Abstract
The majority of acute promyelocytic leukemia (APL) cases are characterized by the presence of a promyelocytic leukemia–retinoic acid receptor alpha(RARA) fusion gene. In a small subset, RARA is fused to a different partner, usually involved in regulating cell growth and differentiation. Here, we identified a novel RARA fusion transcript, BCOR-RARA, in a t(X;17)(p11;q12) variant of APL with unique morphologic features, including rectangular and round cytoplasmic inclusion bodies. Although the patient was clinically responsive to all-trans retinoic acid, several relapses occurred with standard chemotherapy and all-trans retinoic acid. BCOR is a transcriptional corepressor through the proto-oncoprotein, BCL6, recruiting histone deacetylases and polycomb repressive complex 1 components. BCOR-RARA was found to possess common features with other RARA fusion proteins. These included: (1) the same break point in RARA cDNA; (2) self-association; (3) retinoid X receptor alpha is necessary for BCOR-RARA to associate with the RARA responsive element; (4) action in a dominant-negative manner on RARA transcriptional activation; and (5) aberrant subcellular relocalization. It should be noted that there was no intact BCOR found in the 45,-Y,t(X;17)(p11;q12) APL cells because they featured only a rearranged X chromosome. These results highlight essential features of pathogenesis in APL in more detail. BCOR appears to be involved not only in human congenital diseases, but also in a human cancer.
Introduction
Acute promyelocytic leukemia (APL) is a distinct disease entity within the acute myelogenous leukemia (AML).1-3 In the clinic, APL has characteristic morphologic features, including hypergranular promyelocytes, and exhibits a severe bleeding tendency, which is efficiently controlled with all-trans retinoic acid (ATRA) treatment.4 The majority of APLs feature a balanced reciprocal translocation between chromosomes 15q22 and 17q12, which results in the fusion of the promyelocytic leukemia (PML) and retinoic acid receptor alpha (RARA) genes.5 In rare cases, other fusion partners of RARA are found, such as promyelocytic leukemia zinc finger (PLZF),6 nucleophosmin (NPM1),7 nuclear mitotic apparatus protein (NuMA),8 signal transducer and activator of transcription 5b,9 protein kinase A regulatory subunit type 1A,10 and Fip1-like 1.11 All of the RARA fusion proteins comprise all but the first 30 amino acids of RARA fused to a variable partner at its aminoterminus.1-3 It is characteristic that all fusion partners have self-association domains. In the case of PML-RARA and PLZF-RARA, aberrant recruitment of transcriptional repressors, including nuclear corepressor protein/silencing mediator of retinoid and thyroid hormone receptor (NCOR/SMRT), histone deacetylases (HDACs),12,13 and polycomb complexes,14,15 to the retinoic acid responsive element (RARE) leads to ectopic repression of RAR target genes.1,2 In mouse models of RARA fusion proteins, APL-like diseases occur after a long latency, presumably because of the requirement for other cooperative genetic events for overt APL.16 Hence, other mechanisms for leukemogenesis of APL have been advocated, such as the double dominant-negative activity of PML-RARA, which also interferes with a remaining tumor suppressor, PML.1,2
The BCL6 corepressor, BCOR,17 is a ubiquitously expressed nuclear protein, which directly interfaces to proto-oncoprotein BCL6 through the BCOR-BCL6–binding domain (BCORBBD).18 BCOR also associates with HDACs, the polycomb group protein, Polycomb group RING finger protein 1/nervous system polycomb 1 (PCGF1/NSPC1), and the histone demethylase, F-box– and leucine-rich repeat protein 10 (FBXL10), through its C-terminal region,19,20 which implies that it could suppress gene transcription by epigenetic mechanisms.21,22 Recently, it was reported that BCOR is critical for early embryonic development, mesenchymal stem cell function, primitive erythroid differentiation, and lymphoid cell differentiation.22,23 Moreover, BCOR gene mutations have been linked with human inherited diseases—the oculofaciocardiodental and lenz microphthalmia syndromes.24
Here, we report that BCOR is a novel fusion partner of RARA in a variant APL patient with 45,-Y,t(X;17)(p11;q12). The patient demonstrated overt coagulopathy and sensitivity to ATRA, but resistance to arsenic trioxide. The APL cells showed a unique morphology with round inclusions and rectangular cytoplasmic bodies. Dissection of the pathogenesis of such a variant of APL with BCOR-RARA might reveal significant aspects of molecular mechanisms underlying human leukemias.
Methods
Case report
The patient was a 45-year-old man who was admitted because of fever, dyspnea, and bloody phlegm. A full blood count showed a hemoglobin level of 12.1 g/dL, a white blood cell count of 25.3 × 109/L, including 59% promyelocytes, and a platelet count of 116 × 109/L. A bone marrow sample was markedly hypercellular, containing 83% promyelocytes. Reverse-transcription polymerase chain reaction (RT-PCR) analysis failed to detect PML-RARA fusion transcripts from the bone marrow cells. However, coagulopathy was present, with an increased prothrombin time of 1.58 (international normalized ratio), an activated partial thromboplastin time of 33.7 seconds (normal, 26 ∼ 39), decreased fibrinogen of 52 mg/dL (200 ∼ 400), and mildly increased fibrin/fibrinogen degradation products of 50.6 mg/L (0 ∼ 5). Diagnosis of a variant of APL was made according to the morphology, fluorescence in situ hybridization (FISH), and flow cytometric analysis [human leukocyte antigen DR (HLA-DR−)/CD13+/CD33+/CD56+] (Figure 1, supplemental Figure 1 and supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Remission induction therapy was commenced following a Japan Adult Leukemia Study Group APL204 protocol with ATRA (45 mg/m2/d), idarubicin (12 mg/m2/d from days 1 to 3), and cytosine arabinoside (Ara-C; 100 mg/m2/d from days 1 to 7). A complete remission (CR) was documented on day 27 with reference to morphology, cytogenetics, and FISH. A set of consolidation therapies was administered as follows: first course, mitoxantrone 7 mg/m2 × 3 days and Ara-C 200 mg/m2 × 5 days; second course, daunorubicin 50 mg/m2 × 3 days and Ara-C 200 mg/m2 × 5 days; and third course, idarubicin 12 mg/m2 × 3 days and Ara-C 140 mg/m2 × 5 days. Thirty-five months after diagnosis, the patient relapsed with gradually reducing blood counts and overt coagulopathy. A bone marrow sample showed that 90% of the cells were leukemic blasts with the recurring chromosomal translocation, FISH abnormality of RARA, and aberrant expression of CD56 and CD13. A second remission induction therapy, which was the same regimen at the diagnosis, was administered. A second CR was documented on day 32. Two cycles of consolidation therapy were administered. At 41 months after diagnosis, the patient had a second relapse. Arsenic trioxide (0.15 mg/kg) was administered for 16 days, but no response occurred. Then, tamibarotene (6 mg/kg) was given,25 which mildly reduced the leukemia burden. After single chemotherapy with daunorubicin and Ara-C, a cord-blood transplantation was performed with a myeloablative conditioning regimen. After engraftment was achieved, a bone marrow sample showed a third CR.
Cell lines, reagents, and antibodies.
A human AML cell line, EOL1, was maintained in RPMI 1640 (Sigma-Aldrich) supplemented with 10% fetal bovine serum (Equitech-Bio) at 37°C in a humidified atmosphere of 5% CO2. 293T and HepG2 were maintained in Dulbecco modified Eagle medium with 10% fetal bovine serum. Antibodies used were as follows: FLAG (M2; Sigma-Aldrich), Myc (PL14; MBL), BCL6 (N-3; Santa Cruz Biotechnology), and alpha-tubulin (B-7; Santa Cruz Biotechnology).
Cytogenetic studies
Karyotype analysis was performed with regular G-banding by trypsin and Giemsa staining banding (SRL). The Vysis LSI PML/RARA dual-color, dual-fusion translocation probe was used for FISH analysis (Vysis). PML signals were red, and RARA signals were green. The RARA probe covered up to 700-kb pairs, sufficient to surround the RARA gene on 17q12.
Cloning of BCOR-RARA fusion transcripts
Rapid 5′ amplification of cDNA ends (5′-RACE) was performed with a SMARTer RACE cDNA Amplification kit (Clontech), according to the manufacturer's instructions. Clinical samples were collected under institutional review board–approved protocols and with written informed consent in accordance with the Declaration of Helsinki. Briefly, total RNA from bone marrow nuclear cells before treatment was isolated with an RNeasy plus mini kit (QIAGEN). One microgram of the total RNA was subjected to reverse transcription with the 5′-coding sequence primer A. A first polymerase chain reaction (PCR) from the cDNA was performed with the universal primer A mix and RAe3R (5′-GCAAGGCTTGTAGATGCGGGGTAGA-3′), followed by nested PCR with the nested universal primer A and R8R (5′-CAGAACTGCTGCTCTGGGTCTCAAT-3′).26 The PCR products were purified and directly sequenced (ABI 3100; Applied Biosystems).
RT-PCR
The patient's cDNA and EOL1 cDNA as a control were subjected to PCR with the following primers: RARA: first PCR R18F (5′-TGGACAGCAGCTCCAGGACA-3′)/R5R (5′-CCCATAGTGGTAGCCTGAGGACT-3′), followed by nested PCR R19F (5′-CTGTCTAGATGCCAGACTGTCTGCCTC-3′)26 /RAe3R; BCOR: BR12F1 (5′-CTCTGCACGATGCTGTTGAGAACGA-3′)/BR13R1 (5′-GAGCTGCCATAGAAGTCCCAAGTGC-3′); BCOR-RARA: BR12F1/R5R; and RARA-BCOR: first PCR R18F/BR13R1 was followed by nested PCR R19F/ BR13R2 (5′-CATCATTGCGACCCTGGAGGTCATT-3′). All PCR reactions were accomplished under the following conditions. Denaturing, annealing, and extension steps were performed at 95°C for 30 seconds, 58°C for 30 seconds, and 72°C for 1 minute, respectively, for 30 cycles on a PCR thermal cycler (Dice; TaKaRa). For long-distance RT-PCR, we used TaKaLa LA-Taq and the following primers: 5′-BCOR-RARA: BR2F1 (5′-AGACGCTTGAAGCAAAGCTGCCATC-3′)/RAe3R; and 3′-BCOR-RARA: BR12F1/RAUTR (5′-AGTTCTGAGGATGGGGGAGCCAAGT-3′). Touchdown PCR was achieved as denaturing and extension steps were performed at 94°C for 30 seconds and 72°C for 6 minutes, respectively. Annealing steps were 70°C for 30 seconds from the 6th to 10th cycles and 68°C for 30 seconds from the 11th to 30th cycles.
Plasmid construction
Full-length cDNAs of RARA, RXRA, BCOR, BCL6, PML-RARA, and BCOR-RARA were amplified from total RNA of bone marrow samples with Phusion DNA polymerase (Finnzymes). After sequence validation, each cDNA was cloned into the pcDNA3 expression vector (Invitrogen). Either a Myc (EQKLISEEDL) or a FLAG (DYKDDDDK) tag was fused to the aminotermini of BCOR and BCOR-RARA, or the carboxytermini of RARA, RXRA, BCL6, and PML-RARA by PCR. (See Figure 3A for composition of the deletion mutants.)
Coimmunoprecipitation and immunoblotting analysis
For coimmunoprecipitation, expression plasmids of Myc- or FLAG-tagged proteins were transiently transfected with Lipofectamine 2000 (Invitrogen) into 293T. After 48 hours of incubation, cells were harvested, washed once in phosphate-buffered saline and lysed in 150mM NaCl, 50mM Tris [tris(hydroxymethyl)aminomethane]-HCl (pH 7.4), 1mM EDTA (ethylenedimainetetraacetic acid), 1% Triton X-100, and PhosSTOP (Roche) on ice for 20 minutes. After centrifugation, the supernatants were collected. Then, 40 μL of anti-FLAG M2-agarose affinity gel (Sigma-Aldrich) was added to the lysates and incubated at 4°C for 2 hours. The affinity gels were washed 3 times with Tris-buffered saline, and the immunoprecipitated proteins were eluted by incubation with 50 μL of 3× FLAG peptide solution (0.5 mg/mL; Sigma-Aldrich) on ice for 30 minutes. For immunoblotting analysis, extracted proteins were separated in 7.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels and transferred to Immobilon-P (Millipore) at 8 V/cm for 4 hours in Towbin buffer (25mM Tris and 192mM glycine; pH 8.3). The membranes were sequentially incubated with 5% skimmed milk, primary antibodies at a 1:2000 dilution, and secondary antibodies conjugated with horseradish peroxidase at a 1:5000 dilution. Then, the membranes were incubated for chemiluminescence (Luminol Reagent; Santa Cruz Biotechnology), and proteins were detected by X-ray film exposure. The films were visualized with a PCX-101 (Konica Minolta).
Electrophoretic mobility shift assays (EMSAs)
In vitro translated proteins were produced by T7 priming with the TNT Quick Coupled Transcription/Translation System (Promega). Protein expression was confirmed by 35S-methionine incorporation according to the manufacturer's instructions. Oligonucleotides of the RARE-containing DR5G sequence27 were annealed to form a double-stranded probe, radiolabeled with 32P by Klenow fragment (New England Biolabs), and purified through a G-25 Sephadex column (Roche). An equal amount of in vitro translated proteins was incubated on ice for 15 minutes with the radiolabeled probes (20 000 cpm) and 1 μg of poly-dI-dC (Sigma-Aldrich) in binding buffer containing 10mM Tris-HCl (pH 7.4), 10mM MgCl2, and 1mM 1,4-dithioerythritol or with the addition of a 100-fold excess of unradiolabeled probes and anti-RARA antibody to the mixture. Then, the mixture was loaded onto a 6% polyacrylamide gel and run in 0.5% Tris-borate–EDTA buffer. The gels were air dried with a Promega gel drying kit, followed by X-ray film exposures at −80°C overnight before visualization.
Transcription reporter assay
The pcDNA3 expression vectors, 4 times repeats of RARE (DR5G) containing pGL4.26 reporter vector (Promega) and pRL-TK for transfection efficiency control, were transiently transfected into HepG2 cells with Fugene HD (Roche). In addition, ATRA was added at the same time. After 48 hours of incubation, transfected cells were lysed and incubated with Dual-Luciferase Reporter reagent (Promega) according to the manufacturer's instructions. Luciferase activity was analyzed with ARVO X (PerkinElmer) and normalized to the Renilla activity. The results were the means of triplicate wells and representative of 3 independent experiments. The input proteins were confirmed by immunoblotting analysis with anti-FLAG antibody.
Immunofluorescence analysis and microscopy
293T cells were transiently transfected with expression plasmids. The cells were fixed for 30 minutes in methanol at −20°C. Primary antibodies at a 1:50 dilution were incubated for 1 hour at room temperature. Secondary antibodies at a 1:200 dilution, including donkey anti-mouse immunoglobulin –Alexa Fluor 488 and donkey anti-rabbit immunoglobulin–Alexa Fluor 594 (Molecular Probes), were incubated for 1 hour at room temperature. The slides were mounted in Vectashield mounting medium with DAPI (4,6 diamidino-2-phenylindole) (Vector Laboratories). Confocal laser images were captured with an Olympus BX51 microscope. The original magnification was ×400 for all panels.
Results
Unique morphologic and cytogenetic features of t(X;17)(p11;q12) APL blasts, distinct from typical AML M3 or M3 variants
Bone marrow promyelocytes were strongly positive for Sudan black B, myeloperoxidase staining (Figure 1), and naphthol AS-D chloroacetate staining and negative for alpha-naphthyl butyrate staining (data not shown). They were more granular than those of AML M2, but less granular than the classical t(15;17) APL.28 Round inclusions and rectangular cytoplasmic bodies were remarkable morphologic features, instead of typical Auer bodies and faggot cells (Figure 1). Round inclusions have been reported in variants of APL,28 but such rectangular cytoplasmic bodies have not been previously described. They were found positive for periodic acid–Schiff staining (supplemental Figure 1A), suggesting that the cytoplasmic bodies were a kind of polysaccharide. Some leukemia cells showed that the cytoplasm was filled with deposits, and inclusion bodies were forming (supplemental Figure 1B). Moreover, crystallized cytoplasmic bodies were found (supplemental Figure 1C). In a rare leukemia cell population, Auer-like bodies were found (Figure 1B). These results indicated that aberrant deposits could crystallize in the cytoplasm, which were distinct from those found in the classical t(15;17) APL. In this case, the nuclei were regular in shape without the bilobed grooving, which resemble the nuclei of t(11;17)-associated APL.28 Flow cytometric analysis revealed strong expression of CD13, CD33, and CD56, weak expression of CD11c, and lack of HLA-DR and CD7 (supplemental Table 1). This expression pattern is reminiscent of APL harboring PLZF-RARA.28 FLT3–internal tandem duplication was not detected (data not shown). Karyotype analysis detected a novel chromosomal translocation described as 45,-Y, t(X;17)(p11.2;q12)[19]/ 46,XY[1] (Figure 1E). FISH analysis indicated 1 intact and 2 split signals of RARA and 2 intact signals of PML (Figure 1F). These results confirmed t(X;17)(p11;q12) APL blasts to be distinct from typical AML M3 or M3 variants.
Morphology and cytogenetic analysis of a bone marrow sample of t(X;17)(p11;q12) APL. Leukemic promyelocytes before treatment. (A-B) May-Giemsa staining. (C) Sudan black B staining. (D) Myeloperoxidase staining. Original magnification ×400 for all panels. The black, blue, and red arrows show rectangular cytoplasmic bodies, round inclusions, and Auer-like bodies, respectively. (E) Karyotype analysis. 45,−Y,t(X;17)(p11;q12) was detected in 95% of the cells. (F) FISH analysis. One of the RARA signals is split as (s). Other intact signals of RARA and PML are shown as (r) and (p), respectively.
Morphology and cytogenetic analysis of a bone marrow sample of t(X;17)(p11;q12) APL. Leukemic promyelocytes before treatment. (A-B) May-Giemsa staining. (C) Sudan black B staining. (D) Myeloperoxidase staining. Original magnification ×400 for all panels. The black, blue, and red arrows show rectangular cytoplasmic bodies, round inclusions, and Auer-like bodies, respectively. (E) Karyotype analysis. 45,−Y,t(X;17)(p11;q12) was detected in 95% of the cells. (F) FISH analysis. One of the RARA signals is split as (s). Other intact signals of RARA and PML are shown as (r) and (p), respectively.
Molecular cloning of BCOR-RARA fusion transcripts in t(X;17)(p11;q12)
The FISH analysis suggested that one RARA gene was rearranged because the RARA probe covered the entire RARA gene spanning from the 5′- to the 3′-untranslated region. To amplify unknown chimeric fusion transcripts, we performed 5′-RACE, and PCR products derived from the patient bone marrow cells were directly sequenced, revealing BCOR cDNA from exons 9 to 12 to be fused to RARA exon 3 (Figure 2A). By RT-PCR, we confirmed full-length chimeric fusion transcripts spanning from the start codon to 4948 nt of BCOR cDNA (isoform b; reference NM_001123384) fused to RARA cDNA from exon 3 to the stop codon (Figure 2D). The chimeric cDNA had an in-frame codon from BCOR through RARA, creating a 1931 amino acid (aa) fusion protein described as BCOR-RARA (Figure 2B). A reciprocal chimeric cDNA of RARA-BCOR was not detected (Figure 2C). The BCOR-RARA had a BCORBBD (498 ∼ 514 aa), 3 ankyrin repeats (1410 ∼ 1509 aa) of BCOR, a DNA-binding domain (DBD) derived from RARA (1557 ∼ 1622 aa), and a ligand-binding domain of RARA (1669 ∼ 1888 aa; Figure 2B).3,17,18
Molecular analysis of the BCOR-RARA fusion transcript. (A) Sequence analysis of the BCOR-RARA transcript at the junction site. The junction of the BCOR and RARA transcripts is indicated by a bold arrowhead. Rearrangement between BCOR exon 12 and RARA exon 3 resulting in an in-frame BCOR-RARA fusion protein. The DNA and amino acid sequences spanning the junction of the BCOR and RARA genes (in bold and normal characters, respectively) are shown below. (B) Schematic diagram of RARA, BCOR, and the BCOR-RARA fusion protein. The break point is indicated by the black line. Primer locations for RT-PCR are shown as arrows. Domains of BCOR-RARA are indicated as follows: BCOR BCL6-binding domain (BBD), ankyrin repeats (AR), DNA-binding domain (DBD), and ligand-binding domain (LBD). (C) RT-PCR analysis of BCOR-RARA fusion transcripts. BCOR-RARA transcripts were detected in cDNA derived from the patient's bone marrow cells on diagnosis, which also contained normal cells with intact BCOR. The control was cDNA derived from EOL1. Primer pairs are shown in the lower lane. A vertical line was inserted to indicate a repositioned gel lane. (D) Long-distance RT-PCR of BCOR-RARA transcripts. No alternative splicing variant was detected.
Molecular analysis of the BCOR-RARA fusion transcript. (A) Sequence analysis of the BCOR-RARA transcript at the junction site. The junction of the BCOR and RARA transcripts is indicated by a bold arrowhead. Rearrangement between BCOR exon 12 and RARA exon 3 resulting in an in-frame BCOR-RARA fusion protein. The DNA and amino acid sequences spanning the junction of the BCOR and RARA genes (in bold and normal characters, respectively) are shown below. (B) Schematic diagram of RARA, BCOR, and the BCOR-RARA fusion protein. The break point is indicated by the black line. Primer locations for RT-PCR are shown as arrows. Domains of BCOR-RARA are indicated as follows: BCOR BCL6-binding domain (BBD), ankyrin repeats (AR), DNA-binding domain (DBD), and ligand-binding domain (LBD). (C) RT-PCR analysis of BCOR-RARA fusion transcripts. BCOR-RARA transcripts were detected in cDNA derived from the patient's bone marrow cells on diagnosis, which also contained normal cells with intact BCOR. The control was cDNA derived from EOL1. Primer pairs are shown in the lower lane. A vertical line was inserted to indicate a repositioned gel lane. (D) Long-distance RT-PCR of BCOR-RARA transcripts. No alternative splicing variant was detected.
Identification of self-association of BCOR-RARA
One of the consistent features in all known RARA fusion partners is self-association.2,3 So far, there has been no report that BCOR is able to self-associate. To determine whether BCOR and/or BCOR-RARA self-associate, we performed coimmunoprecipitation assays. Consistent with other RARA fusion partners,2,3 Myc-tagged (Mt)-BCOR coimmunoprecipitated with FLAG-tagged (Ft)-BCOR (Figure 3B). Subsequently, we confirmed that Mt-BCOR-RARA also coimmunoprecipitated with Ft-BCOR-RARA. To further investigate which domains of BCOR contribute to the self-association property, we created a set of deletion mutants of BCOR (Figure 3A). Huynh et al previously described that there are 2 transcripts as full-length BCOR and a variant form (BCOR-S).17 As shown in Figure 3B, BCOR-S and NBC-RARA retained the self-association. Surprisingly, CBC-RARA also retained the self-association property. Furthermore, AR-RARA containing AR still possessed the self-association, but with dNRARA, it was lost. These results showed that BCOR-RARA is able to self-associate both through the BCOR-S region and the ankyrin-repeat domain of BCOR. Moreover, BCL6 interacts with BCOR through BCORBBD (Figure 2B).18 We also confirmed that BCOR-RARA associates with BCL6 (Figure 3C).
Identification of BCOR-RARA self-association. (A) Schematic representation of deletion mutants. The filled box is BCOR, and the open box is RARA. AR, ankyrin repeat domain; aa, amino acid. (B) Coimmunoprecipitation in 293T cells between Myc- and FLAG-tagged proteins. (C) Coimmunoprecipitation in 293T cells between Mt-BCL6 and Ft-BCOR-RARA. IP, immunoprecipitation; IB, immunoblotting. *Nonspecific band.
Identification of BCOR-RARA self-association. (A) Schematic representation of deletion mutants. The filled box is BCOR, and the open box is RARA. AR, ankyrin repeat domain; aa, amino acid. (B) Coimmunoprecipitation in 293T cells between Myc- and FLAG-tagged proteins. (C) Coimmunoprecipitation in 293T cells between Mt-BCL6 and Ft-BCOR-RARA. IP, immunoprecipitation; IB, immunoblotting. *Nonspecific band.
The BCOR-RARA/RXRA complex, but not BCOR-RARA by itself, associates with the RARE
RXR recruitment is a critical determinant of transforming the potential of oligomeric RARA fusion proteins.29,30 In addition, RARA fusion proteins gain alternative abilities to associate with RARE.31 To investigate how BCOR-RARA associates with RARE in vitro, we performed EMSA. In the absence of RXRA, PML-RARA associated with DR5G RARE by itself, but BCOR-RARA, CBC-RARA, BCOR, and BCOR-S did not (Figure 4B left). In addition, it was considered that AR-RARA, dNRARA, and RARA faintly associate with RARE. In the presence of RXRA, however, RARA, BCOR-RARA, and the deletion mutants could synergistically associate with RARE (Figure 4B middle). To further confirm that BCOR-RARA deletion mutants lose association with RARE in the absence of RXRA, 4-fold increased input proteins were subjected to EMSA. As shown in Figure 4C, RARA, AR-RARA, and dNRARA clearly interacted with RARE, as verified by addition of anti-RARA antibody inducing supershift. In contrast, CBC-RARA completely lost the interaction with RARE in the absence of RXRA. These data supported the conclusion that RXRA is necessary for BCOR-RARA to associate with RARE, and showed that the BCOR-RARA/RXRA complex associates with at least DR5G RARE in an alternative manner, compared with RARA and PML-RARA.
BCOR-RARA associates with the RARA responsive element with RXRA. (A) 35S-Methionine–labeled input proteins. (B) EMSA analysis with in vitro translated proteins and radiolabeled DR5G RARE probes. Without RXRA, PML-RARA associated with RARE, but BCOR-RARA did not. BCOR and BCOR-S did not associate with RARE, regardless of the presence of RXRA. 100× competitor: 100-fold excess of nonradiolabeled-RARE was additionally incubated with the reaction. (C) CBC-RARA did not associate with RARE without RXRA. In addition, 4-fold increased input proteins in less in vitro translation kit solution were subjected to EMSA to enhance specific bands and reduce nonspecific background. An arrow indicates an input protein/RARE complex included in the upper lane. Control Ab, normal rabbit polyclonal IgG; N.S., nonspecific band.
BCOR-RARA associates with the RARA responsive element with RXRA. (A) 35S-Methionine–labeled input proteins. (B) EMSA analysis with in vitro translated proteins and radiolabeled DR5G RARE probes. Without RXRA, PML-RARA associated with RARE, but BCOR-RARA did not. BCOR and BCOR-S did not associate with RARE, regardless of the presence of RXRA. 100× competitor: 100-fold excess of nonradiolabeled-RARE was additionally incubated with the reaction. (C) CBC-RARA did not associate with RARE without RXRA. In addition, 4-fold increased input proteins in less in vitro translation kit solution were subjected to EMSA to enhance specific bands and reduce nonspecific background. An arrow indicates an input protein/RARE complex included in the upper lane. Control Ab, normal rabbit polyclonal IgG; N.S., nonspecific band.
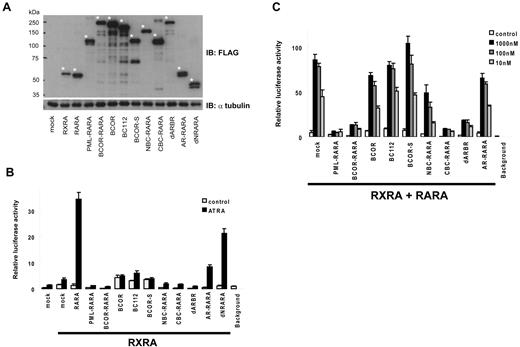
BCOR-RARA inhibits ATRA-induced RARA transcriptional activation in a dominant-negative manner
Deregulation of RARA transcriptional activations has a central role in the pathogenesis of APL. BCOR-RARA and other RARA fusion proteins invariably retain the ability to associate with RARE in the presence of RXRA, which potentially could interfere with RAR/RXR pathways. Therefore, we evaluated ATRA-induced transcriptional activation of 4 × RAREs with a reporter assay in HepG2 cells. At first, we assessed the basal transcriptional activation of a sample with RXRA (Figure 5B). Without ATRA, BCOR-RARA repressed the reporter activity, compared with mock control. With addition of 1μM of ATRA, BCOR-RARA induced transcriptional activation very weakly. However, addition of ATRA did not robustly induce transcriptional activation of PML-RARA, BCOR-RARA, NBC-RARA, CBC-RARA, and dARBR, compared with RARA. In contrast, AR-RARA showed partial transcriptional activation between dNRARA and dARBR. In addition, we confirmed that BCOR and other deletion mutants did not influence Renilla transcriptional activity because BCOR is a transcriptional repressor (supplemental Figure 2). Subsequently, we evaluated the dominant-negative effects of samples in the RARA/RXRA pathway. RARA and RXRA were cotransfected with mock, PML-RARA, BCOR-RARA, or deletion mutants. In contrast to BCOR and BC112, BCOR-RARA obviously inhibited ATRA-induced RARA transcriptional activation as well as PML-RARA (Figure 5C). Furthermore, we asked which domains are sufficient for dominant-negative effects with the deletion mutants. Although AR-RARA had reduced ATRA-induced transcriptional activation by itself, it did not significantly interfere with ATRA-induced RARA transcriptional activation. Interestingly, CBC-RARA suppressed RARA transcriptional activation equivalent to BCOR-RARA. These results indicated that the region spanning from 999 to 1409 aa of BCOR-RARA has pivotal roles in the dominant-negative effects.
BCOR-RARA inhibits RARA transcriptional activation by ATRA in a dominant-negative manner. (A) Input proteins in HepG2 cells. An asterisk indicates a protein included in the lower lane. (B) BCOR-RARA itself induced weak transcriptional activation by ATRA. Luciferase assays were performed on cell extracts from HepG2 transiently transfected with the 4× RAREs containing a luciferase reporter, RXRA and 1μM of ATRA or vehicle control. (C) Dominant-negative effects of BCOR-RARA on RARA transcriptional activation by ATRA. Plasmids of 4× RAREs containing luciferase reporter, RARA, RXRA, and the fusion proteins were cotransfected into HepG2 cells with various concentrations of ATRA.
BCOR-RARA inhibits RARA transcriptional activation by ATRA in a dominant-negative manner. (A) Input proteins in HepG2 cells. An asterisk indicates a protein included in the lower lane. (B) BCOR-RARA itself induced weak transcriptional activation by ATRA. Luciferase assays were performed on cell extracts from HepG2 transiently transfected with the 4× RAREs containing a luciferase reporter, RXRA and 1μM of ATRA or vehicle control. (C) Dominant-negative effects of BCOR-RARA on RARA transcriptional activation by ATRA. Plasmids of 4× RAREs containing luciferase reporter, RARA, RXRA, and the fusion proteins were cotransfected into HepG2 cells with various concentrations of ATRA.
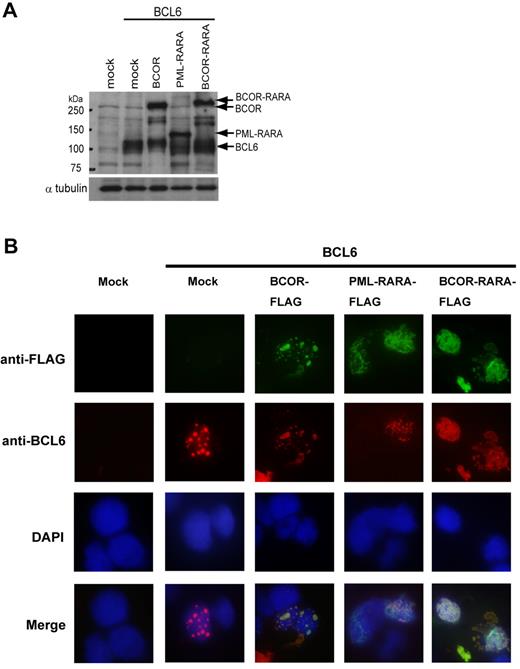
Subcellular localization of BCOR-RARA/BCL6 is distinct from BCOR/BCL6
Correct protein function is highly dependent on intracellular localization. For instance, aberrant subcellular localization of PML-RARA disrupts PML localization within nuclear bodies and delocalizes PML in a microspeckled nuclear pattern mediated through self-association between PML-RARA and PML. In addition, PML-RARA interacts with RXRA, potentially deregulating retinoid signaling by sequestering RXRA from normal compartments.2,3 To investigate the subcellular localization of BCOR-RARA, we performed immunofluorescence analysis in 293T cells. As previously reported, PML-RARA was localized in the nucleus and cytoplasm (data not shown).32 In BCOR-RARA–expressing cells, BCOR-RARA localized as 2 patterns: (1) diffusely in the nucleus as well as PML-RARA in 82% of the cells and (2) diffusely in the nucleus and aggregately in the cytoplasm in 18% of the cells (Figure 6B). The subcellular localization of BCOR-RARA was clearly distinguishable from the punctuate pattern shown in the nucleus of BCOR-expressing cells.17 Although functions of BCL6 in myeloid cells are largely unknown, the expression of BCL6 is robustly induced in granulocytic differentiation.33 To further investigate aberrant localizations of BCOR-related proteins, we performed coimmunofluorescence analysis between BCOR-RARA and BCL6 in 293T cells. As shown in Figure 6B, BCL6 colocalized with BCOR in the nucleus as the punctuate pattern, but did not colocalize with PML-RARA. Interestingly, BCL6 clearly colocalized with BCOR-RARA and relocalized in aberrant subcellular compartments. These results indicated that the subcellular localization of BCOR-RARA/BCL6 is distinct from that of BCOR/BCL6.
Subcellular localizations of BCOR-RARA and BCL6. (A) Input proteins. Immunoblotting was performed with anti-FLAG and BCL6 antibodies. (B) Coimmunofluorescence analysis was performed in 293T cells transfected with BCL6 and one of mock, Ft-BCOR, Ft-PML-RARA, or Ft-BCOR-RARA plasmids using DAPI for nuclear staining. Multiple confocal microscopy images of the given fields are merged in the lower lane. The patterns of BCOR-RARA localization were analyzed for 100 cells counted.
Subcellular localizations of BCOR-RARA and BCL6. (A) Input proteins. Immunoblotting was performed with anti-FLAG and BCL6 antibodies. (B) Coimmunofluorescence analysis was performed in 293T cells transfected with BCL6 and one of mock, Ft-BCOR, Ft-PML-RARA, or Ft-BCOR-RARA plasmids using DAPI for nuclear staining. Multiple confocal microscopy images of the given fields are merged in the lower lane. The patterns of BCOR-RARA localization were analyzed for 100 cells counted.
Discussion
To our knowledge, this is a first report of a novel variant of APL with t(X;17)(p11;q12). The classical APL with PML-RARA typically has hypergranular leukemic promyelocytes with Auer bodies and/or faggot cells and bilobed grooved nuclei.28 In the present case, round inclusions and rectangular cytoplasmic bodies were remarkable morphologic features, instead of Auer bodies. Such round inclusions have previously been reported in cases of NPM1-RARA and PLZF-RARA.28 The morphologic features, including a regular shaped nucleus and less granular cytoplasm than classical APL, were in fact reminiscent of APL with PLZF-RARA.28 This unique profile of leukemia cells might reflect the presence of BCOR-RARA.
The clinical course indicated that coagulopathy on diagnosis could be relieved by administration of ATRA and tamibarotene (synthetic retinoid)25 but not arsenic trioxide. Leukemia cell differentiation was also accomplished with ATRA and tamibarotene. The resistance to arsenic trioxide in this patient was reminiscent of a PLZF-RARA case,34 which can be explained by the fact that arsenic trioxide induces apoptosis by PML-mediated sumoylation.35 Even though the patient was sensitive to retinoic acids, relapse occurred several times, and allogeneic myeloablative transplantation was needed to eradicate the leukemia cells. CD56+ APL is a rare subtype with PML-RARA, PLZF-RARA, or unknown chromosomal alterations.36 The CD56+ APL failed to differentiate in vitro in response to ATRA and prognosis of the CD56+ APL is poor, compared with that of the classical APL.36,37 Our patient was classified as a CD56+ APL subtype, in agreement with the resistance to repeated standard chemotherapy with ATRA.
FISH analysis with dual fusion translocation probes is useful to detect unknown rearrangements of the RARA gene.10 Initially, we failed to detect any karyotype abnormality by regular G-banding and FISH analysis with a PML-RARA dual-color probe, only detecting the 3′ region of the RARA gene (Vysis; data not shown). However, use of a PML-RARA dual fusion translocation probe allowed us to detect a novel rearrangement of the RARA gene. On retrospective reanalysis of the karyotype with G-banding, t(X;17)(p11;q12) was detected. Moreover, spectral karyotyping analysis confirmed that other subtle karyotype abnormalities did not exist in this case (data not shown). For cloning of the fusion transcripts, we used 5′-RACE. The transcripts were, fortunately, short, spanning from BCOR exon 9 to RARA. Long-distance PCR confirmed that there was a full-length BCOR-RARA fusion transcript without any alternative splicing. On the other hand, a reciprocal fusion transcript encoding RARA-BCOR was not detected. Although reciprocal fusion transcripts of RARA-PLZF could play roles in the pathogenesis of rare APLs,38,39 reciprocal fusion transcripts of RARA-PML have been found to be missing in 20%-30% of major APL cases.3 This means the presence of reciprocal fusion proteins is not always indispensable for transformation in APL.
BCOR is a transcriptional corepressor through the proto-oncoprotein, BCL6.17,40 In diffuse large B-cell lymphomas, 3q27 chromosomal translocations involving BCL6 are found in approximately 40% of cases.40 BCL6 acts as a transcriptional repressor by recruiting NCOR/SMRT, mammalian Sin3A, and HDAC1.40 BCOR is a ubiquitously expressed protein originally identified as associating with BCL6.17 BCOR augments BCL6 transcriptional repression by interacting with class I and II HDACs and, potentially, with PCGF1 and FBXL10.17,19,20 PCGF1 is highly homologous with BMI1, which is associated with PLZF-RARA as a repressive chromatin-modification factor.15,21 The association between BCOR-RARA and PCGF1 is functionally coincident with evidence that PCGF1 also acts as a transcriptional repressor of p21Waf1/Cip1 via RARE, which is competitive with RARA/RXR in RA-pathway activation.41 To investigate whether BCOR-RARA associates with PCGF1, we performed a coimmunoprecipitation assay (data not shown). As we expected, BCOR-RARA coimmunoprecipitated with PCGF1 in 293T cells. Nevertheless, these results do not preclude the possibility that the association may be indirect. Since BCOR is a ubiquitously expressed protein, the association between BCOR-RARA and PCGF1 might be mediated through binding to endogenous BCOR.
The self-association domains in known RARA fusion partners are the most conserved features.2 In BCOR-RARA, the domain accounting for self-association has yet to be defined, but we here obtained evidence that BCOR also self-associates, as well as BCOR-RARA, both through the ankyrin repeat domain of BCOR and the region of BCOR-S. Nearly 6% of eukaryotic protein sequences contain AR domains, which consist of several repeats and function in protein-protein binding.42 The AR domain of BCOR lent itself to self-association as well as that reported for transient receptor potential vanilloid 6 and signal-recognition particle 43.43,44 On the other hand, the domains responsible for the self-association mediated with the region of BCOR-S remain to be investigated.
Although the self-association feature is conserved among all RARA fusion proteins, its exact roles in transformation in vivo are still controversial.29 Two recent reports indicated that RXR recruitment is a critical determinant of transforming potential in vivo of oligomeric RARA fusion proteins. The results are consistent with our finding that RXRA is necessary for BCOR-RARA to associate with RARE. In the reporter assay, BCOR-RARA acted in a dominant-negative manner as well as PML-RARA (Figure 5C). This is reasonable, because BCOR-RARA is associated with ectopic transcriptional repressors BCL6 and PCGF1 and, potentially, recruits HDACs. Interestingly, CBC-RARA proved sufficient to induce dominant-negative effects. These results suggest that critical transcriptional repressors might be recruited through the region from 999 to 1409 aa of BCOR. Current dogma holds that PLZF-RARA fails to respond to ATRA because of the RA-insensitive recruitment of NCOR, SMRT, Sin3A, and HDACs to the PLZF-RARA N-terminus.12 Although BCOR-RARA also contains an N-terminal corepressor binding site, the patient's blasts apparently differentiated in response to ATRA. The difference in ATRA sensitivity might be explained by variation in corepressors. Precise mapping of corepressor binding sites more than BCL6 remains to be investigated to explain this issue. RARA fusion to other partners is able to alter binding site specificity.31 In contrast to other RARA fusion proteins,27,45-47 RXRA is necessary for BCOR-RARA association with RARE. The results potentially suggest that the binding-site specificity of BCOR-RARA might be different from those of other RARA fusion proteins, but this remains to be further elucidated in hematopoietic progenitor cells.
While the deregulation of RARA transcriptional activation has a central role for the pathogenesis of APL, other cooperative genetic events are still needed for full-blown transformation.1,16 For instance, PML-RARA is regarded as a dominant-negative mutant for both RARA and PML pathways.1 Indeed, when PML-RARA is expressed in PML-null marrow, leukemogenesis is accelerated.48 To date, there have been no reports that BCOR acts as a tumor suppressor. It is noteworthy that a monoallelic deletion of BCOR was recently found in a male case of AML M7 by genomewide copy-number analysis.49 Because the patient presented here also did not have an intact BCOR, it could be that loss of BCOR is as significant for the pathology of the leukemia as the fact that BCOR is fused to RARA. Another gain-of-function phenotype of BCOR-RARA is relocalization (Figure 6). Other RARA fusion proteins, including PML-RARA and NuMA-RARA, localize in the nucleus/cytoplasm and cytoplasm, respectively.32 The relocalization of BCOR-RARA is consistent with aberrant localization of PML-RARA.32 This feature is able to sequester BCOR-associated proteins, including BCL6, from normal compartments. To date, substantial functions of BCL6 in myeloid malignancies have not been well investigated, and BCL6 transcripts are preferentially detected in differentiated AML, including APL, AML M4, and M5.50 The aberrant relocalization of BCOR-associated proteins potentially cooperates in the leukemogenesis of t(X;17)-associated APL.
In summary, we here identified BCOR-RARA fusion transcripts in a t(X;17)(p11;q12) variant of APL with unique cytoplasmic inclusion bodies. Although the patient was clinically responsive to ATRA, repeated standard chemotherapy did not effect a cure. BCOR-RARA could be shown to possess common features with other RARA fusion proteins, such as: (1) the same break point in RARA cDNA, (2) self-association, (3) RXRA is necessary for BCOR-RARA to associate with RARE; (4) action in a dominant-negative manner on RARA transcriptional activation; and (5) aberrant subcellular relocalization. These results highlight essential features of pathogenesis in APL in more detail. BCOR thus appears to be involved not only in human congenital diseases, but also in a human cancer.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to Dr Hiroki Kurahashi (Department of Molecular Genetics, Fujita Health University) for helpful discussions and Akemi Endo for technical assistance.
This work was supported by a grant-in-aid from Fujita Health University (Y.Y.).
Authorship
Contribution: Y.Y. designed the research; Y.Y. and S.T. performed the research; Y.Y. analyzed the data; M.T., K.H., and Y.I. collected the clinical data; and Y.Y. and E.N. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Yukiya Yamamoto, Department of Hematology, Fujita Health University, 1-98 Dengakugakubo, Kutsukake-cho, Toyoake, Aichi 470-1192, Japan; e-mail: yyukiya@fujita-hu.ac.jp.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal