 2 independent TAC4−/− founders. Chimeras were crossed with C57BL/6 mice to obtain the N1 offspring on a mixed genetic background (129 and C57BL/6). Germline transmission was identified by agouti coat color and confirmed by genotyping. The REDExtract-N-Amp Tissue PCR kit (Sigma-Aldrich) was used for polymerase chain reaction (PCR) genotyping. Southern blot analysis was carried out according to standard procedures. TAC4 gene expression analysis (reverse-transcribed [RT]–PCR and Northern blot analysis) was carried out according to standard procedures (primers: Table 1).
Primers for TAC4 genotyping, generation of probes, and gene expression analysis
| Probe/primer . | Sequence, 5′-3′ . |
|---|---|
| Genotyping PCR | |
| TAC4 forward | CCTGGGGTAAACTAGAATGGT |
| WT reverse | GTACCAGAGCTAAGGGCTGTTC |
| neo reverse | CATCGCCTTCTATCGCCTTCT |
| Southern probe neo | |
| neo2 forward | GAAGAACTCGTCAAGAAGGCG |
| neo1 reverse | AACAAGATGGATTGCACGCAG |
| Southern probe TAC4 | |
| HK1 ex1 forward | TCCCATGAAGCATGGCAAAG |
| HK1 ex1 reverse | ACCAAAAGCCAGTTCCTCT |
| RT-PCR β-actin | |
| β-Actin forward | TCCCTGGAGAAGAGCTACGA |
| β-Actin reverse | ATCTGCTGGAAGGTGGACAG |
| RT-PCR TAC4 | |
| TAC4 forward | CGGGCCATCAGTGTGCACTA |
| TAC4 711 reverse | CGTTGCTACCAGATGCCAAGAG |
| Northern probe TAC4 | |
| TAC4 CM2 forward | GAGGAGATGTGACAGGCAGGTG |
| TAC4 genA1 reverse | ACATATGCCATGACACATGCAGAG |
| Northern probe L32 | |
| L32 forward | GTGAAGCCCAAGATCGTC |
| L32 reverse | AGCAATCTCAGCACAGTAAG |
| Global RT-PCR TAC4 | |
| 3′ forward 2 TAC4 | GAAGACGCTGCATGTATTG |
| 3′ reverse TAC4 | CATATGCCATGACACATGCAG |
| Global RT-PCR NK-1 | |
| 3′ forward TACR1 | CTCCAGTATCGACAGGCTGG |
| 3′ reverse TACR1 | ACAGATGCTTCAGGAATGAC |
| Probe/primer . | Sequence, 5′-3′ . |
|---|---|
| Genotyping PCR | |
| TAC4 forward | CCTGGGGTAAACTAGAATGGT |
| WT reverse | GTACCAGAGCTAAGGGCTGTTC |
| neo reverse | CATCGCCTTCTATCGCCTTCT |
| Southern probe neo | |
| neo2 forward | GAAGAACTCGTCAAGAAGGCG |
| neo1 reverse | AACAAGATGGATTGCACGCAG |
| Southern probe TAC4 | |
| HK1 ex1 forward | TCCCATGAAGCATGGCAAAG |
| HK1 ex1 reverse | ACCAAAAGCCAGTTCCTCT |
| RT-PCR β-actin | |
| β-Actin forward | TCCCTGGAGAAGAGCTACGA |
| β-Actin reverse | ATCTGCTGGAAGGTGGACAG |
| RT-PCR TAC4 | |
| TAC4 forward | CGGGCCATCAGTGTGCACTA |
| TAC4 711 reverse | CGTTGCTACCAGATGCCAAGAG |
| Northern probe TAC4 | |
| TAC4 CM2 forward | GAGGAGATGTGACAGGCAGGTG |
| TAC4 genA1 reverse | ACATATGCCATGACACATGCAGAG |
| Northern probe L32 | |
| L32 forward | GTGAAGCCCAAGATCGTC |
| L32 reverse | AGCAATCTCAGCACAGTAAG |
| Global RT-PCR TAC4 | |
| 3′ forward 2 TAC4 | GAAGACGCTGCATGTATTG |
| 3′ reverse TAC4 | CATATGCCATGACACATGCAG |
| Global RT-PCR NK-1 | |
| 3′ forward TACR1 | CTCCAGTATCGACAGGCTGG |
| 3′ reverse TACR1 | ACAGATGCTTCAGGAATGAC |
Heterozygous N1 mice were backcrossed 4 times to C57BL/6 mice to obtain TAC4−/− mice on a C57BL/6 background (N5). C57BL/6 mice were purchased from The Jackson Laboratory. Mice were housed under pathogen-free conditions. Experiments were performed in compliance with the University Health Network's Institutional Animal Facility.
FACS analysis of total bone marrow
Bone marrow was flushed with sterile phosphate-buffered saline (PBS) containing 3% fetal calf serum (FCS). After red blood cell lysis, bone marrow cells were resuspended in PBS with 3% FCS. Cells were stained for 30 minutes at 4°C with a combination of antibodies commonly used for bone marrow analysis. Antibodies were directly conjugated to fluorescein isothiocyanate (FITC), phycoerythrin (PE), biotin, or allophycocyanin (APC; BD Biosciences). Peridinin chlorophyll protein (PerCP) streptavidin protein was used to stain cells in combination with biotinylated primary antibodies. Fluorescence-activated cell sorter (FACS) analysis was performed on FACSCalibur (BD Biosciences), and data analysis was conducted using CellQuest Pro.
All experiments were performed using female mice, 7 to 10 weeks old. Every FACS experiment included an age-matched set of TAC4−/− and wild-type (WT) controls from the same founder. The 2 independent TAC4−/− mutant mouse founders gave similar results and were combined into 1 group.
Cell sorting for CD19+CD117+HSA+BP.1− “fraction B” pro-B cells
Equal numbers of bone marrow cells harvested from WT and TAC4−/− mice were stained with BP.1 FITC, CD117 PE, HSA biotin, and CD19 APC for 30 minutes at 4°C. After a wash, the cells were stained with PerCP streptavidin for 20 minutes at 4°C, washed again, and subsequently sorted using FACSAria (BD Biosciences). CD19+CD117+HSA+BP.1− “fraction B” pro-B cells were collected into collection tubes containing Opti-MEM with 10% non–heat-inactivated FCS.
In vitro B-cell experiments
For in vitro cultures, cells were seeded into B-cell media (Opti-MEM with 10% non–heat-inactivated FCS (Invitrogen), 1 times penicillin/streptomycin, and 5.5 × 10−5M 2-mercaptoethanol) containing 0.5 ng/mL IL-7 (total bone marrow: 3 × 106 cells in 20 mL media in a 25-cm2 flask; CD19+CD117+HSA+BP.1− cells: 5 × 104 cells in 1 mL of media in 24-well plates). Cells were grown with and without HK-1 (100nM, added daily). On day 4, cells were harvested, counted, and FACS analysis was performed.
Carboxyfluorescein succinimidyl ester (CFSE) labeling was performed according to the manufacturer's instructions (Invitrogen). Cells were cultured in B-cell media containing 0.5 ng/mL IL-7. CFSE fluorescence data were acquired every day, using day zero cells as a reference.
Purification of hematopoietic stem cells
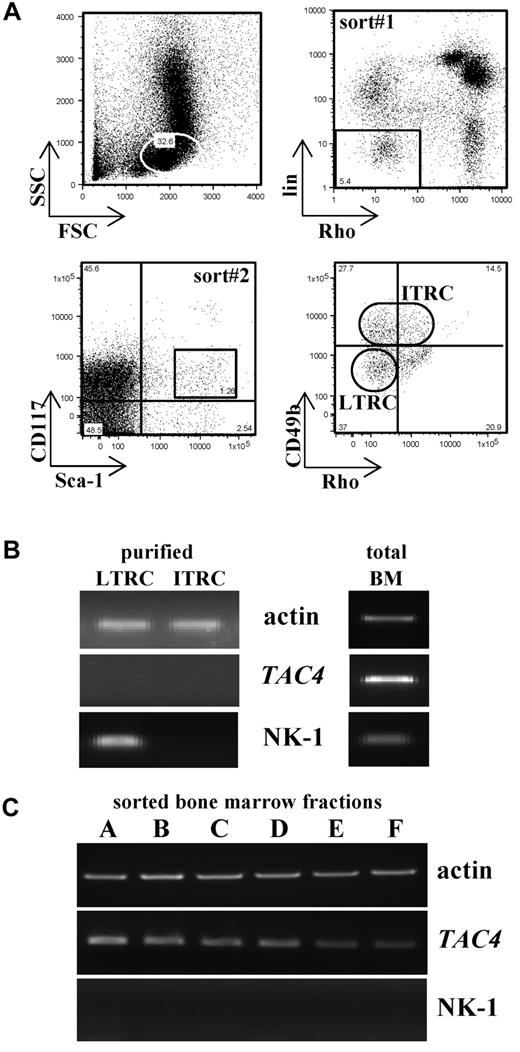
Bone marrow from 4 or 5 mice was pooled and sorted as previously described.14 Briefly, 2 consecutive sorts were performed. The first sort isolated a cell population negative/low for rhodamine-123 (Rho) and the lineage markers B220 and CD3. This negative fraction was further stained and sorted for Sca-1+ and CD117+ cells, which are heterogeneous for the expression of CD49b (see Figure 5A). Based on the expression of CD49b, 2 subsets were defined: long-term reconstituting cells (LTRCs): Sca-1+CD117+lin−Rho−CD49b−/low and intermediate-term reconstituting cells (ITRCs): Sca-1+CD117+lin−Rho−/lowCD49b+/intermediate (Figure 5A). Antibodies were directly conjugated to PE, PECy5, PECy7, and APC (BD Biosciences). Rhodamine-123 was purchased from Eastman-Kodak. Cells were sorted into 100 μL of complete hematopoietic stem cell media. Cell sorting was performed on FACSAria (BD Biosciences), and data analysis was conducted using Cell Quest Pro.
TAC4 and NK-1 receptor transcript expression in LTRCs and ITRCs
First cell divisions
For first cell division studies, purified LTRC and ITRC derived from TAC4−/− and WT bone marrow were plated in 96-well round bottom plates (Nunc) at 1 cell/well as previously described.14 Briefly, single cells were plated in 100 μL complete hematopoietic stem cell media containing recombinant IL-11 (10 ng/mL), recombinant stem cell factor (SCF; 50 ng/mL), IL-7 (1 ng/mL), recombinant Fms-like tyrosine kinase 3 (FLT3; 50 ng/mL), transferrin (5 μg/mL), insulin (5 μg/mL), 4% FCS, and 0.1% bovine serum albumin (BSA). HK-1 was added daily to specified wells (100nM). Cells were allowed to settle, and wells containing only a single cell were identified the next morning. All wells containing 1 cell/well were monitored every 5 hours over a time period of 55 to 60 hours, and cell numbers/well reflecting division rates were recorded.
In vitro cell cultures derived from hematopoietic stem cells
Purified LTRCs and ITRCs were plated in 96-well, round-bottom plates at a density of 100, 30, 10 cells/well (numbers permitting). HK-1 was added daily to specified wells (10nM, 100nM). Cells were fed twice weekly by adding fresh complete hematopoietic stem cell media. Starting on day 16, an aliquot from each well was counted and analyzed for the presence of myeloid cells (CD11b and Ly6GR FITC) and pro-B cells (CD43 PE, B220 PECy5, and IgM APC). FACS analysis was performed every second day until day 24. Antibodies were directly conjugated to FITC, PE, PECy5, and APC and were purchased from BD Biosciences. FACS analysis was performed on FACS Calibur (BD Biosciences), and data analysis was conducted using Cell Quest Pro.
Results
Mice with a targeted disruption of the TAC4 gene are healthy and fertile
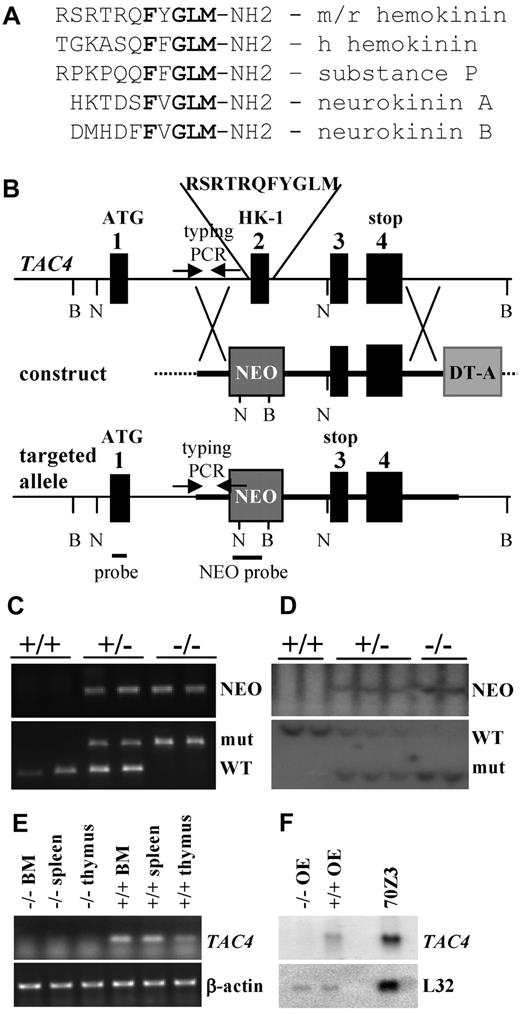
We engineered a TAC4 gene-targeting vector that replaced exon 2 of the TAC4 gene, which encodes the entire HK-1 peptide, with a neomycin resistance gene. The lack of exon 2 led to a premature stop codon in exon 3 of the TAC4 transcript (Figure 1B), resulting in a truncated TAC4 message. TAC4−/− mutant mice containing the desired mutation were confirmed by PCR and Southern blot analysis (Figure 1C-D). TAC4−/− mutant mice were obtained with the expected frequency (∼ 25%). Mutant mice had no gross physical abnormalities, were similar in size and weight to their littermates, and appeared healthy. All animals were fertile and cared for their offspring.
Generation of TAC4−/− mice. (A) Comparison of amino acid sequences of mouse/rat (m/r) HK-1, human (h) HK-1, SP, NKA, and NKB. The tachykinin motif FXGLM is indicated in bold type. (B) Targeted mutagenesis of the TAC4 gene. The map shows the WT locus, the construct, and the targeted allele after homologous recombination. Exons are represented as black boxes. Markers NEO and DT-A are shown as gray boxes. DNA probes for Southern hybridization and primers used for PCR typing (arrows) are indicated. Restriction enzymes used for Southern blot analysis are BglI (B) and NcoI (N). (C) Two TAC4+/+, 2 heterozygote (+/−), and 2 TAC4−/− mice were typed by PCR analysis for the presence of NEO as well as the WT and/or mutant band of TAC4. (D) Two TAC4+/+, 3 heterozygote (+/−), and 2 TAC4−/− mice were typed by Southern blot analysis for the presence of NEO as well as the WT and/or mutant band of TAC4. (E) RT-PCR analysis for TAC4 mRNA expression in bone marrow (BM), spleen, and thymus of a TAC4−/− and TAC4+/+ mouse. β-actin was used to confirm the presence of equal amounts of cDNA. (F) Northern blot analysis for TAC4 mRNA in the olfactory epithelium (OE) of a TAC4−/− and TAC4+/+ mouse. A total of 15 μg of total RNA was loaded per lane. 70Z/3 RNA was used as a positive control for TAC4 expression. L32 was used to assure equal loading of the gel.
Generation of TAC4−/− mice. (A) Comparison of amino acid sequences of mouse/rat (m/r) HK-1, human (h) HK-1, SP, NKA, and NKB. The tachykinin motif FXGLM is indicated in bold type. (B) Targeted mutagenesis of the TAC4 gene. The map shows the WT locus, the construct, and the targeted allele after homologous recombination. Exons are represented as black boxes. Markers NEO and DT-A are shown as gray boxes. DNA probes for Southern hybridization and primers used for PCR typing (arrows) are indicated. Restriction enzymes used for Southern blot analysis are BglI (B) and NcoI (N). (C) Two TAC4+/+, 2 heterozygote (+/−), and 2 TAC4−/− mice were typed by PCR analysis for the presence of NEO as well as the WT and/or mutant band of TAC4. (D) Two TAC4+/+, 3 heterozygote (+/−), and 2 TAC4−/− mice were typed by Southern blot analysis for the presence of NEO as well as the WT and/or mutant band of TAC4. (E) RT-PCR analysis for TAC4 mRNA expression in bone marrow (BM), spleen, and thymus of a TAC4−/− and TAC4+/+ mouse. β-actin was used to confirm the presence of equal amounts of cDNA. (F) Northern blot analysis for TAC4 mRNA in the olfactory epithelium (OE) of a TAC4−/− and TAC4+/+ mouse. A total of 15 μg of total RNA was loaded per lane. 70Z/3 RNA was used as a positive control for TAC4 expression. L32 was used to assure equal loading of the gel.
RT-PCR and Northern blot analysis were used to confirm the lack of TAC4 mRNA in TAC4−/− mice. Whereas the TAC4 message was easily detectable in bone marrow, spleen, and thymus of WT mice, TAC4 mRNA could be not detected in TAC4−/− mice by RT-PCR analysis (Figure 1E). TAC4 mRNA is expressed at very low levels and cannot be detected in most tissues by Northern blot analysis. An exception to this is the olfactory epithelium (A.H.T. et al, manuscript submitted), which contains high levels of TAC4 mRNA. However, in the olfactory epithelium of TAC4−/− mice, no TAC4 mRNA could be detected by Northern blot analysis (Figure 1F).
TAC4 deficiency results in altered frequency of pro-B cells in vivo
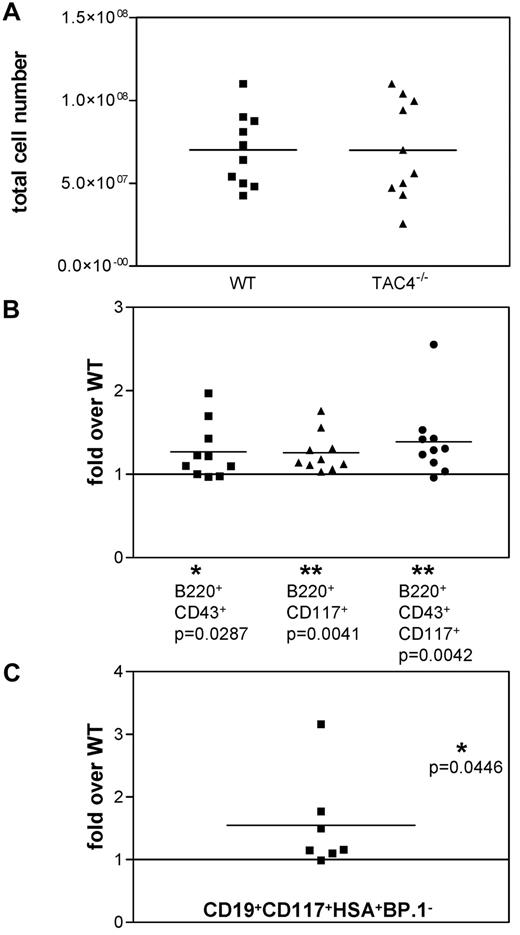
We extracted bone marrow from TAC4−/− and WT mice to determine whether TAC4 deficiency altered the development of the hematopoietic system. Over the course of these experiments, we found no significant differences in total cell numbers of TAC4−/− bone marrow compared with WT bone marrow (Figure 2A). We found that mutant mice exhibited a significant increase in populations containing pro-B-cell “fractions A-C” (identified as B220+CD43+, B220+CD117+, or B220+CD43+CD117+; Figure 2B). B220+CD43+ and B220+CD43+CD117+ cells were also significantly elevated in terms of absolute cell numbers (B220+CD43+: WT, 2.8 ± 0.6 × 106 cells; TAC4−/−, 3.5 ± 0.4 × 106 cells, P = .0351; B220+CD43+CD117+: WT, 1.2 ± 0.2 × 106 cells; TAC4−/−, 1.7 ± 0.3 × 106 cells, P = .0468). No significant differences in pre-B, immature, and mature B-cell populations or myeloid cells were detected (data not shown).
Analysis of TAC4−/− and WT bone marrow. (A) Scatter gram representing the absolute number of bone marrow cells recovered from WT and TAC4−/− mice. (B) Four-color FACS analysis was performed on total bone marrow samples. The fold change of TAC4−/− B-cell populations compared with WT B-cell populations is shown based on percentage. (C) The fold change of the CD19+CD117+HSA+BP.1− “fraction B” pro-B-cell population compared with WT “fraction B” is shown based on the cell number of sorted cells. Means, P values, and levels of significance are indicated (n = 7-10). *P < .05. **P < .005. P values were calculated from percentages using paired 2-tailed t tests (GraphPad Prism software Version 3.0).
Analysis of TAC4−/− and WT bone marrow. (A) Scatter gram representing the absolute number of bone marrow cells recovered from WT and TAC4−/− mice. (B) Four-color FACS analysis was performed on total bone marrow samples. The fold change of TAC4−/− B-cell populations compared with WT B-cell populations is shown based on percentage. (C) The fold change of the CD19+CD117+HSA+BP.1− “fraction B” pro-B-cell population compared with WT “fraction B” is shown based on the cell number of sorted cells. Means, P values, and levels of significance are indicated (n = 7-10). *P < .05. **P < .005. P values were calculated from percentages using paired 2-tailed t tests (GraphPad Prism software Version 3.0).
We further demonstrated that CD19+CD117+HSA+BP.1− cells, corresponding to the “fraction B” of pro-B-cell development, were significantly increased in TAC4−/− bone marrow compared with WT bone marrow (Figure 2C).
TAC4 deficiency leads to increased numbers of pro-B cells in vitro
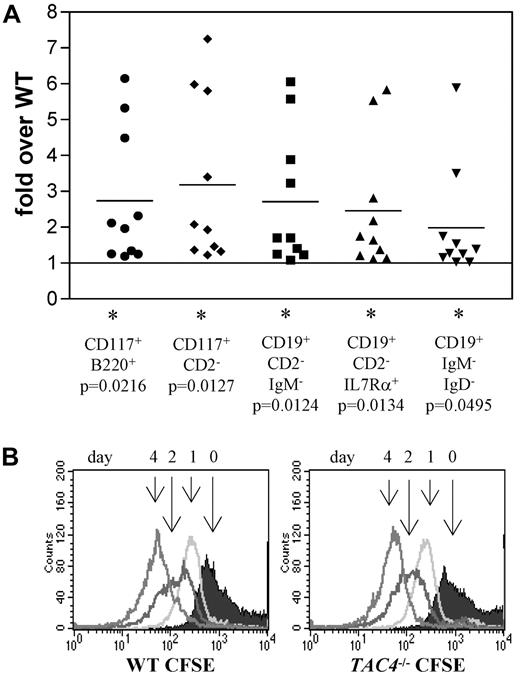
We next tracked the in vitro development of bone marrow cells isolated from WT and TAC4−/− mice. Equal numbers of WT and TAC4−/− bone marrow cells were cultured in the presence of IL-7. After 4 days, nearly all cells were CD19+ B lineage cells. Compared with WT controls, more cells were recovered from TAC4−/− cultures. Specifically, elevated numbers of B220+CD117+, CD2−CD117+, CD19+CD2−IgM−, CD19+CD2−IL7Ra+, and CD19+IgM−IgD− cell populations were found in TAC4−/− cultures. These cell surface traits mark populations containing the “fractions B and/or C” of B lineage populations (Table 2; Figure 3A). Although pro-B-cell numbers were elevated, there was no change in absolute numbers of more mature B-cell populations (Table 2).
FACS analysis of bone marrow cells from WT and TAC4−/− mice cultured in the presence of IL-7 for 4 days
| Population . | B-cell stage . | Fraction . | WT . | TAC4−/− . | P . |
|---|---|---|---|---|---|
| Total cell number | 5.6 ± 0.9 × 106 | 8.2 ± 1.2 × 106 | .0647 | ||
| B220+CD117+ | Unc → pro-B | A → C | 1.7 ± 0.4 × 106 | 3.4 ± 0.7 × 106 | .0216* |
| B220+CD117− | pre-B → mat | C′ → mat | 2.5 ± 0.4 × 106 | 2.8 ± 0.5 × 106 | .5899 |
| CD2−CD117+ | ELP → pro-B | ELP → C | 1.1 ± 0.3 × 106 | 2.4 ± 0.5 × 106 | .0127* |
| CD2−CD117− | pre-B | C′ | 0.7 ± 0.2 × 106 | 1.1 ± 0.2 × 106 | .0729 |
| CD2+CD117− | pre-B → mat | D → mat | 2.6 ± 0.5 × 106 | 2.9 ± 0.6 × 106 | .6453 |
| CD19+CD2−IgM− | pro-B → pre-B | B → C′ | 1.7 ± 0.4 × 106 | 3.4 ± 0.6 × 106 | .0124* |
| CD19+CD2+IgM− | pre-B | D | 1.9 ± 0.3 × 106 | 2.0 ± 0.4 × 106 | .8244 |
| CD19+CD2+IgM+ | imm → mat | E → mat | 0.7 ± 0.1 × 106 | 1.1 ± 0.2 × 106 | .0969 |
| CD19+CD2−IL7Rα+ | pro-B → pre-B | B → C′ | 1.8 ± 0.4 × 106 | 3.6 ± 0.6 × 106 | .0134* |
| CD19+CD2+IL7Rα+ | pre-B → imm | D,E | 1.7 ± 0.3 × 106 | 1.9 ± 0.4 × 106 | .6064 |
| CD19+CD2+IL7Rα− | imm → mat | imm → mat | 0.7 ± 0.2 × 106 | 0.7 ± 0.2 × 106 | .7970 |
| CD19+IgM−IgD− | pro-B → pre-B | B → D | 3.3 ± 0.6 × 106 | 5.3 ± 0.8 × 106 | .0495* |
| CD19+IgM+IgD− | imm | E | 0.7 ± 0.1 × 106 | 0.9 ± 0.2 × 106 | .2613 |
| CD19+IgM+IgD+ | imm → mat | imm → mat | 0.3 ± 0.1 × 106 | 0.4 ± 0.1 × 106 | .0718 |
| Population . | B-cell stage . | Fraction . | WT . | TAC4−/− . | P . |
|---|---|---|---|---|---|
| Total cell number | 5.6 ± 0.9 × 106 | 8.2 ± 1.2 × 106 | .0647 | ||
| B220+CD117+ | Unc → pro-B | A → C | 1.7 ± 0.4 × 106 | 3.4 ± 0.7 × 106 | .0216* |
| B220+CD117− | pre-B → mat | C′ → mat | 2.5 ± 0.4 × 106 | 2.8 ± 0.5 × 106 | .5899 |
| CD2−CD117+ | ELP → pro-B | ELP → C | 1.1 ± 0.3 × 106 | 2.4 ± 0.5 × 106 | .0127* |
| CD2−CD117− | pre-B | C′ | 0.7 ± 0.2 × 106 | 1.1 ± 0.2 × 106 | .0729 |
| CD2+CD117− | pre-B → mat | D → mat | 2.6 ± 0.5 × 106 | 2.9 ± 0.6 × 106 | .6453 |
| CD19+CD2−IgM− | pro-B → pre-B | B → C′ | 1.7 ± 0.4 × 106 | 3.4 ± 0.6 × 106 | .0124* |
| CD19+CD2+IgM− | pre-B | D | 1.9 ± 0.3 × 106 | 2.0 ± 0.4 × 106 | .8244 |
| CD19+CD2+IgM+ | imm → mat | E → mat | 0.7 ± 0.1 × 106 | 1.1 ± 0.2 × 106 | .0969 |
| CD19+CD2−IL7Rα+ | pro-B → pre-B | B → C′ | 1.8 ± 0.4 × 106 | 3.6 ± 0.6 × 106 | .0134* |
| CD19+CD2+IL7Rα+ | pre-B → imm | D,E | 1.7 ± 0.3 × 106 | 1.9 ± 0.4 × 106 | .6064 |
| CD19+CD2+IL7Rα− | imm → mat | imm → mat | 0.7 ± 0.2 × 106 | 0.7 ± 0.2 × 106 | .7970 |
| CD19+IgM−IgD− | pro-B → pre-B | B → D | 3.3 ± 0.6 × 106 | 5.3 ± 0.8 × 106 | .0495* |
| CD19+IgM+IgD− | imm | E | 0.7 ± 0.1 × 106 | 0.9 ± 0.2 × 106 | .2613 |
| CD19+IgM+IgD+ | imm → mat | imm → mat | 0.3 ± 0.1 × 106 | 0.4 ± 0.1 × 106 | .0718 |
Columns WT and TAC4−/− show absolute cell numbers (mean ± SE, n = 10). Surface markers used for FACS analysis and the developmental stage of B-cell populations are indicated: ELP (early lymphoid progenitor), fractions A (uncommitted [unc]), B and C (pro-B), C′ and D (pre-B), E (immature [imm] and mature [mat]). P values were calculated from absolute cell numbers using paired 2-tailed t tests.
Significance (P < .05).
Analysis of IL-7 cultures derived from total bone marrow from WT and TAC4−/− mice. (A) Four-color FACS analysis was performed on bone marrow cells grown in the presence of IL-7 for 4 days. The fold change of TAC4−/− B-cell populations compared with WT B-cell populations is shown based on absolute cell numbers. Data are means, P values, and levels of significance (n = 10). *P < .05. P values were calculated from absolute numbers using paired 2-tailed t tests (GraphPad Prism software Version 3.0). (B) CFSE staining profiles are shown for WT and TAC4−/− total bone marrow cultures over a time course of 4 days. The figure shows 1 representative experiment that has been performed multiple times with similar results.
Analysis of IL-7 cultures derived from total bone marrow from WT and TAC4−/− mice. (A) Four-color FACS analysis was performed on bone marrow cells grown in the presence of IL-7 for 4 days. The fold change of TAC4−/− B-cell populations compared with WT B-cell populations is shown based on absolute cell numbers. Data are means, P values, and levels of significance (n = 10). *P < .05. P values were calculated from absolute numbers using paired 2-tailed t tests (GraphPad Prism software Version 3.0). (B) CFSE staining profiles are shown for WT and TAC4−/− total bone marrow cultures over a time course of 4 days. The figure shows 1 representative experiment that has been performed multiple times with similar results.
CFSE analysis was performed to determine whether faster cell division rates are responsible for the increased number of pro-B cells in TAC4−/− bone marrow cultures. CFSE analysis revealed that the cell division rates were not significantly different in WT and TAC4−/− bone marrow cultures (Figure 3B).
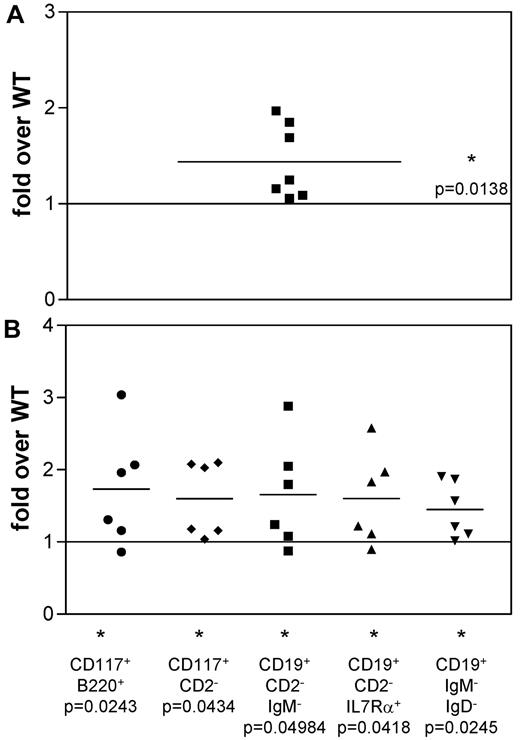
To investigate whether the difference in TAC4−/− cultures is a consequence of more pro-B cells in the starting culture, we purified CD19+CD117+HSA+BP.1− “fraction B” pro-B cells and initiated cultures with equal numbers of these cells. We found that cell recoveries from TAC4−/− cultures were significantly higher than from WT cultures (Table 3; Figure 4A). FACS analysis revealed that TAC4−/− derived pro-B-cell cultures contained significantly higher numbers of pro-B cells compared with WT cultures (Table 3; Figure 4B). However, in contrast to cultures established from total bone marrow that showed a specific increase of pro-B cells, cultures established from pro-B cells also revealed significant increases in IgM+ B-cell populations (Table 3).
FACS analysis of cultures established from “fraction B” pro-B cells from wild-type (WT) and TAC4−/− mice cultured in the presence of IL-7 for 4 days
| Population . | B-cell stage . | Fraction . | WT . | TAC4−/− . | P . |
|---|---|---|---|---|---|
| Total cell number | 8.4 ± 0.1 × 105 | 1.2 ± 0.1 × 106 | .0108* | ||
| B220+CD117+ | unc → pro-B | A → C | 5.2 ± 0.6 × 105 | 7.7 ± 0.5 × 105 | .0243* |
| B220+CD117− | pre-B → mat | C′ → mat | 2.8 ± 0.2 × 105 | 3.2 ± 0.2 × 105 | .0926 |
| CD2−CD117+ | ELP → pro-B | ELP → C | 3.6 ± 0.6 × 105 | 5.4 ± 0.4 × 105 | .0434* |
| CD2−CD117− | pre-B | C′ | 2.7 ± 1.3 × 105 | 1.8 ± 0.3 × 105 | .5888 |
| CD2+CD117− | pre-B → mat | D → mat | 2.7 ± 0.4 × 105 | 3.5 ± 0.3 × 105 | .1208 |
| CD19+CD2−IgM− | pro-B → pre-B | B → C′ | 5.0 ± 0.7 × 105 | 7.1 ± 0.4 × 105 | .0498* |
| CD19+CD2+IgM− | pre-B | D | 2.5 ± 0.4 × 105 | 2.7 ± 0.3 × 105 | .5110 |
| CD19+CD2+IgM+ | imm → mat | E → mat | 4.7 ± 0.6 × 104 | 8.8 ± 1.0 × 104 | .0020** |
| CD19+CD2−IL7Rα+ | pro-B → pre-B | B → C′ | 5.3 ± 0.7 × 105 | 7.6 ± 0.6 × 105 | .0418* |
| CD19+CD2+IL7Rα+ | pre-B → imm | D,E | 2.2 ± 0.3 × 105 | 2.8 ± 0.3 × 105 | .1021 |
| CD19+CD2+IL7Rα− | imm → mat | imm → mat | 3.3 ± 0.7 × 104 | 4.0 ± 0.7 × 104 | .2405 |
| CD19+IgM−IgD− | pro-B → pre-B | B → D | 7.3 ± 0.7 × 105 | 9.9 ± 0.5 × 105 | .0245* |
| CD19+IgM+IgD− | imm | E | 5.7 ± 1.0 × 104 | 8.8 ± 1.0 × 104 | .0012** |
| CD19+IgM+IgD+ | imm → mat | imm → mat | 1.3 ± 0.2 × 104 | 2.4 ± 0.2 × 104 | .0094** |
| Population . | B-cell stage . | Fraction . | WT . | TAC4−/− . | P . |
|---|---|---|---|---|---|
| Total cell number | 8.4 ± 0.1 × 105 | 1.2 ± 0.1 × 106 | .0108* | ||
| B220+CD117+ | unc → pro-B | A → C | 5.2 ± 0.6 × 105 | 7.7 ± 0.5 × 105 | .0243* |
| B220+CD117− | pre-B → mat | C′ → mat | 2.8 ± 0.2 × 105 | 3.2 ± 0.2 × 105 | .0926 |
| CD2−CD117+ | ELP → pro-B | ELP → C | 3.6 ± 0.6 × 105 | 5.4 ± 0.4 × 105 | .0434* |
| CD2−CD117− | pre-B | C′ | 2.7 ± 1.3 × 105 | 1.8 ± 0.3 × 105 | .5888 |
| CD2+CD117− | pre-B → mat | D → mat | 2.7 ± 0.4 × 105 | 3.5 ± 0.3 × 105 | .1208 |
| CD19+CD2−IgM− | pro-B → pre-B | B → C′ | 5.0 ± 0.7 × 105 | 7.1 ± 0.4 × 105 | .0498* |
| CD19+CD2+IgM− | pre-B | D | 2.5 ± 0.4 × 105 | 2.7 ± 0.3 × 105 | .5110 |
| CD19+CD2+IgM+ | imm → mat | E → mat | 4.7 ± 0.6 × 104 | 8.8 ± 1.0 × 104 | .0020** |
| CD19+CD2−IL7Rα+ | pro-B → pre-B | B → C′ | 5.3 ± 0.7 × 105 | 7.6 ± 0.6 × 105 | .0418* |
| CD19+CD2+IL7Rα+ | pre-B → imm | D,E | 2.2 ± 0.3 × 105 | 2.8 ± 0.3 × 105 | .1021 |
| CD19+CD2+IL7Rα− | imm → mat | imm → mat | 3.3 ± 0.7 × 104 | 4.0 ± 0.7 × 104 | .2405 |
| CD19+IgM−IgD− | pro-B → pre-B | B → D | 7.3 ± 0.7 × 105 | 9.9 ± 0.5 × 105 | .0245* |
| CD19+IgM+IgD− | imm | E | 5.7 ± 1.0 × 104 | 8.8 ± 1.0 × 104 | .0012** |
| CD19+IgM+IgD+ | imm → mat | imm → mat | 1.3 ± 0.2 × 104 | 2.4 ± 0.2 × 104 | .0094** |
Columns WT and TAC4−/− show absolute cell numbers (mean ± SE, n = 10). Surface markers used for FACS analysis and the developmental stage of B-cell populations are indicated: ELP (early lymphoid progenitor), fractions A (uncommitted [unc]), B and C (pro-B), C′ and D (pre-B), E (immature [imm] and mature [mat]). P values were calculated from absolute cell numbers using paired 2-tailed t tests.
Significance (P < .05).
Significance (P < .005).
Analysis of IL-7 cultures derived from “fraction B” pro-B cells from WT and TAC4−/− bone marrow. (A) The fold change in absolute cell numbers recovered from TAC4−/− pro-B-cell–derived cultures compared with WT cultures. (B) Four-color FACS analysis was performed on cells grown in the presence of IL-7 for 4 days. The fold change of TAC4−/− pro-B-cell populations compared with WT pro-B-cell populations is shown based on absolute cell numbers. Data are means, P values, and levels of significance (n = 6 or 7). *P < .05. P values were calculated from absolute numbers using paired 2-tailed t tests (GraphPad Prism software Version 3.0).
Analysis of IL-7 cultures derived from “fraction B” pro-B cells from WT and TAC4−/− bone marrow. (A) The fold change in absolute cell numbers recovered from TAC4−/− pro-B-cell–derived cultures compared with WT cultures. (B) Four-color FACS analysis was performed on cells grown in the presence of IL-7 for 4 days. The fold change of TAC4−/− pro-B-cell populations compared with WT pro-B-cell populations is shown based on absolute cell numbers. Data are means, P values, and levels of significance (n = 6 or 7). *P < .05. P values were calculated from absolute numbers using paired 2-tailed t tests (GraphPad Prism software Version 3.0).
We also assessed the effect of HK-1 on cultures derived from total bone marrow and CD19+CD117+HSA+BP.1− “fraction B” cells. Interestingly, daily addition of HK-1 (100nM) had no effect on developing B-cell populations in WT and TAC4−/− cultures (data not shown).
Purification of the hematopoietic stem cell populations LTRCs and ITRCs
The fact that the increase of pro-B cells in TAC4−/− cultures could not be reversed by adding HK-1 suggested that a developmentally earlier event may be responsible for the increased number of pro-B cells. To determine whether precursors upstream of the “fraction B” pro-B-cell stage are altered by the lack of HK-1, we examined cultures established from purified hematopoietic stem cell populations isolated from WT and TAC4−/− bone marrow.
Enrichment for LTRCs and ITRCs was performed according to Benveniste et al14 (Figure 5A). Similar cell numbers (300-1000 LTRCs and ITRCs for a pooled bone marrow sample obtained from 4 or 5 mice) were recovered from TAC4−/− and WT bone marrow.
Properties of LTRCs and ITRCs. (A) Bone marrow cells were enriched first for Rho123−/lowB220−CD3− cells (sort 1) and then sorted for Sca-1+CD117+ cells (sort 2). Sca-1+CD117+ cells were further purified based on the expression of CD49b. Sorted LTRCs are Sca-1+CD117+lin−Rho−CD49b−/low, and sorted ITRCs are Sca-1+CD117+lin−Rho−/lowCD49b+/intermediate. The example given shows a cell sort of WT bone marrow. (B) LTRCs and ITRCs purified from WT bone marrow were analyzed for their expression of TAC4 and NK-1 receptor by global RT-PCR. Total bone marrow cDNA was used as a positive control. β-actin was used to confirm the presence of equal amounts of cDNA. (C) Bone marrow “fractions A-F” were analyzed for their expression of TAC4 and NK-1 receptor mRNA. For B cell “fractions A-F,” bone marrow from 10 mice was pooled and stained with antibodies used to distinguish B-cell fractions. Enrichment was carried out in 2 independent sorts. In the first sort, B220+CD43+ live cells were gated and subsequently sorted for HSA−BP.1− (fraction A), HSA+BP.1− (fraction B), or HSA+BP.1+ (fraction C) populations. In the second sort, B220+CD43− cells were gated and sorted for IgM−IgD− (fraction D), IgM+IgD− (fraction E), or IgM+IgD+ (fraction F) populations.
Properties of LTRCs and ITRCs. (A) Bone marrow cells were enriched first for Rho123−/lowB220−CD3− cells (sort 1) and then sorted for Sca-1+CD117+ cells (sort 2). Sca-1+CD117+ cells were further purified based on the expression of CD49b. Sorted LTRCs are Sca-1+CD117+lin−Rho−CD49b−/low, and sorted ITRCs are Sca-1+CD117+lin−Rho−/lowCD49b+/intermediate. The example given shows a cell sort of WT bone marrow. (B) LTRCs and ITRCs purified from WT bone marrow were analyzed for their expression of TAC4 and NK-1 receptor by global RT-PCR. Total bone marrow cDNA was used as a positive control. β-actin was used to confirm the presence of equal amounts of cDNA. (C) Bone marrow “fractions A-F” were analyzed for their expression of TAC4 and NK-1 receptor mRNA. For B cell “fractions A-F,” bone marrow from 10 mice was pooled and stained with antibodies used to distinguish B-cell fractions. Enrichment was carried out in 2 independent sorts. In the first sort, B220+CD43+ live cells were gated and subsequently sorted for HSA−BP.1− (fraction A), HSA+BP.1− (fraction B), or HSA+BP.1+ (fraction C) populations. In the second sort, B220+CD43− cells were gated and sorted for IgM−IgD− (fraction D), IgM+IgD− (fraction E), or IgM+IgD+ (fraction F) populations.
TAC4 and NK-1 receptor gene expression analysis of various bone marrow fractions
To determine whether LTRCs and ITRCs express TAC4 and NK-1 receptor mRNA, sorted cells were used for gene expression analysis using global RT-PCR amplification. We show here that neither LTRCs nor ITRCs expressed TAC4 message (Figure 5B). However, TAC4 mRNA is abundant in total bone marrow and HK-1 may act on stem cells in a paracrine fashion. Interestingly, NK-1 receptor mRNA could be amplified in LTRCs but not in ITRCs (Figure 5B). Message for NK-2 and NK-3 receptor could not be detected (data not shown).
We further analyzed TAC4 and NK-1 gene expression in sorted B-cell “fractions A-F.” Whereas all sorted B-cell fractions expressed TAC4 mRNA to various degrees, no NK-1 receptor mRNA could be detected (Figure 5C). NK-2 and NK-3 receptor mRNA could not be detected (data not shown).
First cell division rates of hematopoietic stem cells are not altered in TAC4−/− cultures
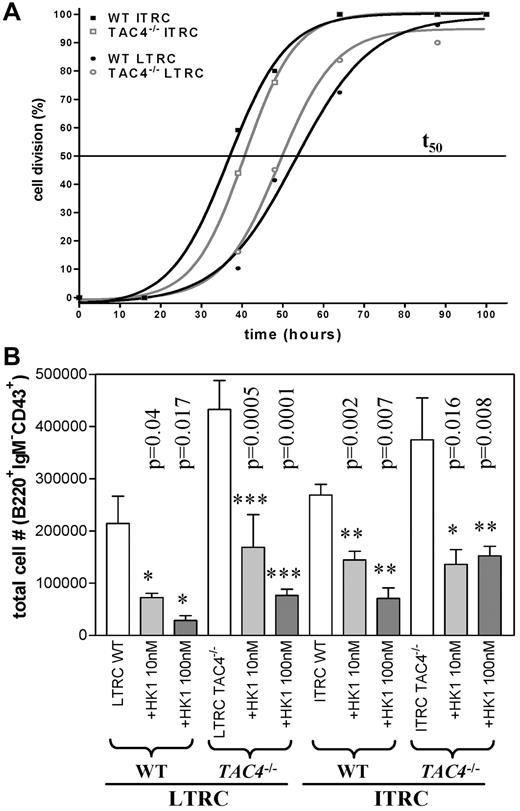
We next examined the kinetics of the first cell division of LTRCs and ITRCs isolated from WT and TAC4−/− bone marrow. Single-cell cultures revealed that first cell division rates were similar in LTRC and ITRC populations purified from WT and TAC4−/− bone marrow (Figure 6A). We also assessed the influence of HK-1 on first cell division rates. However, the addition of HK-1 (100nM) did not significantly affect the kinetics of the first cell division or the survival of these cells (data not shown).
In vitro analysis of LTRCs and ITRCs purified from WT and TAC4−/− bone marrow. (A) The kinetics of the first cell division of purified LTRCs and ITRCs in WT and TAC4−/− bone marrow was determined by single-cell division assays. The time point where 50% of cells from single-cell cultures (1 cell/well) have divided was calculated (t50). The figure shows 1 representative experiment performed 2 times with similar results (n = 25-50 wells). In this experiment, the values for t50 were: LTRCs WT (53 hours), LTRCs TAC4−/− (49 hours), ITRCs WT (36.7 hours), and ITRCs TAC4−/− (40.6 hours). (B) Bar graph showing total numbers of pro-B cells (B220+IgM−CD43+) derived from WT and TAC4−/− LTRC and ITRC cultures harvested after 20 to 22 days. TAC4−/− LTRC-derived cultures contained significantly elevated numbers of pro-B cells (LTRC WT vs TAC4−/−, P = .01; ITRC WT vs TAC4−/−, P = .3). Furthermore, the effect of HK-1 (10nM and 100nM) on B-cell development in LTRC- and ITRC-derived cultures was assessed. HK-1 significantly decreased the total number of pro-B cells in all groups. This figure shows 1 representative experiment. Data are mean ± SD (n = 6). P values and levels of significance are indicated. Experiments were performed at least 3 times with similar results.
In vitro analysis of LTRCs and ITRCs purified from WT and TAC4−/− bone marrow. (A) The kinetics of the first cell division of purified LTRCs and ITRCs in WT and TAC4−/− bone marrow was determined by single-cell division assays. The time point where 50% of cells from single-cell cultures (1 cell/well) have divided was calculated (t50). The figure shows 1 representative experiment performed 2 times with similar results (n = 25-50 wells). In this experiment, the values for t50 were: LTRCs WT (53 hours), LTRCs TAC4−/− (49 hours), ITRCs WT (36.7 hours), and ITRCs TAC4−/− (40.6 hours). (B) Bar graph showing total numbers of pro-B cells (B220+IgM−CD43+) derived from WT and TAC4−/− LTRC and ITRC cultures harvested after 20 to 22 days. TAC4−/− LTRC-derived cultures contained significantly elevated numbers of pro-B cells (LTRC WT vs TAC4−/−, P = .01; ITRC WT vs TAC4−/−, P = .3). Furthermore, the effect of HK-1 (10nM and 100nM) on B-cell development in LTRC- and ITRC-derived cultures was assessed. HK-1 significantly decreased the total number of pro-B cells in all groups. This figure shows 1 representative experiment. Data are mean ± SD (n = 6). P values and levels of significance are indicated. Experiments were performed at least 3 times with similar results.
TAC4 deficiency leads to increased numbers of de novo generated pro-B cells in vitro
We further analyzed the de novo generation of B cells in vitro in cultures established from purified populations of LTRCs and ITRCs grown for 20 to 22 days. In vitro cultures derived from TAC4−/− LTRCs contained a significant increase in absolute numbers of pro-B cells compared with WT cultures (Figure 6B). Although not statistically significant at the P < .05 level, TAC4−/−-derived ITRC cultures also showed a tendency toward increased pro-B-cell numbers. No difference in the number of myeloid cells between WT and TAC4−/− was observed (data not shown).
Addition of HK-1 reversed the increase in pro-B cells in TAC4−/− cultures in a dose-dependent manner
The increased number of TAC4−/− pro-B cells in vivo as well as in vitro suggested that HK-1 may play an inhibitory role in early B-cell development. To test this hypothesis, we added HK-1 to LTRC- and ITRC-derived cultures. We show here that daily addition of HK-1 significantly decreased the number of LTRC- and ITRC-derived pro-B cells in a dose-dependent manner (Figure 6B). The HK-1-induced suppression of pro-B-cell development was similar in LTRC- and ITRC-derived cultures and was seen in WT and TAC4−/− bone marrow (Figure 6B). The absolute number of pro-B cells decreased by 80% to 90% in the presence of 100nM HK-1 (Figure 6B). HK-1 did not induce cell death as determined by daily visual inspection.
Discussion
HK-1 and the endokinins, encoded by the TAC4 gene, are the newest members of the tachykinin family and close relatives to SP. Since the discovery of TAC4 orthologs in mouse, rat, human, and rabbit, the number of tachykinin peptides has more than doubled. In addition to the originally identified mammalian tachykinins SP and NKA (encoded by TAC1) and NKB (encoded by TAC2), the tachykinins now include the TAC4 gene products HK-1 in mouse, rat, and human,7,9 EKA, EKB, EKC, and EKD in human,6 and EK-1 and EK-2 in rabbit.6,8,17
In contrast to other tachykinins and neuropeptides in general, TAC4 is not expressed at high levels in the nervous system and is found widely in the periphery7 and in cells of hematopoietic origin, such as B cells, T cells, monocytes/macrophages, dendritic cells, and platelets.9,11,18-23 Interestingly, various groups have demonstrated that TAC4 gene expression is regulated during hematopoiesis. For example, maturation of B cells, dendritic cells, and monocytes/macrophages resulted in a decrease of TAC4 mRNA.9,18,22,24 The significance of high TAC4 expression in hematopoietic cells during early but not late stages of development remains unknown. These findings suggest that HK-1 probably plays a role during the differentiation of hematopoietic cells, but the physiologic role of HK-1 has yet to be determined.
To further elucidate the role of HK-1, we generated TAC4−/− mice and analyzed the B-cell compartment in the bone marrow of these mice. Based on previous studies that pointed toward an important role for HK-1 at the pro-B to pre-B stage of B-cell development,9,25 we hypothesized that mice carrying a HK-1 deletion would display changes in early B-cell development. Here we show, for the first time, that TAC4−/− bone marrow contains an increased proportion of “fraction B” pro-B cells compared with WT mice, whereas mature B-cell numbers remained normal. We further show that TAC4−/− total bone marrow cells cultured in the presence of IL-7 for 4 days yielded higher cell numbers and contained a significantly higher absolute number of pro-B cells than WT bone marrow cultures. The elevated cell population consisted of cells that were positive for B220, CD43, CD117, CD19, and IL7Rα and negative for CD2, IgM, and IgD, which is consistent with “Hardy fractions B and/or C” of the pro-B-cell stage of B-cell development.
Because CFSE staining revealed that cell division rates were not significantly different in WT and TAC4−/− bone marrow cultures, we think that the observed phenomenon is mainly the result of a higher number of pro-B cells present at the start of the culture. This is consistent with our FACS analysis in TAC4−/− bone marrow.
However, when equal numbers of sorted CD19+CD117+HSA+BP.1− “fraction B” pro-B cells were cultured in the presence of IL-7 for 4 days, we also saw a significant increase in cell numbers in cultures derived from TAC4−/− bone marrow compared with WT cultures. Similar to total bone marrow cultures, it was predominantly “fractions A-C” pro-B-cell populations that were elevated in cultures initiated with TAC4−/− “fraction B” cells. This finding was surprising as we expected that cultures initiated with equal numbers of cells of the same fraction would produce similar numbers of pro-B cells. However, sorted cell populations are never truly homogeneous, and it is possible that TAC4−/− bone marrow contains small cell populations within “fraction B” that have properties different from WT cells. Furthermore, in contrast to total bone marrow cultures, we also detected increased numbers of IgM+ cells in TAC4−/− cultures initiated with sorted pro-B cells. Because the numbers of IgM+ cells were not significantly different in vivo in WT and TAC4−/− bone marrow, this difference may only be revealed in vitro. The reason for the discrepancy between total bone marrow cultures and “fraction B” pro-B-cell cultures remains unknown. Both culture systems were maintained in the presence of IL-7, which favors the growth of IL-7-responsive pro-B cells. Mature cells undergo differentiation, lose IL-7 responsiveness, and die, thus resulting in an enriched population of pro-B cells.
Given the specific increase of pro-B-cell “fractions B/C” in TAC4−/− total bone marrow cultures, it was somewhat surprising that the difference in vivo, although statistically significant, was small. Furthermore, we were surprised to see that the difference did not extend to later stages of B-cell development as might have been expected. However, compensatory mechanisms that maintain these cells at a near-normal state in vivo may account for this phenomenon. Alternatively, the high rate of cell proliferation seen in vitro may not occur under physiologic conditions as tissue culture may have intrinsic properties not easily achieved in vivo. A small difference in vivo may therefore be amplified in vitro.
The fact that TAC4−/− bone marrow contains increased pro-B cells with normal mature B-cell numbers raises the possibility that these cells have the potential to develop into B-cell malignancies. However, to date we have not detected increased incidences of cancer development in these mice.
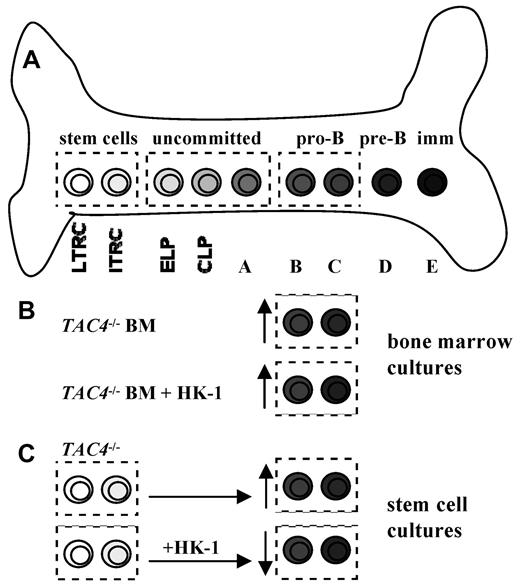
Our findings of an altered development of B lineage cells in bone marrow of TAC4−/− mice with increased numbers of pro-B cells may indicate that HK-1 is a critical cofactor at the pro-B to pre-B checkpoint for transitioning cells, resulting in an incomplete transition and accumulation of pro-B cells. However, the fact that pre-B, immature, and mature B-cell numbers are similar in vivo in WT and TAC4−/− mice suggests that the transition is not compromised in mutant mice. Furthermore, daily addition of HK-1 to TAC4−/− bone marrow cultures could not reverse the increase in TAC4−/− pro-B cells, suggesting that the absence of HK-1 at the pro-B to pre-B transition is not the reason for the elevated levels of pro-B cells in TAC4−/− bone marrow. An alternative and more probable explanation for the increased pro-B-cell population would implicate HK-1 at an earlier developmental stage.
We therefore examined the effect of HK-1 on the earliest hematopoietic precursors in lymphoid development, the hematopoietic stem cell. Hematopoietic stem cells represent a rare subpopulation of undifferentiated cells that maintain their cell number by self-renewal and have the potential to differentiate to replenish all blood and immune cell lineages. There are different populations of hematopoietic stem cells. LTRCs have the ability to reconstitute the hematopoietic system for life as they have the potential to self-renew and differentiate. ITRCs undergo significant self-renewal initially after transplantation but differ from LTRCs in failing to sustain that initial level of self-renewal. STRCs can differentiate but have lost the ability to self-renew.14,26
In the adult mouse, all hematopoietic stem cells are maintained in an undifferentiated state within a bone marrow niche supported by its microenvironment. Hematopoiesis is regulated by various factors present in the bone marrow microenvironment, such as cytokines, chemokines, hormones, neurotransmitters, and neuropeptides. The tachykinin peptides SP and NKA have been shown to be expressed in hematopoietic and stromal cells and have been reported to have opposing effects on hematopoiesis through the induction of cytokines via NK1 and NK2 receptors, respectively.27,28 Whereas SP induced stimulatory factors, such as IL-3, IL-6, GM-CSF, SCF, and FLT3, NKA stimulated the induction of growth-suppressive factors, such as transforming growth factor-β (TGF-β) and macrophage inflammatory protein-1α (MIP-1α).29,30
Although we show here that TAC4 is not expressed in purified LTRCs or ITRCs, TAC4 mRNA is abundant in the bone marrow as it is expressed by many hematopoietic cells. Specifically, TAC4 mRNA is expressed throughout B-cell development and can be detected in B cell “fractions A-F.” Interestingly, NK-1 receptor mRNA is expressed in LTRCs but not ITRCs, which prompted us to investigate the potential influence of HK-1 on these populations. To determine whether the lack of HK-1 affected stem cells, we measured the rate of the first cell division of TAC4−/− and WT LTRCs or ITRCs. Single-cell assays revealed that LTRCs and ITRCs purified from these mice divided at a similar rate. Furthermore, the addition of HK-1 did not affect the kinetics of the first cell division in either TAC4−/− or WT cells, suggesting that the first division of hematopoietic stem cells is not influenced by HK-1 in this in vitro setting.
Benveniste et al have previously demonstrated the ability to generate pro-B cells in vitro derived from hematopoietic stem cells.14,26 We used this system to examine the effect of HK-1 deficiency on pro-B-cell development from these cells. We show here that in vitro cultures derived from TAC4−/− LTRCs contain significantly higher absolute numbers of pro-B cells compared with WT cultures, which is consistent with our observation of more pro-B cells in TAC4−/− bone marrow. Because HK-1 deficiency led to an increase of TAC4−/− pro-B cells in vivo as well as in 3 independent in vitro systems, this suggests that HK-1 may play an inhibitory role in hematopoiesis (Figure 7).
B-cell development and proposed model of action for HK-1. (A) The drawing shows a simplified model of bone marrow B lymphocyte development. B-cell development occurs through several stages in a complex bone marrow microenvironment. ELP indicates early lymphoid progenitor; and CLP, common lymphoid progenitor. (B) The lack of HK-1 in TAC4−/− bone marrow cultures resulted in a specific increase of pro-B cells compared with WT cultures. The addition of HK-1 to these cultures had no effect on the elevated number of pro-B cells, suggesting that an earlier developmental event may be responsible for this phenotype. (C) Cultures derived from purified TAC4−/− hematopoietic stem cells also resulted in an increase of pro-B cells compared with WT control cultures. In contrast to total bone marrow cultures, daily addition of HK-1 to LTRC- and ITRC-derived cultures resulted in significantly decreased numbers of pro-B cells in WT- and HK-1-deficient cultures, suggesting that HK-1 plays an inhibitory role during hematopoiesis.
B-cell development and proposed model of action for HK-1. (A) The drawing shows a simplified model of bone marrow B lymphocyte development. B-cell development occurs through several stages in a complex bone marrow microenvironment. ELP indicates early lymphoid progenitor; and CLP, common lymphoid progenitor. (B) The lack of HK-1 in TAC4−/− bone marrow cultures resulted in a specific increase of pro-B cells compared with WT cultures. The addition of HK-1 to these cultures had no effect on the elevated number of pro-B cells, suggesting that an earlier developmental event may be responsible for this phenotype. (C) Cultures derived from purified TAC4−/− hematopoietic stem cells also resulted in an increase of pro-B cells compared with WT control cultures. In contrast to total bone marrow cultures, daily addition of HK-1 to LTRC- and ITRC-derived cultures resulted in significantly decreased numbers of pro-B cells in WT- and HK-1-deficient cultures, suggesting that HK-1 plays an inhibitory role during hematopoiesis.
To test this hypothesis, we assessed the impact of HK-1 on the de novo generation of pro-B cells derived from LTRC and ITRC cultures. We show that daily addition of HK-1 to these cultures not only reversed the increase of pro-B cells in TAC4−/−-derived cultures but also had a significant inhibitory effect on the development of pro-B cells in WT and TAC4−/− cultures. Addition of HK-1 decreased the total number of pro-B cells in these cultures in a dose-dependent manner without significantly affecting cell survival. Based on our data, we propose a novel model whereby HK-1 acts as an inhibitory factor on developing B cells (Figure 7). The loss of the inhibitory action of HK-1 in TAC4−/− mice may have also resulted in a shift of the tachykinin balance in the bone marrow microenvironment. Consequently, in TAC4−/− mice, the stimulatory peptide SP could play the predominant role in hematopoiesis, accounting for the increased numbers of pro-B cells.
Because we could not detect NK receptor mRNA on pro-B cells, these cells are probably not the target for HK-1. We propose 2 possible models of action for HK-1. Given that addition of HK-1 to stem cell-derived cultures, but not to bone marrow-derived cultures, led to a reversal of the phenotype, our data may suggest a model in which a precursor before the pro-B stage is the direct target of HK-1, ultimately suppressing pro-B-cell proliferation. Detailed studies of NK receptor expression on progenitors and the effect of HK-1 on these cells are warranted to fully understand the role of HK-1 in hematopoiesis and ultimately to find its mechanism of action.
Alternatively, the lack of NK receptor mRNA on pro-B cells could imply an indirect mechanism of action of HK-1 on B cells involving other cell types residing in the bone marrow microenvironment. Other tachykinin peptides, such as SP and NKA, have previously been shown to induce the release of stimulatory and growth-suppressive factors into the microenvironment, thereby indirectly influencing hematopoiesis.29,30 Our findings on the effect of HK-1 on pro-B cells may therefore suggest a model in which HK-1, secreted by early B cells, elicits the release of growth-suppressive factors from the milieu, consequently resulting in a decrease of pro-B-cell proliferation. The exact nature of the NK receptor–expressing target cell that is involved in this feedback regulation remains to be investigated. It is widely accepted that NK receptors are expressed in the bone marrow. For example, myeloid cells have been reported to express NK-1 receptor mRNA and functional protein.18,31-35 Because cultures established from LTRCs and ITRCs contain a large proportion of myeloid cells, this may be the population responsible for the response elicited by HK-1 in this in vitro system. Total bone marrow cultured with IL-7 for 4 days contained few myeloid cells, as nearly all cells were CD19+ B lineage cells, offering an explanation as to why HK-1 had no effect on these cultures.
Our findings on the phenotype of TAC4−/− mice presented in this study confirm the involvement of HK-1 in early B lymphocyte development. Based on our data, we propose that HK-1 is one of many factors secreted into the bone marrow microenvironment, which supports and fine-tunes hematopoietic development. We hypothesize that, by deleting TAC4, we have created a bone marrow microenvironment that lacks the inhibitory action of HK-1, thus altering the outcome of lymphocyte development and consequently leading to increased numbers of pro-B cells.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by the National Cancer Institute of Canada (Terry Fox Program Project Grant 015005) and the Canadian Institute of Health Research (grant 9862). The 129-mouse BAC library screen for TAC4 was performed by the Genome Resource Facility (Hospital of Sick Children, Toronto).
Authorship
Contribution: A.B. designed and performed research, analyzed data, and wrote the paper; P.B designed and performed research and analyzed data; S.A.C., M.B., and A.W. performed research; A.W. and T.W.M. contributed new reagents/tools; A.H.T. contributed to writing the paper; and N.N.I. and C.J.P. designed research and contributed to writing the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alexandra Berger, Department of Stem Cell and Developmental Biology, Ontario Cancer Institute, Princess Margaret Hospital, 610 University Ave, M5G 2M9 Toronto, ON, Canada; e-mail: aberger@uhnres.utoronto.ca.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal