Abstract
The primitive erythroid (EryP) lineage is the first to differentiate during mammalian embryogenesis. Eklf/Klf1 is a transcriptional regulator that is essential for definitive erythropoiesis in the fetal liver. Dissection of the role(s) of Eklf within the EryP compartment has been confounded by the simultaneous presence of EryP and fetal liver–derived definitive erythroid (EryD) cells in the blood. To address this problem, we have distinguished EryP from their definitive counterparts by crossing Eklf+/− mutant and ϵ-globin::histone H2B-GFP transgenic mice. Eklf-deficient EryP exhibit membrane ruffling and a failure to acquire the typical discoidal erythroid shape but they can enucleate. Flow cytometric analyses of H2B-GFP+ EryP revealed that Eklf heterozygosity results in the loss of Ter119 surface expression on EryP but not on EryD. Null mutation of Eklf resulted in abnormal expression of a range of surface proteins by EryP. In particular, several megakaryocyte markers were ectopically expressed by maturing Eklf-null EryP. Unexpectedly, the platelet tetraspanin CD9 was detected on nucleated wild-type EryP but not on mature EryD and thus provides a useful marker for purifying circulating EryP. We conclude that Eklf gene dosage is crucial for regulating the surface phenotype and molecular identity of maturing primitive erythroid cells.
Introduction
Primitive erythroid cells (EryP) are the first hematopoietic cells to appear and mature in the developing embryo.1-3 Before the formation of the adult hematopoietic system from hematopoietic stem cells, the yolk sac–derived EryP number in the millions and are the dominant cell type of the mid-gestation mouse embryo circulation.4,5 Larger than their definitive erythroid (EryD) counterparts, EryP express a unique set of globin genes and circulate, for much of their existence, as nucleated cells.3 Shortly after midgestation in the mouse embryo, EryD are produced in enormous numbers by progenitors in the fetal liver (FL) and rapidly outnumber EryP. By birth, circulating EryP comprise only a tiny fraction of cells in the blood, and the transition to the definitive erythroid system is complete.
The genetic programs that regulate the development and maturation of the primitive erythroid compartment are being elucidated. However, this analysis is complicated by the simultaneous presence of definitive and primitive erythroid cells in the embryonic circulation after mid-gestation. To identify and monitor EryP, we have previously capitalized upon human ϵ-globin regulatory elements to drive expression of fluorescent reporters solely within this population in transgenic mice.4,6 Here, we have asked whether null mutation of Eklf, a gene reported to play a critical role in erythropoiesis during embryonic development, has distinct functions in the primitive and definitive erythroid lineages.
The erythroid kruppel-like factor (Eklf, also known as Klf1)7 is the prototype of the Krüppel-like family of zinc finger–containing transcription factors.8 Klf family members have been shown to play crucial roles in diverse biologic processes ranging from hematopoiesis to maintenance of embryonic stem cells.8 Originally isolated from a mouse erythroleukemia cell cDNA library7 and proposed to regulate the fetal-to-adult globin gene switch,9 Eklf is transcriptionally restricted to the erythroid lineage from as early as embryonic day 7.5 (E7.5) in the mouse.10 Targeted null mutation of Eklf results in severe anemia and death in utero due to defective erythropoiesis.11,12 Eklf-deficient EryD progenitors are generated in normal numbers but fail to mature and enucleate effectively.12,13 Analysis of Eklf mutant embryos has led to the identification of a large group of target genes (in addition to globins) in erythroid cells (reviewed in references13-15 ). Erythroid cells lacking Eklf exhibit severely ruffled plasma membranes, suggesting defects in the expression and/or function of erythroid membrane and cytoskeletal proteins.13
It was not previously possible to focus specifically on the EryP lineage in wild-type (WT) or mutant embryos due to the lack of lineage-restricted markers that could be used to distinguish them from definitive erythroid cells. To analyze the impact of loss of one or both alleles of Eklf during primitive erythropoiesis, we have crossed the ϵ-globin:H2B-GFP transgenic mouse6 onto an Eklf mutant background.12 Haploinsufficiency of Eklf had a profound effect on the surface phenotype of EryP but not EryD, indicating that the differentiation of EryP is more sensitive to Eklf gene dosage than that of EryD. Null mutation resulted in a partial “identity crisis” within maturing EryP in the circulation, with up-regulation of a range of megakaryocyte-related surface antigens and transcriptional regulators. Therefore, Eklf regulates EryP maturation and molecular identity.
Methods
Generation of human ϵ-globin::H2B-EGFP transgenic mice
The entire insert from pCX-ϵ-globin::H2B-EGFP (see supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article), containing the human ϵ-globin::H2B-EGFP cassette, was excised and gel-purified to remove prokaryotic sequences. Pronuclear injections into C57BL/6 × C3H (B6C3) F1 hybrid embryos were performed by the Mount Sinai Mouse Genetics Shared Resource Facility using standard methods.16 Two transgenic founder animals were obtained, genotyped by polymerase chain reaction (PCR), and screened for enhanced green fluorescent protein (EGFP) expression using fluorescence microscopy and flow cytometric analysis (fluorescence-activated cell sorting [FACS]). Line #38 was used for the studies reported here. The mice were maintained as homozygotes (termed Tg/Tg) on an ICR (outbred; Taconic) background, following Mount Sinai institutional guidelines and approval by the Mount Sinai Institutional Animal Care and Use Committee.
The Eklf-null mouse line has been reported previously.12 Eklf heterozygotes were obtained from Dr David Bodine (National Human Genome Research Institute, Bethesda, MD) and were maintained on an ICR background. Genotyping of the targeted Eklf allele in embryos and adults was performed as described.12 The ϵ-globin::H2B-EGFP transgenic mouse line was crossed with Eklf+/− mice to obtain animals bearing the H2B-EGFP reporter and one null allele of Eklf. To generate timed Eklf-null embryos carrying the H2B-EGFP reporter allele, male and female Eklf+/−;ϵ::H2B-GFP (Tg/Tg) mice were intercrossed. The morning of detection of the vaginal plug was taken as day 0.5 of gestation.
Flow cytometry and sorting
Embryos from Eklf+/−;ϵ::H2B-GFP intercrosses were isolated at the indicated times, and blood was collected by exsanguination into FACS buffer (phosphate-buffered saline [PBS]; Gibco-BRL, Invitrogen), containing 10% fetal calf serum (Hyclone) and heparin (12.5 μg/mL; Sigma-Aldrich).4 FLs were dissected, triturated, and filtered through a 40-μm cell strainer to produce a single-cell suspension. Cells from embryonic peripheral blood (PB) and FL were washed and stained for flow cytometric analysis.4 The directly conjugated monoclonal antibodies used in this study are listed in Table 1. Biotinylated antibody binding was detected using streptavidin conjugated to phycoerythrin-Cy7 (eBioscience). Samples were analyzed using an LSRII cell analyzer (Becton Dickinson). Data were analyzed using the FlowJo Version 8.8.6 software package (TreeStar). Cell sorting was performed using either the Vantage Cell Sorter (Becton Dickinson) or a Mo-Flo cell sorter (DakoCytomation). Analytical flow cytometry and cell sorting were performed at the Mount Sinai Flow Cytometry Core Facility.
Antibodies used for flow cytometric analyses in this study
| Surface marker: fluorochrome . | Clone . | Source . |
|---|---|---|
| Ter119: PE, Pacific Blue, PE-Cy7 | Ter119 | eBioscience |
| α2 integrin: APC | DX5 | BioLegend |
| α4 integrin: PE | R1-2 | eBioscience |
| α5 integrin: biotin | MFR5 | BD Biosciences |
| αV integrin: PE | RMV-7 | eBioscience |
| β1 integrin: APC | eBioHmb1-1 | eBioscience |
| β3 integrin: biotin | 2C9.G3 | eBioscience |
| CD9: PE | eBioKMC8 | eBioscience |
| CD24: Pacific Blue | M1/69 | BioLegend |
| CD31: PE-Cy7 | 390 | BioLegend |
| CD34: PE-Cy5 | MEC14.7 | BioLegend |
| CD41: biotin/PE-Cy7 | eBioMWReg30 | eBioscience |
| CD42: APC | 1B5 | Dr Barry Coller, Rockefeller University |
| CD44: PE | IM7 | BD Biosciences |
| CD55: PE | RIKO-5 | BD Biosciences |
| CD71: PE | R17217 | eBioscience |
| CD117/c-kit: APC | 2B8 | BioLegend |
| CD147: PE | RL73 | eBioscience |
| Surface marker: fluorochrome . | Clone . | Source . |
|---|---|---|
| Ter119: PE, Pacific Blue, PE-Cy7 | Ter119 | eBioscience |
| α2 integrin: APC | DX5 | BioLegend |
| α4 integrin: PE | R1-2 | eBioscience |
| α5 integrin: biotin | MFR5 | BD Biosciences |
| αV integrin: PE | RMV-7 | eBioscience |
| β1 integrin: APC | eBioHmb1-1 | eBioscience |
| β3 integrin: biotin | 2C9.G3 | eBioscience |
| CD9: PE | eBioKMC8 | eBioscience |
| CD24: Pacific Blue | M1/69 | BioLegend |
| CD31: PE-Cy7 | 390 | BioLegend |
| CD34: PE-Cy5 | MEC14.7 | BioLegend |
| CD41: biotin/PE-Cy7 | eBioMWReg30 | eBioscience |
| CD42: APC | 1B5 | Dr Barry Coller, Rockefeller University |
| CD44: PE | IM7 | BD Biosciences |
| CD55: PE | RIKO-5 | BD Biosciences |
| CD71: PE | R17217 | eBioscience |
| CD117/c-kit: APC | 2B8 | BioLegend |
| CD147: PE | RL73 | eBioscience |
Conjugated antibodies were purchased from eBioscience, BioLegend, or BD Biosciences. PE indicates phycoerythrin; and APC, allophycocyanin.
Quantitative real-time RT-PCR
Total RNA was extracted from sorted cells using an RNeasy Mini Kit (QIAGEN), and cDNA was prepared using the Superscript III RT first-strand synthesis system (Invitrogen) according to the manufacturer's instructions. Gene expression was analyzed by quantitative real-time reverse transcription PCR (qRT-PCR) using SYBR-green (SYBR-green PCR master mix; Applied Biosystems), following the protocol recommended by the supplier. Ubiquitin b (Ubb) or 18S ribosomal RNA (Rn18s) genes were used as internal controls for normalization. Values obtained for target gene expression were normalized to the control gene and then expressed relative to the levels in Eklf WT samples, which were set at a value of 1.0. The sequences of the primers used in this study are shown in Table 2.
Quantitative RT-PCR primers used in this study
| Target gene . | Accession no. . | Primer sequences . | Details . |
|---|---|---|---|
| Klf3 | NM_008453 | Forward: 5′-GAAATGTCACCCCCTTTAATGAAC-3′; reverse: 5′-CACGATGACGGAAGGATGGT-3′ | Ref. 19 |
| Gypa | NM_010369 | Forward: 5′-GCCGAATGACAAAGAAAAGTTCA-3′; reverse: 5′-TCAATAGAACTCAAAGGCACACTGT-3′ | ID 328119a1* |
| Gabpa | NM_008065 | Forward: 5′-AGCGCATCTCGTTGAAGAAG-3′; reverse: 5′-TCCTGCTCTTTTCTGTAGCCT-3′ | ID 4328119a1* |
| Fli-1 | NM_008026 | Forward: 5′-ATGGACGGGACTATTAAGGAGG-3′; reverse: 5′-GAAGCAGTCATATCTGCCTTGG-3′ | ID 6679807a1* |
| Ubb | NM_011664 | Forward: 5′-TGGCTATTAATTATTCGGTCTGCAT-3′; reverse: 5′-GCAAGTGGCTAGAGTGCAGAGTAA-3′ | Ref. 39 |
| 18S | NM_011664 | Forward: 5′-CACGGCCGGTACAGTGAAAC-3′; reverse: 5′-AGAGGAGCGAGCGACCAA-3′ | Ref. 19 |
| Eklf | NM_010635 | Quantitect primer assay; QIAGEN | Cat. no. QT01743777 |
| CD41 | NM_010575 | Quantitect primer assay; QIAGEN | Cat. no. QT00117992 |
| CD9 | NM_007657 | Quantitect primer assay; QIAGEN | Cat. no. QT01752513 |
| ϵy | NM_008221 | Forward: 5′-GCCCTCTCTCTAGCTGTCCA-3′; reverse: 5′-TGCTTTCAAGGAACAGTCCA-3′ | Ref. 40 |
| Target gene . | Accession no. . | Primer sequences . | Details . |
|---|---|---|---|
| Klf3 | NM_008453 | Forward: 5′-GAAATGTCACCCCCTTTAATGAAC-3′; reverse: 5′-CACGATGACGGAAGGATGGT-3′ | Ref. 19 |
| Gypa | NM_010369 | Forward: 5′-GCCGAATGACAAAGAAAAGTTCA-3′; reverse: 5′-TCAATAGAACTCAAAGGCACACTGT-3′ | ID 328119a1* |
| Gabpa | NM_008065 | Forward: 5′-AGCGCATCTCGTTGAAGAAG-3′; reverse: 5′-TCCTGCTCTTTTCTGTAGCCT-3′ | ID 4328119a1* |
| Fli-1 | NM_008026 | Forward: 5′-ATGGACGGGACTATTAAGGAGG-3′; reverse: 5′-GAAGCAGTCATATCTGCCTTGG-3′ | ID 6679807a1* |
| Ubb | NM_011664 | Forward: 5′-TGGCTATTAATTATTCGGTCTGCAT-3′; reverse: 5′-GCAAGTGGCTAGAGTGCAGAGTAA-3′ | Ref. 39 |
| 18S | NM_011664 | Forward: 5′-CACGGCCGGTACAGTGAAAC-3′; reverse: 5′-AGAGGAGCGAGCGACCAA-3′ | Ref. 19 |
| Eklf | NM_010635 | Quantitect primer assay; QIAGEN | Cat. no. QT01743777 |
| CD41 | NM_010575 | Quantitect primer assay; QIAGEN | Cat. no. QT00117992 |
| CD9 | NM_007657 | Quantitect primer assay; QIAGEN | Cat. no. QT01752513 |
| ϵy | NM_008221 | Forward: 5′-GCCCTCTCTCTAGCTGTCCA-3′; reverse: 5′-TGCTTTCAAGGAACAGTCCA-3′ | Ref. 40 |
Cytologic analysis
Collection of embryonic blood, cytocentrifugation onto glass slides, and Giemsa staining was performed as previously described.4 For immunostaining, PB blood cells from E14.5 embryos were harvested and washed with bovine serum albumin (BSA, 0.3% wt/vol; Sigma-Aldrich)/PBS and resuspended in the same buffer. The cells were then incubated for 1 hour at room temperature with 10 μg/mL of rat anti–mouse CD9 monoclonal antibody (KMC8; BD Pharmingen; Table 1) or rat immunoglobulin G2a (IgG2a) isotype control (cat. no. 553926; BD Pharmingen) using a rotator. After washing 3× with 0.3% BSA/PBS, samples were incubated with Alexa Fluor 568–conjugated goat anti–rat IgG (Invitrogen) for 1 hour at room temperature and then washed twice with 0.3% BSA/PBS to remove unbound secondary antibody. The stained cells were centrifuged onto glass slides using a Cytospin3 cytocentrifuge (Shandon), air-dried, and mounted in Vectashield containing DAPI (4′,6-diamidino-2-phenylindole; Vector Laboratories). Images were acquired using an Axioplan 2 Epifluorescence microscope (Zeiss). Lens specifications are provided in the figure legends.
Isolation of cells from BM
Single-cell suspensions were prepared from bone marrow (BM) by dissecting the femora of 6- to 8-week–old female ICR mice. Femora were dissected and flushed with 3 mL Dulbecco modified Eagle medium (Gibco BRL) using a 5-mL syringe fitted with a 20-G needle. Cells were dispersed by trituration, filtered through a 40-μm cell strainer (BD Falcon) and pelleted by centrifugation. Cells were then counted using a hemocytometer. For flow cytometry, 1 × 106 cells were stained with fluorescently conjugated antibodies and then analyzed.
Results
Anomalous maturation of primitive erythroid cells lacking Eklf
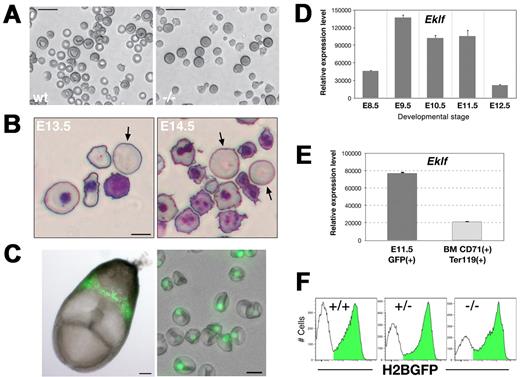
Although several morphologic and molecular defects have been reported for Eklf-null mutant erythroid cells, it has not previously been possible to focus specifically on the primitive erythroid lineage. We were able to overcome this obstacle by analyzing an Eklf mutation on the background of our ϵ-globin:H2B-GFP transgenic mouse, in which expression of a histone H2B-GFP reporter is restricted to EryP.6 Visual inspection revealed that the Eklf-null embryos were severely anemic at mid-gestation (supplemental Figure 1A), as reported previously.11,12 Normal EryP maturation is characterized by a stepwise developmental progression that can be monitored by cytology4 and is characterized by a series of discrete observable features. These include progressive loss of basophilia, loss of nucleoli, nuclear condensation, acquisition of a typical pink cytoplasmic staining, and nuclear extrusion, a hallmark of terminal erythroid differentiation.4 Microscopic inspection of PB from Eklf-null mutant embryos revealed irregularly shaped EryP, with a wrinkled, nonspherical morphology, as early as E11.5 (supplemental Figure 1B-C). In contrast, EryP from WT littermates were round, with smooth contours (supplemental Figure 1C). Differentiation of Eklf heterozygous cells was indistinguishable from WT (not shown). The abnormal Eklf-null mutant EryP phenotype was particularly striking in unfixed wet preparations, with misshapen macrocytic cells that failed to attain the discoidal shape typical of stage-matched, mature WT EryP (Figure 1A).
Eklf-deficient primitive erythroblasts show dramatic alterations in cell morphology but can complete their maturation. (A) Wet preparations of freshly harvested E14.5 PB cells reveal large Eklf-null mutant EryP that fail to achieve the discoidal shape observed for WT cells. (B) Detail of Giemsa-stained cytospin preparations of PB from E13.5 and E14.5 mutant embryos, showing some macrocytic cells that lack nuclei (arrows). (C) Left panel, H2B-GFP expression in an E8.0 transgenic embryo. Right panel, wet preparation of PB from an E14.5 ϵ-globin::H2B-GFP embryo. An overlay of bright field and green fluorescence is shown. Expression of the fluorescent reporter is restricted to the EryP nucleus. (D) Expression of Eklf mRNA in developing EryP cells. GFP+ cells were sorted from E8.5-12.5 embryos and real-time RT-PCR analysis of Eklf was performed; expression was normalized to that of ubiquitin b. (E) Eklf expression in definitive versus primitive erythroid cells. GFP+ EryP from E11.5 embryos or CD71+Ter119+ definitive erythroid cells from adult mouse BM were sorted and the expression of Eklf mRNA assessed by real time RT-PCR; expression levels are relative to ubiquitin b. A representative experiment (3 independent samples) is shown. (F) Histograms of ϵ-globin::H2B-GFP fluorescence intensity in E13.5 PB cells harvested from Eklf WT, Eklf+/−or Eklf−/− embryos. Images were acquired in AxioVision v4.6 using a Zeiss AxioCam digital color camera mounted on a Zeiss Axiovert 25 inverted microscope outfitted with a 20× (LD-A-Plan/0.3 NA) or 32× (LD-A-Plan/0.4 NA) objective (A-C right panel) or on a Leica MZ12 stereomicroscope outfitted with a plan apo 1.0× objective (C, left panel). Scale bars: A, 20 μm; B, 10 μm; C, left panel, 100 μm, and right panel, 10 μm.
Eklf-deficient primitive erythroblasts show dramatic alterations in cell morphology but can complete their maturation. (A) Wet preparations of freshly harvested E14.5 PB cells reveal large Eklf-null mutant EryP that fail to achieve the discoidal shape observed for WT cells. (B) Detail of Giemsa-stained cytospin preparations of PB from E13.5 and E14.5 mutant embryos, showing some macrocytic cells that lack nuclei (arrows). (C) Left panel, H2B-GFP expression in an E8.0 transgenic embryo. Right panel, wet preparation of PB from an E14.5 ϵ-globin::H2B-GFP embryo. An overlay of bright field and green fluorescence is shown. Expression of the fluorescent reporter is restricted to the EryP nucleus. (D) Expression of Eklf mRNA in developing EryP cells. GFP+ cells were sorted from E8.5-12.5 embryos and real-time RT-PCR analysis of Eklf was performed; expression was normalized to that of ubiquitin b. (E) Eklf expression in definitive versus primitive erythroid cells. GFP+ EryP from E11.5 embryos or CD71+Ter119+ definitive erythroid cells from adult mouse BM were sorted and the expression of Eklf mRNA assessed by real time RT-PCR; expression levels are relative to ubiquitin b. A representative experiment (3 independent samples) is shown. (F) Histograms of ϵ-globin::H2B-GFP fluorescence intensity in E13.5 PB cells harvested from Eklf WT, Eklf+/−or Eklf−/− embryos. Images were acquired in AxioVision v4.6 using a Zeiss AxioCam digital color camera mounted on a Zeiss Axiovert 25 inverted microscope outfitted with a 20× (LD-A-Plan/0.3 NA) or 32× (LD-A-Plan/0.4 NA) objective (A-C right panel) or on a Leica MZ12 stereomicroscope outfitted with a plan apo 1.0× objective (C, left panel). Scale bars: A, 20 μm; B, 10 μm; C, left panel, 100 μm, and right panel, 10 μm.
Despite their abnormal morphology, Eklf-null primitive erythroid cells were able to condense their nuclei and displayed eosinophilic cytoplasmic staining with Giemsa (supplemental Figure 1C), suggesting that the molecular programs leading to chromatin condensation and hemoglobin production, respectively, were not signficantly impaired, despite reduced transcription of the ϵY-globin gene (see below). By E13.5-E14.5, enucleated EryP (identified by size and morphology) were detected in the PB of mutant embryos, indicating that nuclear extrusion can occur in the absence of Eklf (Figure 1B).
Monitoring primitive erythroid maturation in the absence of EKLF
In the studies that follow, we used the ϵ-globin::H2B-EGFP transgenic mouse line (hereafter termed ϵ::H2B-GFP; supplemental Figure 2A) in which EryP nuclei are labeled by a histone H2B-EGFP fusion protein that localizes to the nucleus.6 Extensive characterization of the GFP-expressing cells in this transgenic mouse line confirmed the specificity of the fluorescent reporter for the primitive erythroid lineage.4,6 The bright nuclear fluorescence in this line (Figure 1C) makes it highly amenable for imaging and flow cytometry. GFP-labeled EryP will hereafter be termed EryP/H2B-GFP.
We first examined expression of Eklf in WT EryP sorted from embryonic stages E8.5-E12.5. Real-time RT-PCR analysis revealed that Eklf is up-regulated in EryP from E8.5-E9.5 and is maintained at high levels until E11.5, when the nucleus of the EryP is condensed; transcription of Eklf is significantly reduced by E12.5 (Figure 1D). compared with definitive erythroid cells, circulating EryP (from E11.5) were found to express Eklf mRNA at higher levels (approximately 3.6-fold) than do EryD at a comparable stage of development (CD71+Ter119+, sorted from adult BM; Figure 1E).
To evaluate the impact of the Eklf-null mutation on EryP development, Eklf+/− and ϵ::H2B-GFP mice were mated. Eklf+/−;ϵ::H2B-GFP mice were then intercrossed to generate Eklf-null mutant EryP/H2B-GFP. The frequency of EryP/H2B-GFP and fluorescence intensity of the H2B-GFP reporter were unaffected by the loss of either one or both alleles of Eklf (Figure 1F; the transgene contains only a human ϵ-globin promoter but not additional upstream regulatory sequences). In contrast, transcription of the endogenous ϵY-globin gene was modestly decreased in Eklf heterozygous EryP/H2B-GFP and, as reported by others,17 was reduced to approximately 50% of WT levels in null mutant EryP (supplemental Figure 2B). Therefore, expression of the endogenous ϵY-globin gene is regulated, in part, by Eklf in the EryP lineage. Whether this regulation is direct or is mediated indirectly, through additional Eklf target genes, remains to be evaluated. EKLF has been implicated in direct regulation of the human ϵ-globin gene.18
As the Eklf-null mutation resulted in embryonic lethality by E14.5, analyses were performed before this developmental stage. EryP/H2B-GFP were sorted from WT, heterozygous, or null mutant embryos and gene expression was examined. Eklf expression was reduced by nearly one-half in heterozygous EryP and was absent in Eklf−/− embryos, as expected (supplemental Figure 2C). Klf3/Bklf is a direct target of Eklf, and its expression is greatly reduced in Eklf mutant FL cells.19 In EryP, the levels of Klf3/Bklf mRNA were reduced to 70% (heterozygotes) and 20% (null mutant) of those seen in WT embryos (supplemental Figure 2D).
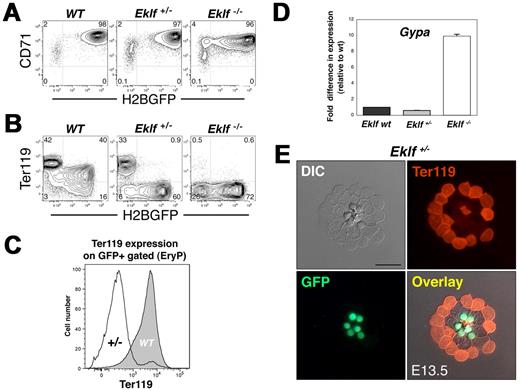
Haploinsufficiency of Eklf results in the absence of surface Ter119 expression on EryP but not on EryD
The maturation of erythroblasts to enucleated reticulocytes can be monitored by examining the combined expression of the transferrin receptor CD71 and Ter119, a heavily glycosylated surface protein that is up-regulated during both primitive4 and definitive20 erythroid maturation. Loss of both alleles of Eklf results in the complete ablation of Ter119 surface expression on circulating blood or FL cells.9,14 We, therefore, evaluated expression of CD71 and Ter119 in the EryP compartment of WT, heterozygous, and Eklf-null embryos. CD71 expression was not affected by the loss of one Eklf allele but was modestly reduced (approximately 4-fold, based on analysis of fluorescence intensity) in Eklf−/− EryP (Figure 2A), as reported by others.14 Some heterogeneity in expression of the ϵ::H2B-GFP transgene in Eklf-null mutant EryP was evident (Figures 1F and 2A) but did not compromise our analyses.
Haploinsufficiency of Eklf results in the absence of Ter119 in the primitive but not the definitive erythroid lineage. (A) FACS analysis of CD71 expression on WT, Eklf+/−, and Eklf−/− embryos. CD71 is expressed on the Eklf mutant EryP, though at modestly reduced levels (approximately 4-fold) on null mutant cells. (B) Eklf dose-dependent expression of Ter119 on E14.5 EryP. WT GFP+ cells (EryP) expressed Ter119 at higher levels than WT GFP− cells (EryD). In contrast, deletion of one allele of Eklf results in loss of expression of Ter119 on GFP+ EryP but not on the GFP− EryD. Ter119 is absent from any of the null mutant cells. (C) Representative FACS histogram of Ter119 fluorescence intensity on GFP+ EryP from Eklf WT or Eklf+/− PB, gated from samples in panel B. Expression of Ter119 on the surface of Eklf heterozygous cells was greatly reduced, compared with WT EryP. (D) Expression of Glycophorin A (Gypa) in E13.5 Eklf+/+, Eklf+/−, and Eklf−/− EryP/H2B-GFP as measured using real-time RT-PCR. mRNA expression was normalized using 18S RNA as a control and was then normalized again, setting WT Eklf levels at 1.0. (E) Ter119 staining of PB cells from E13.5 ϵ-globin:H2B-GFP; Eklf+/− embryos. Cells were cytospun onto glass slides, fixed, and permeabilized before immunostaining. No Ter119 reactivity was detected in EryP (green fluorescent nuclei) compared with the smaller Ter119+ enucleated definitive erythrocytes. Images were acquired using a Zeiss AxioCam camera mounted on a Zeiss Axioplan 2 microscope outfitted with a 63×/Plan neofluar/1.25 NA oil objective. Scale bar, 20 μm.
Haploinsufficiency of Eklf results in the absence of Ter119 in the primitive but not the definitive erythroid lineage. (A) FACS analysis of CD71 expression on WT, Eklf+/−, and Eklf−/− embryos. CD71 is expressed on the Eklf mutant EryP, though at modestly reduced levels (approximately 4-fold) on null mutant cells. (B) Eklf dose-dependent expression of Ter119 on E14.5 EryP. WT GFP+ cells (EryP) expressed Ter119 at higher levels than WT GFP− cells (EryD). In contrast, deletion of one allele of Eklf results in loss of expression of Ter119 on GFP+ EryP but not on the GFP− EryD. Ter119 is absent from any of the null mutant cells. (C) Representative FACS histogram of Ter119 fluorescence intensity on GFP+ EryP from Eklf WT or Eklf+/− PB, gated from samples in panel B. Expression of Ter119 on the surface of Eklf heterozygous cells was greatly reduced, compared with WT EryP. (D) Expression of Glycophorin A (Gypa) in E13.5 Eklf+/+, Eklf+/−, and Eklf−/− EryP/H2B-GFP as measured using real-time RT-PCR. mRNA expression was normalized using 18S RNA as a control and was then normalized again, setting WT Eklf levels at 1.0. (E) Ter119 staining of PB cells from E13.5 ϵ-globin:H2B-GFP; Eklf+/− embryos. Cells were cytospun onto glass slides, fixed, and permeabilized before immunostaining. No Ter119 reactivity was detected in EryP (green fluorescent nuclei) compared with the smaller Ter119+ enucleated definitive erythrocytes. Images were acquired using a Zeiss AxioCam camera mounted on a Zeiss Axioplan 2 microscope outfitted with a 63×/Plan neofluar/1.25 NA oil objective. Scale bar, 20 μm.
Others have reported that unfractionated embryonic blood cells from Eklf heterozygous embryos (containing a mixture of EryP and EryD) show “intermediate” levels of Ter119 expression compared with stage-matched WT littermates.14 To determine whether this intermediate population might in fact be heterogeneous, we took advantage of the ϵ-globin::H2B-EGFP transgene to measure Ter119 levels on each of the 2 Eklf+/− erythroid lineages. Expression of Ter119 protein on Eklf+/− EryD (GFP-negative) was normal but was almost completely absent from Eklf+/− EryP/H2B-GFP (Figure 2B-C). These results suggest a role for Eklf in the maturation of EryP that is distinct from its function in EryD. Both Eklf-null mutant EryD and EryP lacked surface expression of Ter119 (Figure 2B; Eklf−/−).
Ter119 is thought to be Glycophorin A (GPA, encoded by Gypa)21 or a molecule that associates directly with GPA.20 We, therefore, asked whether transcription of Gypa in EryP is altered by loss of one or both copies of Eklf. Gypa mRNA levels were similar in EryP from WT and Eklf+/− embryos (Figure 2D). Strikingly, however, Gypa mRNA levels were approximately 10-fold higher in Eklf−/− EryP.
Thus, Eklf+/− EryP express normal levels of Gypa mRNA but not surface Ter119, whereas transcription of the gene is greatly up-regulated in Eklf−/− EryP, on which surface Ter119 is absent. The uncoupling between transcription and Ter119 protein expression is intriguing and raises the possibility that the absence of surface Ter119 on Eklf+/− and Eklf−/− EryP might be due to aberrant protein trafficking to the plasma membrane. To address this question, we examined expression of Ter119 in permeabilized cells. In contrast with WT and Eklf+/− EryD, which express Ter119 on the cell surface (Figure 2B), no significant intracellular or extracellular signal was detected for permeabilized Eklf+/− EryP stained with Ter119 (Figure 2E; negative control shown in supplemental Figure 2E).
Eklf-null mutation results in aberrant expression of a variety of surface proteins by primitive erythroid cells
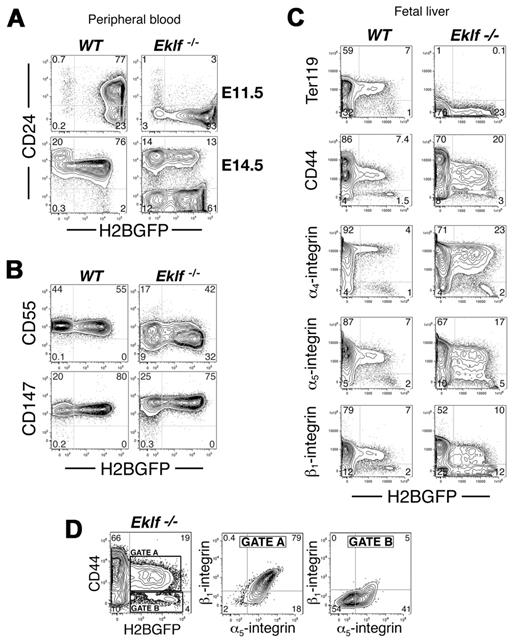
As Eklf-null EryP expressed CD71 but failed to express Ter119, we examined the expression of other proteins that we have found on the surface of EryP during their development.4 Expression of the glycosylphosphatidylinositol (GPI)–anchored glycoprotein, CD24, is up-regulated on EryP as they mature.4 In contrast to WT EryP, on which CD24 is expressed at E11.5, Eklf−/− EryP lacked expression of CD24 at this stage (Figure 3A). By E14.5, only a subpopulation (∼ 20%) of EryP/H2B-GFP had activated expression of CD24 (Figure 3A). CD24 expression on Eklf+/− EryP was normal (not shown).
Aberrant surface protein expression on Eklf-null mutant EryP. (A) CD24 is not expressed on Eklf−/− EryP at E11.5 (top panel). By E14.5, a subpopulation of EryP/H2B-GFP have activated expression of CD24 to levels seen on EryD, while the remaining EryP/H2B-GFP fail to express CD24. (B) CD55 was modestly down-regulated on Eklf-null mutant EryP, while CD147 was moderately up-regulated. (C) FACS analysis of FL cells isolated from WT and Eklf mutant embryos. In comparison with the uniform levels of expression of α4, α5, and β1 integrins and CD44 on WT EryP/H2B-GFP, heterogeneous expression was seen on Eklf mutant EryP. (D) EryP/H2B-GFP Eklf−/− FL cells from panel C were gated according to CD44 surface expression (gate A, expressed; gate B, not expressed). The corresponding levels of integrins α5 and β1 are shown.
Aberrant surface protein expression on Eklf-null mutant EryP. (A) CD24 is not expressed on Eklf−/− EryP at E11.5 (top panel). By E14.5, a subpopulation of EryP/H2B-GFP have activated expression of CD24 to levels seen on EryD, while the remaining EryP/H2B-GFP fail to express CD24. (B) CD55 was modestly down-regulated on Eklf-null mutant EryP, while CD147 was moderately up-regulated. (C) FACS analysis of FL cells isolated from WT and Eklf mutant embryos. In comparison with the uniform levels of expression of α4, α5, and β1 integrins and CD44 on WT EryP/H2B-GFP, heterogeneous expression was seen on Eklf mutant EryP. (D) EryP/H2B-GFP Eklf−/− FL cells from panel C were gated according to CD44 surface expression (gate A, expressed; gate B, not expressed). The corresponding levels of integrins α5 and β1 are shown.
Several GPI-anchored molecules are lost from the surface of erythrocytes in human paroxysmal nocturnal hemoglobinuria (PNH),22 suggesting a generalized defect in GPI-protein production. We asked whether an analogous defect might characterize Eklf-null mutant EryP. CD55, a GPI-anchored molecule missing from the surface of paroxysmal nocturnal hemoglobinuria erythrocytes,22 was expressed on both WT and null mutant EryP (Figure 3B). In contrast, the GPI-anchored protein CD147 was expressed at approximately 2.5-fold higher levels (as determined from mean fluorescence intensity from histograms) on Eklf-null mutant than on WT cells (Figure 3B). Therefore, Eklf does not appear to regulate the pathway by which GPI-proteins are modified and inserted into the cell membrane.
We have reported that EryP are found in the parenchyma of the FL, in contact with the central macrophages of erythroblastic islands, where they apparently complete their maturation and enucleate.6 The FL phase of EryP development is accompanied by the up-regulation of several adhesion molecules, including CD44, α4-, α5-, and β1-integrins, on the surface of the cells.6 As Eklf-null mutant EryP display several alterations in cell morphology and surface antigen expression,14 we asked whether they can migrate to FL and whether they up-regulate surface antigen expression.6 EryP/H2B-GFP were present in the FL of Eklf−/−;ϵ::H2B-GFP embryos (Figure 3C and data not shown), suggesting that, despite their morphologic abnormalities, EryP can migrate to this tissue. All GFP+ cells from WT FL expressed CD44 and α4-, α5-, and β1-integrins at uniform levels. However, EryP/H2B-GFP from Eklf-null mutant FL displayed a heterogeneous pattern of adhesion molecule expression (Figure 3C), with a distinct population that had not up-regulated any of these proteins (Figure 3D). For example, the cells that lacked CD44 also lacked or expressed only low levels of the other adhesion molecules examined. These results strongly suggest that Eklf regulates, directly or indirectly, the expression of adhesion molecules that may be required for the final stages of EryP maturation.6
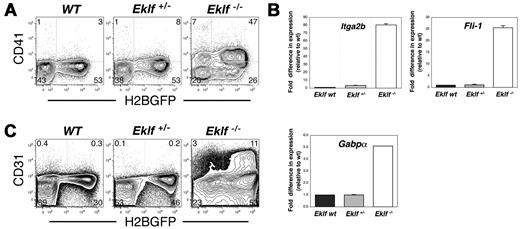
Loss of Eklf results in aberrant expression of megakaryocyte surface proteins and transcription factors in maturing primitive erythroid cells
Eklf has been proposed to regulate a switch between the erythroid and megakaryocytic lineages.23 Eklf-null mutant FL cells produce greater numbers of megakaryocyte progenitors (as detected using colony assays in vitro) than do WT embryos.23 However, it is unclear whether this apparent lineage shift occurs solely at the level of the progenitor or whether maturing EryP/H2B-GFP also exhibit a phenotypic shift toward the megakaryocytic lineage. We addressed this issue by examining the expression of molecular hallmarks of megakaryotic differentiation in Eklf-null mutant EryP. The megakaryocyte/platelet marker CD41 (Itga2b) was aberrantly expressed on the surface of circulating E13.5 Eklf-null EryP (Figure 4A). Expression of Itga2b mRNA, as evaluated using real-time RT-PCR, was consistent with that of CD41/Itga2b protein, with an increase in the number of Itga2b transcripts of at least 80-fold over that measured for WT or Eklf+/− cells (Figure 4B). Eklf is thought to negatively regulate expression of the transcription factor Fli-1.23 A 25-fold increase in the level of Fli-1 mRNA was detected in EryP lacking both alleles of Eklf (Figure 4B). We also observed a 5-fold increase in GA repeat binding protein α (Gabpα), which encodes a transcription factor important in the early stages of megakaryopoiesis (Figure 4B). We analyzed expression of additional megakaryocyte-related proteins on Eklf-null mutant EryP/H2B-GFP using FACS. CD42 (platelet glycoprotein IX), α2 integrin, αVβ3 integrin, c-kit, and CD34 were not expressed on these cells (data not shown). However, Pecam-1/CD31, which is up-regulated on FL cells in the absence of Eklf,23 was readily detected on Eklf-null mutant EryP (Figure 4C). Together, these results suggest that the cellular identity of Eklf-deficient EryP is altered and that specification to the erythroid lineage is compromised in favor of the megakaryocyte program.
Aberrant expression of megakaryocyte-related adhesion proteins and transcription factors in Eklf-null mutant EryP. CD41 (A) and CD31 (Pecam-1, C) protein were up-regulated on EryP/H2B-GFP from Eklf-null E13.5 embryos. (B) Genes encoding the integrin CD41 (Itga2b) and the megakaryocyte transcription factors Fli-1 and Gabpa were strongly up-regulated in EryP sorted from Eklf-null mutant E13.5 embryos, as analyzed using real-time RT-PCR.
Aberrant expression of megakaryocyte-related adhesion proteins and transcription factors in Eklf-null mutant EryP. CD41 (A) and CD31 (Pecam-1, C) protein were up-regulated on EryP/H2B-GFP from Eklf-null E13.5 embryos. (B) Genes encoding the integrin CD41 (Itga2b) and the megakaryocyte transcription factors Fli-1 and Gabpa were strongly up-regulated in EryP sorted from Eklf-null mutant E13.5 embryos, as analyzed using real-time RT-PCR.
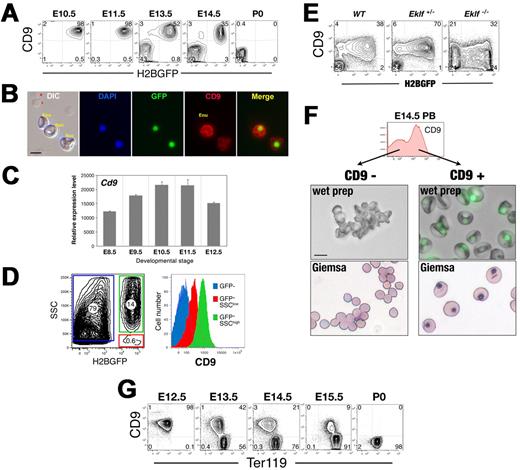
The tetraspanin CD9 is expressed by circulating nucleated WT primitive erythroid cells
In the course of examining Eklf-null mutant EryP for surface megakaryocyte markers, we made the unexpected discovery that the tetraspanin CD9, found on megakaryocytes, platelets, and several other cell types,24 is also expressed on the surface of WT EryP. CD9 was detected on EryP/H2B-GFP as early as E8.5 (data not shown) and was maintained on EryP/H2B-GFP through the period of rapid enucleation (until E14.5; Figure 5A). CD9 was not found on enucleated (GFP-negative) EryP (Figure 5B and data not shown). Cd9 mRNA was detected in EryP sorted from embryos from E8.5-12.5 (Figure 5C).
CD9 surface expression identifies circulating nucleated primitive erythroid cells. (A) The tetraspanin CD9 is expressed on circulating EryP/H2B-GFP. (B) Immunocytochemical staining of PB from 13.5 embryos using anti-CD9. (C) Expression of Cd9 in EryP throughout development. GFP+ cells were sorted from E8.5-12.5 embryos and real-time RT-PCR analysis of Cd9 mRNA expression performed. (D) CD9 is redistributed onto expelled EryP nuclei. Side scatter (SSC) and GFP can be used to identify 3 populations: SSC(high) GFP+ EryP (green), SSC(low) GFP+ free nuclei (red) and SSC(low) GFP− enucleated cells (blue) (left panel). CD9 expression on each population is shown (right panel). CD9 is present on nucleated EryP and at lower levels on free nuclei; it is not expressed on enucleated EryD. (E) CD9 expression is altered by loss of Eklf. While all E13.5 WT and heterozygous EryP express CD9, Eklf-null GFP+ cells show more heterogeneous CD9 expression, with a CD9-negative population. (F) Nucleated EryP can be isolated on the basis of CD9 expression. CD9+ and CD9− cells were sorted from E14.5 blood. Both wet preparations (middle panels) and Giemsa stains (bottom panels) clearly document that the CD9+ cells are EryP. (G) Developmental progression of CD9 expression on circulating WT erythroid cells. CD9 is expressed on EryP but not on definitive erythroid cells. Images were captured in AxioVision using a Zeiss AxioCam camera mounted on a Zeiss Axioplan 2 microscope and outfitted with a 63×/Plan neofluar/1.25 NA oil objective (B) or a Zeiss Axiovert 25 inverted microscope outfitted with a 32×/LD-A-Plan/0.3 NA objective (F). Scale bars, 10 μm.
CD9 surface expression identifies circulating nucleated primitive erythroid cells. (A) The tetraspanin CD9 is expressed on circulating EryP/H2B-GFP. (B) Immunocytochemical staining of PB from 13.5 embryos using anti-CD9. (C) Expression of Cd9 in EryP throughout development. GFP+ cells were sorted from E8.5-12.5 embryos and real-time RT-PCR analysis of Cd9 mRNA expression performed. (D) CD9 is redistributed onto expelled EryP nuclei. Side scatter (SSC) and GFP can be used to identify 3 populations: SSC(high) GFP+ EryP (green), SSC(low) GFP+ free nuclei (red) and SSC(low) GFP− enucleated cells (blue) (left panel). CD9 expression on each population is shown (right panel). CD9 is present on nucleated EryP and at lower levels on free nuclei; it is not expressed on enucleated EryD. (E) CD9 expression is altered by loss of Eklf. While all E13.5 WT and heterozygous EryP express CD9, Eklf-null GFP+ cells show more heterogeneous CD9 expression, with a CD9-negative population. (F) Nucleated EryP can be isolated on the basis of CD9 expression. CD9+ and CD9− cells were sorted from E14.5 blood. Both wet preparations (middle panels) and Giemsa stains (bottom panels) clearly document that the CD9+ cells are EryP. (G) Developmental progression of CD9 expression on circulating WT erythroid cells. CD9 is expressed on EryP but not on definitive erythroid cells. Images were captured in AxioVision using a Zeiss AxioCam camera mounted on a Zeiss Axioplan 2 microscope and outfitted with a 63×/Plan neofluar/1.25 NA oil objective (B) or a Zeiss Axiovert 25 inverted microscope outfitted with a 32×/LD-A-Plan/0.3 NA objective (F). Scale bars, 10 μm.
Expression of CD9 was associated with nucleated but not enucleated EryP (Figure 5B). To determine whether, like several integrins,6 CD9 might be redistributed onto the membrane surrounding the extruded nucleus, we used a flow cytometric assay that we had developed previously in which free nuclei are identified as Side Scatter (SSC)lowH2B-GFPhigh structures in the circulation.6 CD9 was indeed identified on SSClowH2B-GFPhigh structures, although at lower levels than on nucleated EryP and was not expressed on enucleated EryP (Figure 5D). In the absence of Eklf, CD9 expression was heterogeneous, with approximately 50% of Eklf−/− EryP negative for surface protein (Figure 5E). Eklf+/− EryP exhibited WT levels of CD9 (Figure 5E).
CD9 surface expression distinguishes circulating nucleated primitive erythroid cells from circulating definitive erythroid cells
To determine whether CD9 might serve as a useful marker of nucleated primitive erythroblasts, CD9+ and CD9− cells were sorted from PB of WT E14.5 ϵ::H2B-GFP embryos. At this stage, the blood contains a mixture of nucleated and enucleated EryP and FL-derived, enucleated EryD.4 As shown in Figure 5F, the CD9+ cell fraction consisted entirely of nucleated EryP, whereas the CD9-negative fraction contained both enucleated EryP and EryD. Therefore, CD9 marks nucleated EryP in the circulation. Moreover, in combination with either the ϵ::H2B-GFP transgene (Figure 5A) or with Ter119 (Figure 5G), CD9 clearly identifies both EryP and EryD in PB through E15.5.
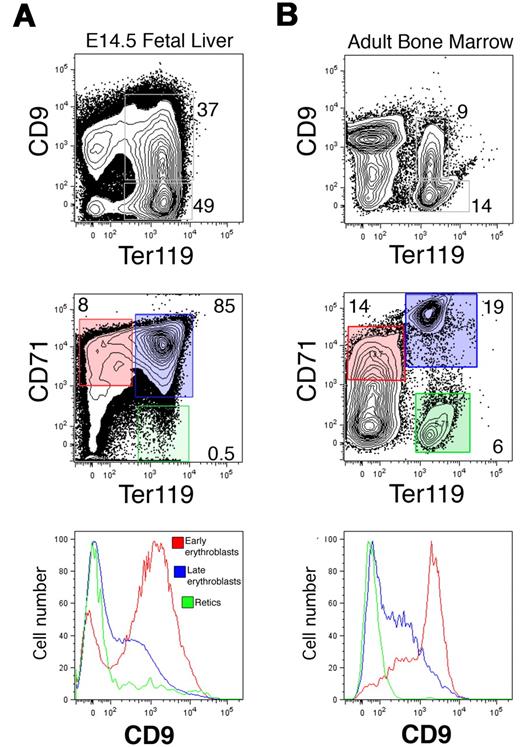
CD9 expression on FL and adult BM erythroid lineages
To determine whether CD9 is expressed on nucleated EryD, we examined Ter119+ cells from E14.5 FL (Figure 6A) and adult BM (Figure 6B). CD9 was found on approximately 40% of the Ter119+ cells in each population (Figure 6A). To identify the developmental stages at which CD9 is expressed during definitive erythropoiesis, we used simultaneous FACS analysis of CD71 and Ter119, a combination of markers that has been used to define maturing erythroid populations based on progressive up-regulation first of CD71 and then Ter119, followed by loss of CD71.25 For both FL and BM, CD9 was expressed on the majority of CD71+Ter119− proerythroblasts and was rapidly down-regulated as these cells matured to CD71+Ter119+ erythroblasts (Figure 6). By the reticulocyte and erythrocyte (CD71−Ter119+) stages, CD9 was absent from the cell surface. Therefore, CD9 is expressed on the nucleated erythroblasts of both the primitive and definitive erythroid lineages but not on mature enucleated cells.
CD9 is expressed on definitive erythroblasts in the FL and BM. CD9 expression was measured on Ter119+ cells from E14.5 FL (A, top panel) or adult BM (B, top panel) using analytical flow cytometry. Approximately 40% of Ter119+ cells from both tissues express CD9. (Middle panels) The developmental progression of erythroid cells from FL or adult BM was identified using CD71 and Ter119 surface expression. (Bottom panels) CD9 was present on most CD71+;Ter119− (immature) erythroid cells (red), was down-regulated on CD71+;Ter119+ erythroblasts (blue), and was absent from mature CD71−;Ter119+ reticulocytes and erythrocytes (green) from both FL and adult BM.
CD9 is expressed on definitive erythroblasts in the FL and BM. CD9 expression was measured on Ter119+ cells from E14.5 FL (A, top panel) or adult BM (B, top panel) using analytical flow cytometry. Approximately 40% of Ter119+ cells from both tissues express CD9. (Middle panels) The developmental progression of erythroid cells from FL or adult BM was identified using CD71 and Ter119 surface expression. (Bottom panels) CD9 was present on most CD71+;Ter119− (immature) erythroid cells (red), was down-regulated on CD71+;Ter119+ erythroblasts (blue), and was absent from mature CD71−;Ter119+ reticulocytes and erythrocytes (green) from both FL and adult BM.
Discussion
Delineation of transcription factor function in cells of the primitive erythroid lineage has been hampered by the simultaneous presence of primitive and definitive erythroid cells in the embryonic circulation. The generation of fluorescent transgenic reporter mouse lines that express cytoplasmic4,26 or nuclear6 GFP in a lineage-restricted manner has allowed us to segregate these 2 closely related erythroid lineages. It was initially believed that the primitive erythroid lineage was less severely affected than the definitive lineage by ablation of Eklf.11,12 By midgestation, the blood of Eklf-null mutant embryos lacks mature, enucleated definitive erythrocytes and contains largely nucleated, blastlike EryD and misshapen EryP.13,14 Previous studies had analyzed whole embryonic blood containing a mixture of both lineages, so that specific analysis of EryP was not possible. Here, we have examined in detail the consequences for development of EryP of loss of one or both alleles of Eklf. We find that a full complement of Eklf is required for normal maturation and identity restriction of primitive erythroid cells.
Eklf haploinsufficiency alters the phenotype of primitive erythroid cells
The primitive and definitive erythroid lineages are cytologically distinct. At the molecular level, the most prominent difference is in their globin gene and protein expression. Here we report additional differences between the lineages. By analyzing PB cells for expression of Ter119 and H2B-GFP together, so that EryP and EryD can be distinguished, we found that EryP lacking a single copy of Eklf fail to express Ter119, while Eklf heterozygous EryD expressed Ter119 at normal levels. Therefore, the Ter119 “intermediate” population reported by others14 in fact comprises Ter119+ EryD and Ter119− EryP. This finding highlights the utility of the ϵ-globin::histone H2B-GFP transgenic mouse line and suggests that at least some functions of Eklf are distinct in the 2 erythroid lineages and that EryP are more sensitive to Eklf gene dosage than are EryD. It is worth noting that Runx1-null mutant EryP, which are essentially haploinsufficient for Eklf (expressed at 50% of WT levels), also show reduced Ter119 protein expression.27 In EryP, Runx1 may activate Eklf, which in turn would regulate (directly or indirectly) expression of Ter119. No change in transcription of Gypa (the gene believed to encode Ter119) was detected in Eklf heterozygous EryP. However, when both copies of Eklf were ablated, Ter119 protein was absent and the numbers of Gypa transcripts actually increased, suggesting that the failure of Eklf mutant EryP to express surface Ter119 might result from aberrant posttranslational processing and/or defects in protein trafficking to the cell membrane. Immunostaining of permeabilized Eklf+/− EryP failed to reveal intracellular Ter119 protein. While this observation suggests that the absence of expression of Ter119 on the surface of Eklf+/− EryP/H2B-GFP is not due to aberrant trafficking, we cannot rule out the possibility that incomplete processing of Ter119 in Eklf+/− EryP results in the production of protein lacking the epitope recognized by the Ter119 antibody. It is also possible that Gypa does not encode Ter119 (ie, Ter119 is not the mouse equivalent of human GPA); a formal demonstration that Ter119 is encoded by Gypa would require expression cloning of Ter119. Uncoupling of Gypa mRNA and surface protein expression has been reported for definitive erythroid cells.28
CD9 and erythropoiesis
In examining the megakaryocyte-related markers that are aberrantly up-regulated in the absence of Eklf, we found that the tetraspanin CD9 is expressed not only on some Eklf-null EryP but also on WT EryP. Surface expression of CD9 correlated strongly with the presence of a nucleus in EryP. Using CD9 surface expression alone, we were able to identify and purify nucleated EryP from the PB of embryos at E14.5, a stage when approximately half of circulating cells are FL-derived EryD.4 The transgenic reporters are still required for isolation of EryP from FL, a more complex milieu in which CD9 is expressed on multiple cell types, including nucleated EryD (unpublished observations). To our knowledge, CD9 is the first marker that distinguishes between nucleated EryP and definitive erythrocytes in the circulation. It will be of interest to determine whether CD9 is also expressed on developing human EryP or on maturing definitive human erythroblasts. CD9 is known to play an important role in the regulation of cell fusion and cell-cell interactions in other systems. Our work may have implications for interactions between erythroid cells and stromal or other supportive cells as well as engulfment of extruded erythroid nuclei by macrophages.
The rapid decrease in CD9 surface expression during EryP maturation suggested that, like several adhesion molecules,6 this protein might also be lost during enucleation. Only approximately 20% of the CD9 measured on nucleated EryP is later found on the membrane surrounding the expelled nucleus. Therefore, an additional mechanism, such as exosomal release, must regulate the loss of CD9 from EryP. Indeed, CD9 is one of the most common components of exosomes.29
With the exception of an erythroid-specific tetraspanin, Penumbra (Pen),30 the functions of this superfamily in erythroid cells have not been well described. Tetraspanins organize microdomains of signaling proteins, such as integrins and antigen receptor proteins. α4, α5, and β1-integrins and CD44 are expressed on the surface of maturing EryP and are redistributed onto the membrane surrounding their extruding nuclei.4,6 Perhaps CD9 assists in the formation of adhesion molecule-rich signaling complexes that mediate interactions between EryP and FL macrophages,6 allowing them to complete their maturation and to enucleate.
A molecular “identity crisis” in primitive erythroid cells lacking Eklf
Several megakaryocyte markers (eg, CD41 and Pecam-1/CD31) were inappropriately activated in Eklf-deficient mutant EryP. In both primitive and definitive hematopoiesis, the erythroid and megakaryocyte lineages share a common progenitor termed the megakaryocyte-erythroid progenitor.31 Eklf has been shown to regulate a switch between the erythroid and megakaryocytic lineages.23 The megakaryocyte program is promoted in the absence of Eklf. For example, Eklf−/− FL cells produce significantly more megakaryocyte progenitors and express the integrin CD41 and a variety of megakaryocyte genes at higher levels than do WT FL cells.23 CD41 is also expressed on cells from the PB of Eklf-null mutant embryos.32 This phenotype is thought to result from an imbalance between Eklf and Fli-1, a transcriptional regulator essential for megakaryocyte commitment and early differentiation.23,33
The aberrant expression of CD41, CD31/Pecam-1, and Gabpα observed for Eklf-null mutant EryP might arise, at least in part, from the increased expression of Fli-1 that we detected in Eklf mutant EryP. Alternatively, or in addition, it could result from derepression, either due to the absence of Eklf/Klf1 itself or, indirectly, through the loss of an Eklf/Klf1-regulated repressor such as Klf3.14,34 Indeed, in Eklf-null mutant EryP, Klf3 transcription was significantly reduced. Using an in silico approach (see supplemental Figure 3 legend), we have identified several potential Klf binding sites within the promoter regions of CD41/Itga2b, CD31/Pecam-1, and Gabpα (supplemental Figure 3). Therefore, Klf3 may repress their transcription in WT cells. It is conceivable that Eklf/Klf1 also plays a role in the negative regulation of these genes, as it is known to function not only as an activator but also as a repressor.35-37 A potential binding site for Fli-1 is present upstream of Itga2b/CD41 and Gabpα but not CD31/Pecam-1 (supplemental Figure 3) and Fli-1 may ectopically activate Itga2b and Gabpα in Eklf-null mutant EryP.
The expression of megakaryocyte markers on mutant EryP was not promiscuous: in addition to CD41 and CD31, other megakaryocyte-related proteins such as CD42, α2-, αV-, and β3-integrins were not detected on these cells. Moreover, they lack the erythroid lineage-specific marker Ter119. These observations suggest that Eklf regulates cell identity not only in megakaryocyte-erythroid progenitors but throughout EryP maturation. One of the morphologic hallmarks of Eklf-null mutant EryP—a ruffled cell membrane with occasional protrusions—may result from aberrant expression of megakaryocyte adhesion molecules and, therefore, alterations in the protein composition of the EryP membrane. Megakaryocytes give rise to platelets by membrane demarcation and budding.38 The protrusions seen on Eklf-deficient EryP may reflect failed attempts at platelet production.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr David Bodine (National Institutes of Health) for the Eklf+/− mouse line and Dr Barry Coller (Rockefeller University) for the APC-conjugated 1B5 anti-CD42 monoclonal antibody. We are grateful to Dr James Bieker and Dr Michael Dyer for insightful comments on the manuscript. We acknowledge the Mount Sinai Flow Cytometry Shared Resource Facility for assistance with cell sorting. Transgenic mice were produced by the Mount Sinai Mouse Genetics Shared Research Facility (National Institutes of Health/National Cancer Institute grant R24 CA88302).
This work was supported by grants to M.H.B. from the National Institutes of Health (nos. RO1 HL62248, DK52191, and EB02209) and the Roche Foundation for Anemia Research, and by a postdoctoral fellowship to J.I. from the Cooley's Anemia Foundation.
National Institutes of Health
Authorship
Contribution: J.I., S.T.F., Z.H., and M.H.B. designed the experiments and prepared the figures; J.I., S.T.F., and Z.H. performed the experiments; H.Z. performed in silico analysis and prepared the supplementary figure; and J.I., S.T.F., and M.H.B. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for S.T.F. is Discipline of Physiology, School of Medical Sciences, University of Sydney, Camperdown, Australia.
Correspondence: Margaret H. Baron, Mount Sinai School of Medicine, Box 1079, 1468 Madison Ave, Annenberg 24-04E, New York, NY 10029-6574; e-mail: margaret.baron@mssm.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal