Abstract
Sprouting of developing blood vessels is mediated by specialized motile endothelial cells localized at the tips of growing capillaries. Following behind the tip cells, endothelial stalk cells form the capillary lumen and proliferate. Expression of the Notch ligand Delta-like-4 (Dll4) in tip cells suppresses tip cell fate in neighboring stalk cells via Notch signaling. In DLL4+/− mouse mutants, most retinal endothelial cells display morphologic features of tip cells. We hypothesized that these mouse mutants could be used to isolate tip cells and so to determine their genetic repertoire. Using transcriptome analysis of retinal endothelial cells isolated from DLL4+/− and wild-type mice, we identified 3 clusters of tip cell–enriched genes, encoding extracellular matrix degrading enzymes, basement membrane components, and secreted molecules. Secreted molecules endothelial-specific molecule 1, angiopoietin 2, and apelin bind to cognate receptors on endothelial stalk cells. Knockout mice and zebrafish morpholino knockdown of apelin showed delayed angiogenesis and reduced proliferation of stalk cells expressing the apelin receptor APJ. Thus, tip cells may regulate angiogenesis via matrix remodeling, production of basement membrane, and release of secreted molecules, some of which regulate stalk cell behavior.
Introduction
During sprouting angiogenesis, growing capillaries are spearheaded by specialized endothelial cells (ECs) termed tip cells.1 In the postnatal mouse retina, tip cells guide outgrowing capillaries toward gradients of matrix-bound vascular endothelial growth factor (VEGF)164 produced by hypoxic astrocytes. Tip cells explore their environment by extending motile slender filopodia, which express VEGF receptor (R)-2 and -3 and transduce the migratory signal initiated by VEGF binding.1,2 Time-lapse videomicroscopy on developing zebrafish embryos has shown that tip cell filopodial movement is essential for capillary extension and that anastomosis of tip cell filopodia leads to the formation of new vessel branches.3 Tip cells are also formed during pathologic processes such as tumor angiogenesis and may be preferential targets for antiangiogenic treatments using VEGF blocking agents. Identifying the signaling molecules expressed by endothelial tip cells could ultimately allow to guide vessels away from a tumor or, on the contrary, toward an ischemic area.
To establish a properly patterned vascular tree, tip cells must be followed by morphologically distinct EC, which form a lumenized tube growing behind the tip cell. In contrast to the tip cells, these follower cells termed stalk cells do not extend filopodia but exhibit proliferative behavior.1 We and others have previously shown that stalk cells are inhibited from becoming tip cells by the Notch signaling pathway.4-7 Hypoxia-driven VEGF signaling induces expression of the Notch ligand Delta-like-4 (Dll4) in tip cells. Dll4 signaling to Notch expressed in adjacent ECs suppresses tip cell fate in the stalk cells, at least in part through alterations in the levels of VEGF receptor expression. Consequently, pharmacologic blockade of Notch signaling using γ-secretase inhibitors or inducible endothelial-specific deletion of the Notch-1 receptor both lead to the formation of excessive tip cell numbers. Similarly, blockage of Dll4 signaling using an extracellular Dll4-Fc trap construct or heterozygous deletion of one DLL4 allele also leads to formation of excessive tip cell numbers. Our previous analysis of DLL4+/− mouse retinal vessels had shown that virtually every EC, with the exception of the main arteries, extends filopodia. Expression of the few known tip cell markers PDGFB and the axon guidance receptor UNC5B, normally restricted to the front of the developing retinal vascular tree, was observed throughout the retinal vascular plexus in DLL4+/− retinas.7 Together, these observations indicate that isolation of ECs from DLL4+/− retinas should allow purification of enriched tip cell numbers and characterization of their gene expression profile using microarray analysis.
Methods
For more details, see supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Mice
Mice were maintained and experiments performed according to the guidelines of the French Ministry of Health. The DLL4+/− (CD1), APLN−/− (C57BL/6N), and APJ−/− (C57BL/6N) mice used in the experiments have been previously described.8-10 To inhibit the mammalian target of rapamycin (mTOR) pathway, 5 mg/kg rapamycin (Invitrogen) dissolved in dimethyl sulfoxide (DMSO) was intraperitoneally injected into mice at P6. Injection with DMSO was used as a negative control. Mice in the C57BL/6N background were killed at P7 and mice in the CD1 background at P5. Retinas were isolated for analysis unless otherwise stated.
Purification of endothelial cells from retinas
Fresh retinas were minced and incubated in 5 mL Dulbecco modified Eagle medium containing 200 U/mL collagenase I (Invitrogen) for 45 minutes at 37°C with occasional shaking followed by filtering through a 40-μm nylon mesh. The cells were then centrifuged at 94g for 5 minutes at 4°C, resuspended in Buffer 1 (0.1% bovine serum albumin, 2mM EDTA (ethylenediaminetetraacetic acid), pH 7.4 in phosphate-buffered saline [PBS]), and incubated with anti rat immunoglobulin G-coated magnetic beads (Invitrogen) precoupled with rat anti–mouse platelet/endothelial cell adhesion molecule-1 (PECAM-1; MEC13.3, BD Pharmingen) for 30 minutes at 4°C in an overhead shaker. Beads were separated from the solution with a magnetic particle concentrator (Dynal MPC-S, Invitrogen), and the cells in the supernatant were kept as the non-EC fraction. The beads were washed 5× with Buffer 1 and centrifuged for 5 minutes at 3400g, and the supernatant removed. The purified ECs were then shock-frozen in liquid nitrogen and stored at −80°C until further use. Total RNA was isolated using the RNAeasy kit from QIAGEN.
Microarray analysis
Retinal ECs isolated from 6 wild-type and dll4+/− mice were hybridized in triplicate. The quality of the RNA was analyzed by using an Agilent Bioanalyzer for microarray analysis. Total RNA (100 ng) was amplified with the Amino allyl messageAmp II aRNA amplification kit (Ambion) to ensure complete synthesis of virtually full-length cDNA. A cRNA in vitro transcription was performed incorporating a modified nucleotide to couple dyes. The quality of the labeled cRNA with nucleotides coupled to fluorescent dye Cy3 (reference sample) or Cy5 (EC sample) (Amersham) was verified in an Agilent Bioanalyzer. Cy5-labeled cRNA (825 ng) from each sample and Cy3-labeled cRNA (825 ng) were hybridized to Agilent Whole Mouse genome slides in 1 × 44 K format. The hybridized arrays were then washed (Gene expression hybridization and wash buffer kits, Agilent) and scanned (Genepix 4000; Molecular Devices), and the data were extracted from the scanned images using Genepix Pro 6.0 (Molecular Devices) and Bioconductor script. The data were normalized (lowess normalization), and 2.5- and higher fold change was used as a cutoff for up-regulated genes and 0.5- and lower fold change as a cutoff for down-regulated genes. The complete dataset from this analysis is available at Array Express with accession number E-MTAB-347.
Quantitative polymerase chain reaction (PCR) analysis
Real-time quantitative PCR (qPCR) reactions were performed in duplicate using the MyIQ real-time PCR system (Biorad). Each 25-μL reaction contained 5 ng cDNA, 12.5 μL iQ SYBR Green Supermix (Biorad), 250 nM forward and reverse primers, and nuclease free water. Fold changes were calculated using the comparative CT method.
Statistical analyses
Gene set enrichment analysis (GSEA) was performed in the R statistical environment. A gene set was generated from the data in the microarray by selecting probes expressed at least 2.5-fold higher in DLL4+/− versus wild-type samples. Agilent ratio data for tip and stalk cells from Strasser et al11 (GSE19284) were filtered to include only those probes for which at least 7 samples contained data (were not NAs). The data were log2 transformed, and any remaining missing values were imputed for each probe by calculating the median for that particular probe. The nsFilter function was used to filter the remaining probes. The geneset was then tested for enrichment in the Strasser et al11 data using the GSEAlm package (1000 permutations). Mann-Whitney tests were used to assess the statistical differences between measurements (GraphPad Prism 4, GraphPad Software) in all the experiments.
Results
Isolation of retinal endothelial cells and microarray analysis
ECs were isolated from P5 retinas by labeling with an antibody directed against the pan-endothelial marker PECAM-1 followed by Dynabead sorting. Semiquantitative reverse transcription PCR confirmed the expression of PECAM-1 and VE-CADHERIN in endothelial-enriched, but not in nonendothelial populations (supplemental Figure 1). Amplification of astrocyte-specific GFAP and pericyte markers NG2, PDGFRB, and smooth-muscle actin SMA showed expression in non-ECs but only weak expression in the endothelial-enriched population, indicating that the purification procedure had significantly enriched for ECs (data not shown). Microarray analysis was performed with total RNA extracted from EC and non-EC populations from wild-type and DLL4+/− mice. 158 genes were found down-regulated 0.5-fold in DLL4+/− ECs compared with wild-type ECs. A total of 411 genes was identified that showed more than 2.5-fold up-regulated expression in ECs of DLL4+/− compared with wild-type mice (supplemental Table 1). Approximately 35 selected up-regulated genes were cloned and analyzed by in situ hybridization (ISH) and/or antibody staining on retinal whole mounts to confirm the up-regulation in DLL4+/− retinas and to localize their expression in the retina (Table 1; see Figure 2 and supplemental Figures 2-3).
List of genes up-regulated in ECs from dll4+/− compared with wild-type retinas verified by ISH or immunohistochemistry
| Gene name . | Protein name . | Fold change . | Method . | Expression in WT retinas . | Expression in dll4+/− retinas . |
|---|---|---|---|---|---|
| ECM/BM molecules | |||||
| NID1 | Nidogen-1 | 4.26 | ISH | Tip cell–enriched and plexus | Vascular plexus higher than in WT |
| IHC | All vessels | All vessels | |||
| NID2 | Nidogen-2 | 5.49 | ISH | Vascular front enriched | Vascular front and plexus |
| PLAUR | uPAR | 5.23 | ISH | Tip cell–enriched | Tip cells and plexus |
| LOXL2 | Lysyl oxidase-like 2 | 3.15 | |||
| TGFBI | TGF-β–induced | 7.63 | ISH | Veins | Veins, signal stronger than in WT |
| ITGB1 | Integrinβ1 | 4.02 | IHC | Tip cells and plexus | Tip cells and plexus |
| LAMB1 | Lamininb1 | 5.44 | |||
| Secreted molecules | |||||
| APLN | Apelin | 3.72 | ISH | Tip cell–enriched | Tip cells and plexus |
| ESM1 | Endothelial-specific molecule 1 | 11.07 | ISH | Tip cell–enriched | Tip cells and plexus |
| IHC | Tip cells/stalk cells | Tip cells and plexus | |||
| ANGPT2 | Angiopoietin-2 | 4.64 | ISH | Tip cells | Tip cells |
| IGFBP3 | Insulin-like growth factor–binding protein-3 | 4.08 | IHC | Vascular front | Vascular front and plexus |
| GMFG | Glia maturation factor g | 5.86 | |||
| TGF-β pathway | |||||
| ACVRL1 | ALK1 | 2.6 | ISH | Arteries/veins | Arteries/veins and plexus |
| TGFB1 | TGFβ1 | 2.88 | ISH | Vascular front | Vascular front and plexus |
| TGFBR2 | TGFβR2 | 3.05 | ISH | Strongest in arteries (also in vascular front and plexus) | Strongest in arteries (also in vascular front and plexus) |
| Transcription factors | |||||
| ETS1 | Ets-1 | 3.40 | ISH | Stronger in veins than arteries, plexus, high in stalk cells, no tip cells | Same as WT, but signal stronger in vascular plexus |
| MEF2C | Mef2c | 2.91 | |||
| KLF2 | Krueppel-like factor (KLF)-2 | 3.2 | ISH | Arteries/veins | Arteries/veins |
| KLF4 | KLF4 | 2.49 | |||
| FOXC1 | FoxC1 | 3.51 | |||
| SOX7 | Sox7 | 4.60 | |||
| SOX17 | Sox17 | 4.64 | |||
| SNAI2 | Slug | 3.16 | |||
| Others | |||||
| SPRY1 | Sprouty1 | 2.72 | ISH | Pericytes | Pericytes |
| AQP1 | Aquaporin-1 | 6.37 | ISH | Not detectable | Arteries |
| CAV1 | Caveolin-1 | 3.46 | ISH | Veins | Veins and vascular plexus |
| IHC | All vessels | All vessels | |||
| CAV2 | Caveolin-2 | 5.18 | |||
| EDG1 | S1PR1 | 2.23 | |||
| LEPR | Leptin receptor | 3.55 | |||
| B3GNT1 | UDP-GlcNAc:βGal β-1,3-N-acetylglucosaminyltransferase1 | 6.92 | |||
| CD38 | CD38 | 7.13 | |||
| RASGRP3 | RAS guanyl releasing protein 3 | 5.71 | |||
| TYROBP | TYRO protein tyrosine kinase binding protein/DAP12 | 3.35 | |||
| SYK | Syk | 2.29 |
| Gene name . | Protein name . | Fold change . | Method . | Expression in WT retinas . | Expression in dll4+/− retinas . |
|---|---|---|---|---|---|
| ECM/BM molecules | |||||
| NID1 | Nidogen-1 | 4.26 | ISH | Tip cell–enriched and plexus | Vascular plexus higher than in WT |
| IHC | All vessels | All vessels | |||
| NID2 | Nidogen-2 | 5.49 | ISH | Vascular front enriched | Vascular front and plexus |
| PLAUR | uPAR | 5.23 | ISH | Tip cell–enriched | Tip cells and plexus |
| LOXL2 | Lysyl oxidase-like 2 | 3.15 | |||
| TGFBI | TGF-β–induced | 7.63 | ISH | Veins | Veins, signal stronger than in WT |
| ITGB1 | Integrinβ1 | 4.02 | IHC | Tip cells and plexus | Tip cells and plexus |
| LAMB1 | Lamininb1 | 5.44 | |||
| Secreted molecules | |||||
| APLN | Apelin | 3.72 | ISH | Tip cell–enriched | Tip cells and plexus |
| ESM1 | Endothelial-specific molecule 1 | 11.07 | ISH | Tip cell–enriched | Tip cells and plexus |
| IHC | Tip cells/stalk cells | Tip cells and plexus | |||
| ANGPT2 | Angiopoietin-2 | 4.64 | ISH | Tip cells | Tip cells |
| IGFBP3 | Insulin-like growth factor–binding protein-3 | 4.08 | IHC | Vascular front | Vascular front and plexus |
| GMFG | Glia maturation factor g | 5.86 | |||
| TGF-β pathway | |||||
| ACVRL1 | ALK1 | 2.6 | ISH | Arteries/veins | Arteries/veins and plexus |
| TGFB1 | TGFβ1 | 2.88 | ISH | Vascular front | Vascular front and plexus |
| TGFBR2 | TGFβR2 | 3.05 | ISH | Strongest in arteries (also in vascular front and plexus) | Strongest in arteries (also in vascular front and plexus) |
| Transcription factors | |||||
| ETS1 | Ets-1 | 3.40 | ISH | Stronger in veins than arteries, plexus, high in stalk cells, no tip cells | Same as WT, but signal stronger in vascular plexus |
| MEF2C | Mef2c | 2.91 | |||
| KLF2 | Krueppel-like factor (KLF)-2 | 3.2 | ISH | Arteries/veins | Arteries/veins |
| KLF4 | KLF4 | 2.49 | |||
| FOXC1 | FoxC1 | 3.51 | |||
| SOX7 | Sox7 | 4.60 | |||
| SOX17 | Sox17 | 4.64 | |||
| SNAI2 | Slug | 3.16 | |||
| Others | |||||
| SPRY1 | Sprouty1 | 2.72 | ISH | Pericytes | Pericytes |
| AQP1 | Aquaporin-1 | 6.37 | ISH | Not detectable | Arteries |
| CAV1 | Caveolin-1 | 3.46 | ISH | Veins | Veins and vascular plexus |
| IHC | All vessels | All vessels | |||
| CAV2 | Caveolin-2 | 5.18 | |||
| EDG1 | S1PR1 | 2.23 | |||
| LEPR | Leptin receptor | 3.55 | |||
| B3GNT1 | UDP-GlcNAc:βGal β-1,3-N-acetylglucosaminyltransferase1 | 6.92 | |||
| CD38 | CD38 | 7.13 | |||
| RASGRP3 | RAS guanyl releasing protein 3 | 5.71 | |||
| TYROBP | TYRO protein tyrosine kinase binding protein/DAP12 | 3.35 | |||
| SYK | Syk | 2.29 |
WT indicates wild-type; IHC, immunohistochemistry; ALK1, activin receptor-like kinase 1; FoxC1, forkhead box C1; and UDP-GlcNAc, uridine diphosphate N-acetylglucosamine.
As the Dll4-Notch pathway is implicated in other angiogenesis processes besides tip cell formation, including arterialization and blood vessel maturation,8,12 we expected that not all of the genes identified were tip cell–enriched genes but that some might be regulated by the Dll4-Notch signaling pathway in other regions of the vasculature. Indeed, of the genes confirmed to be up-regulated in DLL4+/− deficient retinal ECs, not all were tip cell–enriched (Table 1 and supplemental Figure 2). Among several transcription factors tested, ETS-1 showed the most intriguing expression pattern, with high expression in veins, lower expression in capillaries and arteries, and virtually no expression in tip cells (supplemental Figure 2A,C). Comparison of wild-type and dll4+/− retinas showed increased ets-1 signal in DLL4+/− vessels, but the tip cell region remained devoid of ets-1 expression (supplemental Figure 2B,D). Mechanistically, DLL4+/− vessels show reduced Notch signaling in stalk cells, implying that Notch could potentially regulates ets-1 activity in stalk cells, although this remains to be further investigated. A second group of genes up-regulated in DLL4+/−, but with no obvious tip cell expression, were transforming growth factor (TGF)-β signaling molecules TGFBR2, TGFB1, and ACVRL1 (Table 1 and supplemental Figure 2E-H). These observations suggest a possible link between the TGF-β and the Notch signaling pathway in angiogenesis, as previously indicated by in vitro studies,13,14 that remains to be further investigated.
A recent study identified tip cell markers by laser capture microdissection of retinal tip cells.11 Examination of the relationship between the 2 microarray datasets using GSEA showed that tip cell markers from the Strasser publication were significantly enriched (P = .001) in our microarray data. Genes that were highly enriched in tip cell populations in both datasets include molecules involved in extracellular matrix (ECM) degradation, basement membrane (BM) deposition, and secreted molecules, which were subsequently studied.
Molecules involved in extracellular matrix degradation
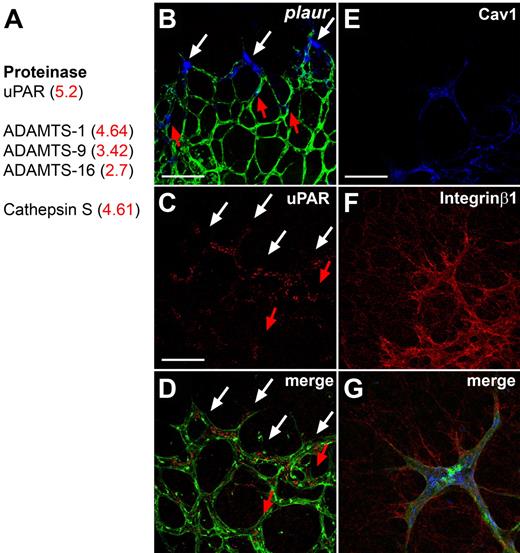
The first group encodes ECM-degrading enzymes, including cathepsin S, a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 1 (ADAM-TS) family members, and urokinase-plasminogen-activated receptor (uPAR), which were up-regulated in DLL4+/− compared with wild-type ECs (Figure 1A). We confirmed preferential expression of uPAR mRNA and protein in tip cells by ISH and immunohistochemistry (Figure 1B-D). uPAR triggers cell migration by converting urokinase into active uPA, which converts plasminogen to plasmin, which in turn breaks down ECM components and releases proangiogenic factors such as VEGF or basic fibroblast growth factor from the matrix.15 In addition, uPAR is known to regulate cell migration by interacting with integrinα5β1 and caveolin,15,16 which are also present on the list of up-regulated genes and expressed at high levels in tip cells (Figure 1E-G). The tip cell expression of uPAR could therefore participate to the degradation of the ECM and facilitate sprout growth in the correct direction. Functionally, uPAR-deficient mice develop normal retinal vasculature (data not shown) and survive into adulthood, which may be due to other proteinases that can substitute for lack of uPAR.17,18 Nevertheless, these results indicate that preferential expression of ECM degrading enzymes by tip cells should enable them to migrate toward the periphery of the retina and to form a functional vascular network.
Tip cell–enriched genes include proteases. (A) The proteases up-regulated in DLL4+/− retinas by microarray analysis include uPAR, members of the ADAMTS family and Cathepsin S. Fold change in DLL4+/− compared with wild-type is shown in red. (B) Whole mount ISH for plaur (blue) on retinas counterstained for isolectinB4 (green). Plaur shows preferential expression in tip cells (white arrows). Some stalk cells also express PLAUR (red arrows). (C-D) Immunohistochemistry confirms an enhanced expression of uPAR in tip cells (white arrows) and some uPAR positive stalk cells (red arrows). (E-G) Antibody staining shows that Caveolin-1 (blue) and Integrinβ1 (red) are also expressed by IsolectinB4 positive tip cells (green). The pictures represent 1 of 3 independent experiments with similar results. Scale bars: (B-D) 50 μm; (E-G) 5 μm. Images of antibody stainings were acquired on a Leica TCS SP5 confocal microscope using a 63×/1.4 NA objective. Images of ISH were acquired on an Olympus BX50 microscope using a 20×/0.05 NA objective and captured using a Coolsnap camera (Roper) through IPLab software version 3.2.4 (BioVision Technologies). Vessels were visualized with IsolectinB4 directly conjugated to AlexaFluor488 and the ISH signal by bright-field microscopy. Single channels were converted to grayscale (ISH in blue) in Photoshop CS2 (Adobe Systems) and merged in ImageJ (National Institutes of Health).
Tip cell–enriched genes include proteases. (A) The proteases up-regulated in DLL4+/− retinas by microarray analysis include uPAR, members of the ADAMTS family and Cathepsin S. Fold change in DLL4+/− compared with wild-type is shown in red. (B) Whole mount ISH for plaur (blue) on retinas counterstained for isolectinB4 (green). Plaur shows preferential expression in tip cells (white arrows). Some stalk cells also express PLAUR (red arrows). (C-D) Immunohistochemistry confirms an enhanced expression of uPAR in tip cells (white arrows) and some uPAR positive stalk cells (red arrows). (E-G) Antibody staining shows that Caveolin-1 (blue) and Integrinβ1 (red) are also expressed by IsolectinB4 positive tip cells (green). The pictures represent 1 of 3 independent experiments with similar results. Scale bars: (B-D) 50 μm; (E-G) 5 μm. Images of antibody stainings were acquired on a Leica TCS SP5 confocal microscope using a 63×/1.4 NA objective. Images of ISH were acquired on an Olympus BX50 microscope using a 20×/0.05 NA objective and captured using a Coolsnap camera (Roper) through IPLab software version 3.2.4 (BioVision Technologies). Vessels were visualized with IsolectinB4 directly conjugated to AlexaFluor488 and the ISH signal by bright-field microscopy. Single channels were converted to grayscale (ISH in blue) in Photoshop CS2 (Adobe Systems) and merged in ImageJ (National Institutes of Health).
Molecules involved in ECM formation
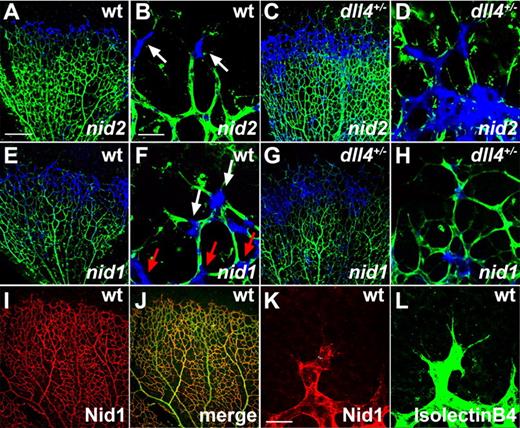
The second cluster of tip cell selective genes encoded BM proteins including Nidogen-1 and -2. Nidogen-1 and -2 are secreted proteins that stabilize the BM by interacting with other ECM proteins such as laminin, collagen IV, perlecan, and fibulin.19 ISH on wild-type retinas showed that Nidogen-2 mRNA was transcribed at highest levels in tip cells and at much lower levels in the rest of the vasculature, including stalk cells (Figure 2A-B). Comparison with DLL4+/− retinas showed that, as expected from the microarray data, Nidogen-2 mRNA was expressed over a wider area in DLL4+/−, with no obvious difference in expression levels between tip and stalk cells (Figure 2C-D). Nidogen-1 mRNA was expressed at higher levels, but not exclusively, in capillary tips (Figure 2E-F) and was also expressed more uniformly over a wider area of the vascular front in DLL4+/− mice (Figure 2G-H). Immunohistochemistry on whole mount retinas confirmed the expression of Nidogen-1 by ECs (Figure 2I-J). The antibody staining revealed a homogenous deposition of Nidogen-1 in the BM of all vessels, which extended up to the posterior pole of the tip cells but did not cover the filopodial-extending pole of tip cells (Figure 2K-L). Nidogens are therefore mainly produced by the tip cells and thereafter deposited in the BM. Nidogen-1– and Nidogen-2–deficient mice do not show a vascular phenotype, probably due to redundant expression.20,21 However, double knockout mice die shortly after birth because of defects in heart and lungs.22 In addition to Nidogens, a number of other genes involved in ECM production were found to be up-regulated in the DLL4+/− retinal vasculature, including perlecan, laminin α5, laminin β1, and Lysyl oxidase-like-2, indicating that tip cells are the main source of ECM components of blood vessels in the mouse retina. Taken together, these results indicate that a balance of ECM formation and degradation by tip cells is necessary to enable the cells to migrate toward the periphery of the retina and to form a functional vascular network.
Tip cells express basement membrane components. (A-B) Whole mount ISH (blue) confirmed enhanced Nidogen-2 (nid2) expression in tip cells of wild-type mice (white arrows) and a broader expression in DLL4+/− mice (C-D). (E-F) Nidogen-1 (nid1) mRNA (blue) is expressed in the vascular front of wild-type mice, including tip cells (white arrows) and stalk cells (red arrows). (G-H) Nidogen-1 mRNA is up-regulated in DLL4+/− mice. (I-L) Whole mount immunohistochemistry revealed Nidogen-1 protein (red) in the basement membrane of all retinal vessels in wild-type mice, at the exclusion of filopodia (K-L). IsolectinB4 was used to counterstain vessels (green). The pictures represent 1 of 3 independent experiments with similar results. Scale bars: (A,C,E,G,I,J) 250 μm; (B,D,F,H) 50 μm; (K-L) 40 μm. Images of antibody stainings were acquired on a Leica TCS SP5 confocal microscope using a 10×/0.3 or 63×/1.4 NA objective. Images of ISH were acquired on an Olympus BX50 microscope using a 4×/0.16 NA or 20×/0.05 NA objective and captured using a Coolsnap camera through IPLab software version 3.2.4. Vessels were visualized with IsolectinB4 directly conjugated to AlexaFluor488 and the ISH signal by bright-field microscopy. Single channels were converted to grayscale in Photoshop CS2 and merged in ImageJ (ISH in blue).
Tip cells express basement membrane components. (A-B) Whole mount ISH (blue) confirmed enhanced Nidogen-2 (nid2) expression in tip cells of wild-type mice (white arrows) and a broader expression in DLL4+/− mice (C-D). (E-F) Nidogen-1 (nid1) mRNA (blue) is expressed in the vascular front of wild-type mice, including tip cells (white arrows) and stalk cells (red arrows). (G-H) Nidogen-1 mRNA is up-regulated in DLL4+/− mice. (I-L) Whole mount immunohistochemistry revealed Nidogen-1 protein (red) in the basement membrane of all retinal vessels in wild-type mice, at the exclusion of filopodia (K-L). IsolectinB4 was used to counterstain vessels (green). The pictures represent 1 of 3 independent experiments with similar results. Scale bars: (A,C,E,G,I,J) 250 μm; (B,D,F,H) 50 μm; (K-L) 40 μm. Images of antibody stainings were acquired on a Leica TCS SP5 confocal microscope using a 10×/0.3 or 63×/1.4 NA objective. Images of ISH were acquired on an Olympus BX50 microscope using a 4×/0.16 NA or 20×/0.05 NA objective and captured using a Coolsnap camera through IPLab software version 3.2.4. Vessels were visualized with IsolectinB4 directly conjugated to AlexaFluor488 and the ISH signal by bright-field microscopy. Single channels were converted to grayscale in Photoshop CS2 and merged in ImageJ (ISH in blue).
Secreted molecules
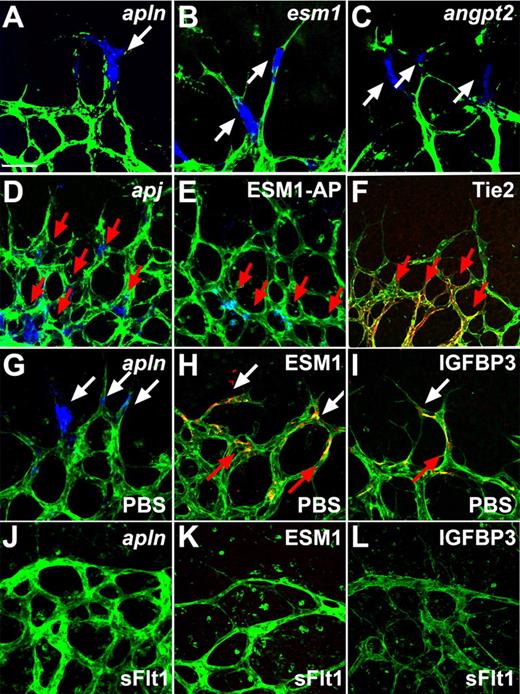
The third and largest cluster of tip cell–enriched genes identified in the microarray encodes secreted molecules, including various chemokines, adrenomedullin, angiopoietin-2 (ang-2), apelin, insulin-like growth factor binding protein 3 (IGFBP-3), and endothelial-specific molecule 1 (ESM-1) (supplemental Table 1, Figure 3A-C, and supplemental Figure 3A-E). Among those, ISH showed that IGFBP-3, ESM-1, ang-2, and apelin are all highly selective for tip cells (Figure 3A-C and supplemental Figure 3A-E). While IGFBP3, ESM1, and APELIN mRNA was also found on some stalk cells, expression of ang-2 was exclusively found in tip cells and did not label any stalk cells. In addition, counting of ang-2–positive tip cells revealed that only 71% of the tip cells expressed ang-2, indicating that ang-2 production might mark newly formed tip cells or some as yet undetermined stage of tip cell formation. Immunostaining for ESM-1 and IGFBP-3 showed expression around stalk cells as well as tip cells, suggesting that secreted protein is retained in the BM and remains around the stalk cells following tip cell-stalk cell conversion (Figure 3H-I). Ang-2 and apelin are known to bind to cognate receptors, the tyrosine kinase with immunoglobulin-like and EGF-like domains 2 (Tie2) receptor tyrosine kinase and the G-protein coupled receptor APJ, respectively.23,24 Immunohistochemistry and ISH on whole mount retinas revealed that Tie2 and APJ are both expressed by stalk cells and are not detectable in tip cells (Figure 3D,F), indicating that secretion of the ligands by tip cells might act in a paracrine fashion on stalk cells. Because it is not known which receptor/cells ESM-1 binds to, alkaline phosphatase (AP)-tagged ESM-1 was incubated with whole mount retinas to determine binding sites of ESM-1 in the retinal vasculature. ESM-1–AP selectively bound to the stalk cells (Figure 3E). Taken together, these results indicate that tip cells express secreted molecules including ESM-1, ang-2, and apelin, which in turn might regulate stalk cell behavior via signaling through their cognate receptors on these cells.
Tip cells express secreted molecules that bind to stalk cells and are regulated by VEGF. (A-C) Whole mount ISH revealed tip cell expression (white arrows) of apelin (APLN), ESM1, and ANGPT2 (blue). (D) Whole mount ISH revealed expression of apj in stalk cells (red arrows). (E) ESM-1-AP fusion protein binds to stalk cells (red arrows). (F) Tie2 was detectable by antibody staining in stalk cells (red arrows) but not in tip cells. (G-L) Compared with PBS control injection (G-I), sFlt injection (J-L) leads to a decrease in APLN, ESM-1, and IGFBP3 expression. Vessels were stained with IsolectinB4 (green). The pictures represent 1 of 3 independent experiments with similar results. Scale bar 50 μm. Images of antibody stainings were acquired on a Leica TCS SP5 confocal microscope using a 63×/1.4 NA objective. Images of ISH were acquired on an Olympus BX50 microscope using a 20×/0.05 NA objective and captured using a Coolsnap camera through IPLab software version 3.2.4. Vessels were visualized with IsolectinB4 directly conjugated to AlexaFluor488 and the ISH and alkaline phosphatase signal by brightfield microscopy. Single channels were converted to grayscale in Photoshop CS2 and merged in ImageJ (ISH in blue).
Tip cells express secreted molecules that bind to stalk cells and are regulated by VEGF. (A-C) Whole mount ISH revealed tip cell expression (white arrows) of apelin (APLN), ESM1, and ANGPT2 (blue). (D) Whole mount ISH revealed expression of apj in stalk cells (red arrows). (E) ESM-1-AP fusion protein binds to stalk cells (red arrows). (F) Tie2 was detectable by antibody staining in stalk cells (red arrows) but not in tip cells. (G-L) Compared with PBS control injection (G-I), sFlt injection (J-L) leads to a decrease in APLN, ESM-1, and IGFBP3 expression. Vessels were stained with IsolectinB4 (green). The pictures represent 1 of 3 independent experiments with similar results. Scale bar 50 μm. Images of antibody stainings were acquired on a Leica TCS SP5 confocal microscope using a 63×/1.4 NA objective. Images of ISH were acquired on an Olympus BX50 microscope using a 20×/0.05 NA objective and captured using a Coolsnap camera through IPLab software version 3.2.4. Vessels were visualized with IsolectinB4 directly conjugated to AlexaFluor488 and the ISH and alkaline phosphatase signal by brightfield microscopy. Single channels were converted to grayscale in Photoshop CS2 and merged in ImageJ (ISH in blue).
Regulation of tip cell–enriched genes by VEGF
The retinal vasculature is preceded by hypoxic astrocytes that express VEGF, resulting in a VEGF gradient that attracts vessel growth toward the hypoxic tissue. VEGF binds to its receptors on the tip cells, thereby conveying signals inside the cell. We therefore tested whether the expression of the identified tip cell markers is regulated by VEGF. ISH and immunohistochemistry showed that the expression of ESM-1, IGFBP3, and apelin was completely abolished 24 hours after VEGF blocking by intraocular injection with soluble fms-like tyrosine kinase-1 (sFlt1) (Figure 3J-L) compared with PBS-injected eyes (Figure 3G-I). Moreover, ESM1, IGFBP3 and APELIN mRNA, and ESM-1 protein levels were significantly increased when we subjected human umbilical vein endothelial cells to VEGF treatment (supplemental Figure 4A-C).25 Taken together, these data illustrate that VEGF positively regulates tip cell gene expression of these secreted molecules.
Impaired retinal vessel outgrowth in apelin and APJ knockout mice
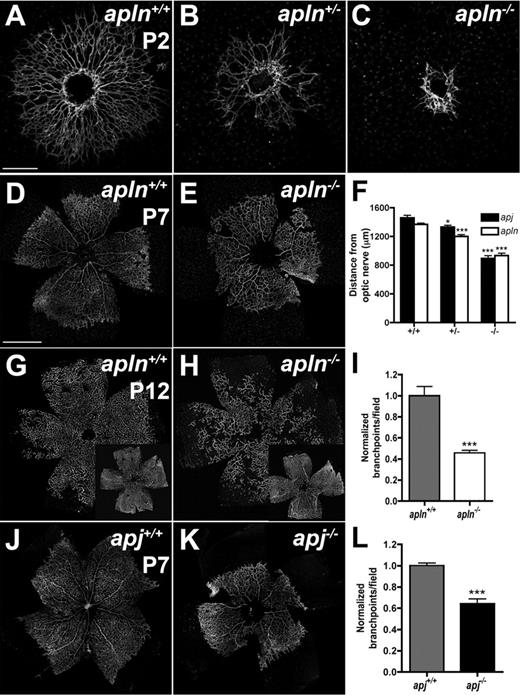
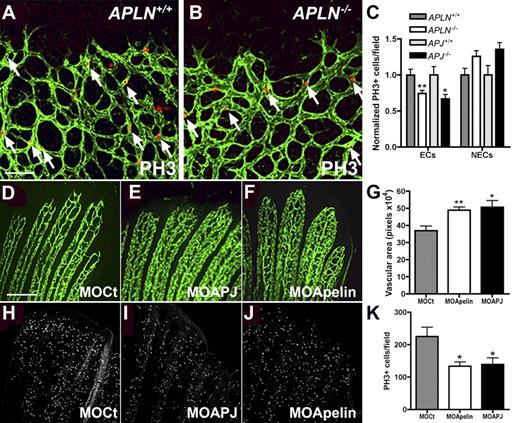
To test the hypothesis that secreted molecules produced by tip cells might act in a paracrine manner on receptor-expressing stalk cells, we analyzed mice deficient for apelin or its APJ receptor. Both APLN- and APJ-deficient mice are viable and fertile,9,10 and pups show normal weight gain during the first postnatal week. However, APLN−/− as well as APJ−/− mice show strikingly delayed retinal vessel outgrowth compared with their wild-type littermates (Figure 4). At P2, wild-type ECs have started to invade the retina through the optic nerve and form a vascular plexus that has not yet differentiated into morphologically distinct arteries and veins (Figure 4A). The APLN−/− endothelial sprouts are still located close to the optic nerve head at P2 (Figure 4C). APLN+/− mice also exhibit a delayed vessel growth compared with wild-type, but less severe than APLN−/− mice (Figure 4B). By P7, APLN−/− vessels have started to grow over the retinal surface but still lag behind their wild-type littermates (Figure 4D-E). Measuring the distance from the optic nerve covered by vessels shows a significant reduction in the outgrowth of the vasculature of APLN−/− mice compared with wild-type (Figure 4F). In addition, the vasculature in APLN−/− mice is also less complex and exhibits a reduced number of branch points (Figure 4I). At P12, APLN−/− retinal vessels have reached the retinal periphery, but sprouting into the deeper layers of the retina remains delayed (Figure 4G-H). Consistent with the phenotype of APLN−/− mice, the outgrowth and complexity of the retinal vasculature in APJ−/− mice is also strongly reduced (Figure 4F,J-L). Similar defects between APLN−/− and APJ−/− vessels support a model whereby tip cell apelin signals to stalk cell APJ receptor to regulate vessel growth and branching.
apln- and apj-deficient mice have impaired retinal vessel outgrowth. IsolectinB4 (green) staining visualizes the impaired retinal vessel outgrowth in APLN+/− and APLN−/− mice compared with wild-type mice at P2 (A-C) and P7 (D-E). (F) Quantification of vessel outgrowth in wild-type, APJ-, and apln-deficient mice at P7. (G-H) Vessel growth into deeper layers at P12 is delayed in apln−/− mice compared with wild-type mice. (J-K) Delay in vessel outgrowth in apj-deficient mice. (I,L) Quantification of branchpoints in APLN−/− and APJ−/− retinas compared with wild-type littermates, respectively. ***P < .001. *P < .05. Mann-Whitney test was used. n = 6 mice per group. Error bars indicate SEM. Scale bars indicate 300 μm (A-C); 600 μm (D,E,G,H,J,K). Images of antibody stainings were acquired on a Leica TCS SP5 confocal microscope using a 10×/0.3 NA objective.
apln- and apj-deficient mice have impaired retinal vessel outgrowth. IsolectinB4 (green) staining visualizes the impaired retinal vessel outgrowth in APLN+/− and APLN−/− mice compared with wild-type mice at P2 (A-C) and P7 (D-E). (F) Quantification of vessel outgrowth in wild-type, APJ-, and apln-deficient mice at P7. (G-H) Vessel growth into deeper layers at P12 is delayed in apln−/− mice compared with wild-type mice. (J-K) Delay in vessel outgrowth in apj-deficient mice. (I,L) Quantification of branchpoints in APLN−/− and APJ−/− retinas compared with wild-type littermates, respectively. ***P < .001. *P < .05. Mann-Whitney test was used. n = 6 mice per group. Error bars indicate SEM. Scale bars indicate 300 μm (A-C); 600 μm (D,E,G,H,J,K). Images of antibody stainings were acquired on a Leica TCS SP5 confocal microscope using a 10×/0.3 NA objective.
Apelin stimulates endothelial cell proliferation
To understand the mechanism(s) leading to reduced vessel growth and branching in APLN−/− and APJ−/− vessels, we performed whole mount VEGF ISH and hypoxiprobe staining. The reduced vessel coverage in APLN−/− retinas was accompanied by a corresponding increase in the area of hypoxic, VEGF expressing astrocytes ahead of the vascular plexus. However, levels of hypoxiprobe staining and VEGF mRNA were similar in the astrocytes of APLN−/− retinas compared with wild-type retinas (supplemental Figure 5A-B and data not shown), indicating that APLN−/− retinas show reduced vessel growth and branching in spite of intact hypoxia and VEGF gradients.
We next analyzed expression of various molecular markers, including markers for astrocytes (GFAP), pericytes (NG2), BM (Coll IV), ECs (VWF, Nrp2, VEGFR1-3, Robo4), tip cells (ESM-1, Dll4, uPAR), stalk cells (Tie2, Jag-1), and EC junctions (ZO1, claudin5, VE-cadherin). No differences between wild-type and APLN−/− vessels were observed (data not shown). To determine if the delay in vessel outgrowth in APLN−/− and APJ−/− retinas might be due to a defect in EC replication, we performed immunohistochemistry for the proliferation marker phosphohistone H3 (PH3). Quantification of positive ECs revealed that replication of APLN−/− and APJ−/− retinal ECs is significantly reduced compared with wild-type (Figure 5A-C). Because APJ expression is EC specific, we expected effects on replication to be selective for ECs in APLN−/− and APJ−/− retinas. Indeed, counting the number of replicating non-ECs in the retinal periphery showed no differences between wild-type and knockout littermates (Figure 5C). Similar results were obtained by counting KI-67 positive proliferating endothelial and non-ECs (data not shown). Taken together, these observations suggest that the retinal vessel growth defect in APLN- and APJ-deficient mice is due to reduced EC division.
Endothelial cell proliferation is reduced in the absence of Apelin or APJ. (A-B) Whole mount immunohistochemistry for PH3 (red) on APLN+/+ and APLN−/− retinas counterstained with IsolectinB4 (green). Quantification in panel C shows that there are less PH3-positive ECs in APLN−/− and APJ−/− retinas compared with wild-type littermates. The number of PH3-positive cells in the non-ECs (NECs) is not significantly different from the wild-type. N = 7 mice for APLN+/+ and APLN−/− and N = 3 for APJ+/+ and APJ−/−. (D-K) apelin and apj (apj1A and B) were knocked down in zebrafish using specific morpholinos (MO). After 3 days of remodelling, the vascular area in the bony rays was quantified (G). (H-K) Quantification of PH3 positive ECs showed less ECs undergoing mitosis in the caudal fin electroporated with MOs against APLN and APJ. **P < .01. *P < .05. N = 4 experiments, 3 fish per group. Mann-Whitney test was used. Error bars indicate SEM. Scale bars indicate 80 μm (A-B) and 100 μm (D-F, H-J). Images of retinas were acquired on a Leica TCS SP5 confocal microscope using 10×/0.3 NA objective and images of zebrafish fins using a 10×/0.25 NA objective.
Endothelial cell proliferation is reduced in the absence of Apelin or APJ. (A-B) Whole mount immunohistochemistry for PH3 (red) on APLN+/+ and APLN−/− retinas counterstained with IsolectinB4 (green). Quantification in panel C shows that there are less PH3-positive ECs in APLN−/− and APJ−/− retinas compared with wild-type littermates. The number of PH3-positive cells in the non-ECs (NECs) is not significantly different from the wild-type. N = 7 mice for APLN+/+ and APLN−/− and N = 3 for APJ+/+ and APJ−/−. (D-K) apelin and apj (apj1A and B) were knocked down in zebrafish using specific morpholinos (MO). After 3 days of remodelling, the vascular area in the bony rays was quantified (G). (H-K) Quantification of PH3 positive ECs showed less ECs undergoing mitosis in the caudal fin electroporated with MOs against APLN and APJ. **P < .01. *P < .05. N = 4 experiments, 3 fish per group. Mann-Whitney test was used. Error bars indicate SEM. Scale bars indicate 80 μm (A-B) and 100 μm (D-F, H-J). Images of retinas were acquired on a Leica TCS SP5 confocal microscope using 10×/0.3 NA objective and images of zebrafish fins using a 10×/0.25 NA objective.
To confirm these results in another in vivo model, we studied the effects of apelin and APJ morpholino (MO) knockdown during zebrafish caudal fin regeneration. Caudal fins of adult FLI1− green fluorescent protein zebrafish were electroporated with MOs directed against apelin or APJ, and the distal part of the fins was sectioned and allowed to regenerate for 3 days.25 Vessel remodeling was delayed in caudal fins electroporated with apelin and APJ MO compared with caudal fins transfected with control MO (Figure 5D-F). Caudal fins of zebrafish transfected with control MO remodeled more rapidly, and the vessels in the bony rays were more structured, leading to a smaller vascular area compared with caudal fins of fish transfected with MO directed against apelin or APJ (Figure 5D-G).26 Quantification of PH3 positive cells on the caudal fins after 3 days of regeneration showed that apelin and APJ MO treated fins have significantly reduced numbers of replicating cells compared with control-MO treated fish (Figure 5H-K), confirming the results obtained in the mouse retina and indicating that apelin and APJ are necessary for EC division.
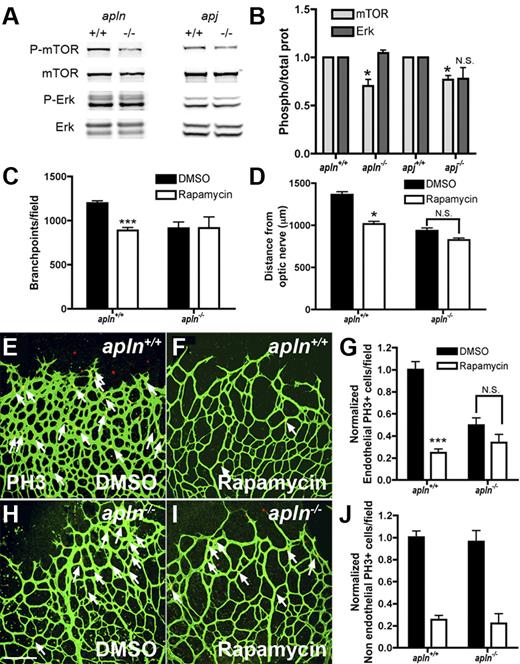
Apelin regulates retinal endothelial cell proliferation through the mTOR pathway
The APJ receptor has been shown to regulate EC proliferation in vitro via mTOR signaling.27 To investigate whether this is also the case in retinal ECs in vivo, protein extracts from whole retinas were analyzed for mTOR phosphorylation by Western blot analysis. mTOR phosphorylation was significantly reduced in APLN- and APJ-deficient retinas compared with those of wild-type mice, while ERK1/2-phosphorylation was similar between both genotypes (Figure 6A-B). To further confirm the effect of apelin/APJ signaling on the mTOR pathway, mice were treated by intraperitoneal injection with the mTOR inhibitor rapamycin,28 and the effect on retinal vascularization was analyzed 24 hours later. Treatment of wild-type mice with rapamycin leads to a significant reduction in vessel outgrowth, number of vessel branch points, and EC replication compared with DMSO-treated controls (Figure 6C-I). Rapamycin treatment induces no changes in any of these parameters in APLN−/− mice (Figure 6C-I). Thus, consistent with reduced mTOR phosphorylation seen in APLN−/− mice, mTOR inhibition has no effect on APLN−/− ECs. In contrast, quantification of the number of replicating non-ECs showed significant inhibition by rapamycin in both wild-type and APLN−/− mice, indicating that mTOR acts downstream of apelin/APJ in ECs, but not in other retinal cells, consistent with the EC-selective expression of APJ (Figure 6J).
Apelin regulates retinal endothelial cell proliferation through the mTOR pathway. (A) Protein extracts from APLN- and APJ-deficient retinas were analyzed by Western blot for mTOR and ERK-phosphorylation. Total ERK and mTOR were used as loading controls. (B) Quantification of band intensities from Western blots (4 different protein extractions). The ratio of P-mTOR/total mTOR and P-Erk/total Erk in APLN+/+ and APJ+/+ was set at 1. (C) Quantification of the branch points in the retinal vasculature following rapamycin injection in wild-type or apln-deficient mice. (D) Quantification of the distance from the optic nerve following rapamycin injection in wild-type or apln-deficient mice. (E-J) PH3 staining (red) of the retina after rapamycin injection shows a decrease in proliferating ECs in wild-type mice, whereas the APLN−/− ECs are not further affected. Vessels were stained with isolectinB4 (green). The number of PH3 stained NECs is decreased in both wild-type and APLN−/− mice. ***P < .001. *P < .05. Mann-Whitney test was used. Error bars indicate SEM. Scale bar indicates 80 μm. Images of antibody stainings were acquired on a Leica TCS SP5 confocal microscope using a 20×/0.7 NA objective.
Apelin regulates retinal endothelial cell proliferation through the mTOR pathway. (A) Protein extracts from APLN- and APJ-deficient retinas were analyzed by Western blot for mTOR and ERK-phosphorylation. Total ERK and mTOR were used as loading controls. (B) Quantification of band intensities from Western blots (4 different protein extractions). The ratio of P-mTOR/total mTOR and P-Erk/total Erk in APLN+/+ and APJ+/+ was set at 1. (C) Quantification of the branch points in the retinal vasculature following rapamycin injection in wild-type or apln-deficient mice. (D) Quantification of the distance from the optic nerve following rapamycin injection in wild-type or apln-deficient mice. (E-J) PH3 staining (red) of the retina after rapamycin injection shows a decrease in proliferating ECs in wild-type mice, whereas the APLN−/− ECs are not further affected. Vessels were stained with isolectinB4 (green). The number of PH3 stained NECs is decreased in both wild-type and APLN−/− mice. ***P < .001. *P < .05. Mann-Whitney test was used. Error bars indicate SEM. Scale bar indicates 80 μm. Images of antibody stainings were acquired on a Leica TCS SP5 confocal microscope using a 20×/0.7 NA objective.
Discussion
Based on the observation that DLL4+/− mice present excessive numbers of tip cells in their retinal vessels, we hypothesized that these mice could be used to isolate large numbers of these cells and to identify novel tip cell markers by transcriptome analysis. A recent study using laser capture microdissection to isolate retinal tip cells identified 197 genes up-regulated in tip cells.11 There was a significant correlation between the 2 gene sets, confirming that at least part of the genes up-regulated in DLL4+/− mice are novel tip cell markers. Microarray comparison of retinal ECs from wild-type and DLL4+/− mice led to the identification of 3 clusters of tip cell-enriched genes: proteases, BM components, and secreted molecules.
Proteases up-regulated in DLL4+/− retinas included uPAR, ADAM-TS family members, and cathepsin S, which are known to facilitate cell migration and angiogenesis via degradation of the ECM.15,29-31 Expression of uPAR was enriched in tip cells, suggesting that production and release of uPAR by tip cells might facilitate tip cell migration. Lack of uPAR alone did not impair postnatal retinal angiogenesis, suggesting functional redundance with other proteases, but which combination of proteases is required for retinal angiogenesis remains to be determined. uPAR-deficient mice, however, show defects in neovascularization during oxygen-induced retinopathy, indicating that uPAR requirement may be enhanced in a pathologic context.32 uPAR has also been implicated in tumor angiogenesis.15 Interestingly, the set of proteases up-regulated in angiogenic tumor vessels, including notably matrix metalloproteinases, appears to be different from the proteases found in the current screen, suggesting that protease requirement may depend on the tissue context.33
The second group of tip cell–enriched genes encoded BM components. To our knowledge, this is the first report describing specific production of mRNA encoding for BM components in tip cells in vivo. Deposition of BM components by tip cells could serve as a template for growth of follower stalk cells. Previous experiments have shown laminin deposition by tip cells growing from sprouting embryoid bodies in vitro.34 Deletion of the laminin γ1 gene in stem cells resulted in complete lack of laminin deposition and increased the lumen diameter in angiogenic sprouts. Enlarged vessels were also seen using 3D EC culture when BM deposition was inhibited34 and in vivo in laminin α4–deficient mice.35 These data suggest that in addition to stabilizing the vasculature, BM components could regulate vessel patterning and branching, although the molecular mechanisms remain unknown. Pericyte recruitment to vessels has been shown to stimulate BM formation.36 Pericyte association with tip cells might restrict BM formation to the posterior pole of the tip cell, so accounting for the observation that BM proteins are excluded from the filopodial-extending pole of the tip cell and are preferentially localized toward the posterior pole of the tip cell, which is in contact with the stalk cell as well as with the pericyte. Taken together, these results indicate that during the formation of the retinal vasculature, tip cells reorganize the ECM by degrading some components and secreting new components for the cells to migrate on.
The largest cluster of up-regulated genes in DLL4+/− mice encoded for secreted molecules. This was a surprise, as tip cells function to guide outgrowth of developing capillary sprouts, and we had therefore expected to find receptors for extracellular cues to be enriched on the tip cell. This expectation was also fuelled by the analogy between endothelial tip cells and the axonal growth cone, which guides outgrowing axons via expression of guidance receptors able to detect secreted molecules produced in the axonal environment during navigation toward its target.37,38 Tip cells are known to express receptors for axon guidance cues, such as the Netrin receptor UNC5B and Neuropilins,39 but except for UNC5B, no other guidance receptor was found in the list of up-regulated genes. Rather, we found many secreted molecules, including chemokines, adrenomedullin, ang-2, ESM-1, IGFBP-3, and apelin. Depending on expression of the cognate receptors for these secreted molecules, they could potentially regulate many aspects of vascular development such as interaction with astrocytes, microglial cells, pericytes, or stalk cells and have a multitude of consequences for vascular development.
We analyzed binding of the secreted molecule ESM-1 using AP-conjugated protein to detect as yet unknown receptors in the retina. ESM-1-AP bound to stalk cells at the vascular front, suggesting that ESM-1 may selectively act on stalk cells to regulate angiogenesis. The function of ESM-1 is unknown, but it has been shown to be highly up-regulated in tumor vessels and to enhance VEGF- and basic fibroblast growth factor-induced signaling in ECs.40 Expression analysis of the Tie2 receptor for ang-2 also showed Tie2 expression on neighboring stalk cells. ang-2 maintains an activated state of growing vessels.41 A higher concentration of ang-2 at the vascular front may therefore keep these cells in an angiogenic state before they differentiate into arteries and veins and become quiescent. Finally, expression of the APJ receptor for apelin also showed high expression in stalk cells.42 Thus, at least 3 of the secreted molecules produced by tip cells have receptors expressed on neighboring stalk cells, suggesting that tip cells may guide behavior of follower stalk cells by release of these secreted molecules.
We proved this functionally by analyzing mice deficient for apelin and its receptor APJ. Mice deficient for both genes are viable and fertile,9,10 indicating that apelin and APJ are not required for vascular development but may modulate angiogenesis. Several studies have reported actions of apelin-APJ in angiogenesis,43-46 but we here show for the first time symmetrical phenotypes in APLN- and APJ-deficient mice and zebrafish. Our data show that both APLN- and APJ-deficient mouse retinas exhibit a delay in vascular outgrowth and branching due to a delay in EC proliferation. Similar defects are observed during angiogenesis accompanying zebrafish tail regeneration following MO-mediated knockdown of APLN and APJ. Reduced EC proliferation in APLN- and APJ-deficient mice is seen despite intact hypoxia and VEGF gradients and normal expression of VEGF receptors and members of the Notch signaling pathway, indicating that apelin-APJ acts independently of these pathways to signal EC proliferation. A previous study reported induction of apelin signaling following activation of the Tie2 receptor,46 indicating that both Tie2 and APJ signaling may occur preferentially in stalk cells, where these receptors are coexpressed. However, we did not find evidence of altered vessel caliber in APLN- or APJ-deficient retinal vessels or in the zebrafish model, suggesting that the previously reported vessel caliber regulation by apelin46 may be tissue- or stage-specific. Nevertheless, in agreement with previous in vitro and in vivo studies,47 our studies reveal an important role of apelin-APJ in endothelial proliferation and support a model whereby tip cell apelin acts via stalk cell APJ to signal EC division. mTOR phosphorylation levels were reduced in APLN- and APJ-deficient mouse retinas, and mTOR inhibition by rapamycin treatment did not affect APLN deficient mice, indicating that mTOR acts downstream of apelin-APJ in endothelial stalk cells to signal cell division. Proliferation of ECs was not completely blocked in the absence of APLN or APJ but almost completely abolished after mTOR treatment, indicating that additional factor(s) signal through mTOR to regulate EC proliferation.
Apelin-APJ are highly expressed in the tumor vasculature,44,48 suggesting that blocking of this ligand-receptor interaction might slow tumor growth. As apelin expression is induced by hypoxia and VEGF, anti-angiogenic therapies blocking VEGF signaling might be expected to decrease expression of apelin and other tip cell markers described here, suggesting that these markers might be useful to predict response to anti-angiogenic treatment of tumor patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Kristell Wanherdrick for help with the microarray analysis, and Jérémie Teillon and Liz Jones for help with imaging.
This work was supported by grants from Inserm, Agence Nationale de la Recherche (ANR Blanc), Institut National du Cancer. This work was also supported by Fondation Bettencourt, Fondation Leducq and Fondation pour la Recherche Médicale (FRM). R.d.T. was supported by grants from Fundación Marcelino Botin and FRM. C.P. was supported by Association sur la Recherche contre le Cancer and Deutsche Forschungsgemeinschaft.
Authorship
Contribution: R.d.T. and C.P. designed and performed the research, analyzed data, and wrote the paper; T.M., G.S., B.L., J.S.K., and C.B. performed research; A.D., N.B., A.F., and J.P. contributed mice; and A.E. designed the research, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no completing financial interests.
Correspondence: Anne Eichmann, Inserm U833, Collège de France, 11 Place Marcelin Berthelot, 75005 Paris, France; e-mail: anne.eichmann@college-de-france.fr.
References
Author notes
R.d.T. and C.P. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal