Abstract
Abstract 942
Emerging evidence suggests that microRNAs (miRNAs) are critical in cancer and adult leukemia by functioning as tumor suppressors and/or oncogenes. Zhang et al identified 32 pediatric acute myeloid leukemia (AML)-specific miRNA patterns by analysis of bone marrow (BM) samples (1). They also established potential miRNAs as biomarkers for predicting CNS-relapse in pediatric acute lymphocytic leukemia (ALL). Altered miRNA expression disrupts normal hematopoiesis and might play a role in niche-induced oncogenesis. Dysfunction of mesenchymal stromal cells induces formation of myeloid sarcomas that infiltrate in the surrounding tissues (2). Previously, we described that mesenchymal stem cells (MSC) of the BM microenvironment participate in leukemic stem cell regulation in an in vivo model of the childhood AML stem cell niche(3). These human MSC niches, created in ectopic bioengineered 3D scaffolds, supported leukemogenesis in NOD/SCID mice. Pediatric AML engrafted at 1 month in the MSC-coated scaffolds in the mice and was retained in the niche up to 4 months, after which distant seeding to murine BM, liver and spleen occurred. The bioengineered niche created a sanctuary for quiescent leukemia cells and at 4 months the AML cells exited the niche and spread hematogenously, mimicking leukemia relapse. Analysis of miRNA patterns in our leukemia niche model could provide novel directions for individual risk-adapted therapy in childhood leukemia.
To analyze miRNA expression patterns of pediatric AML after exposure to the niche microenvironment at different time points.
miRNAs were obtained from primary CD34+ selected AML cells at (d0) Day0= no niche exposure, (1Mo) 1 month =niche engraftment, (4Mo) 4 months=hematogenous spread with leukemic exit from the niche. miRNAs were isolated from single cell suspensions with the mirVana miRNA isolation kit (Ambion) and analyzed on an Ilumina MicroRNA Expression Profiling single Beadchip (#RNA probes = 1145).
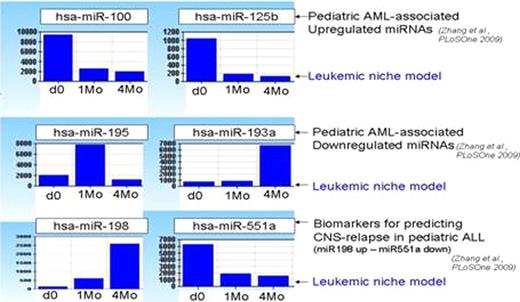
498/1145 miRNAs expression profiles were selected with a detection p value < 0.00001. Out of 498 miRNAs expressed in the leukemic niche model, 23 were previously described as AML-specific miRNAs (2). 10/23 miRNAs were significantly upregulated and 13/23 were downregulated. Pediatric AML-specific miRNAs – miR100 and miR125b had high expression profile at baseline, but were down regulated upon contact with the niche. AML miR195 and miR193a had low expression at baseline, but miR195 was upregulated on engraftment while miR193a only upregulated at the time of hematogenous spread (niche exit). CNS-relapse in ALL might represent a physiological mechanism of leukemic exit of dormant cells from the niche sanctuary. Consistent with this notion, the same expression profile that was found in CNS-relapse in ALL patients (miR198 up – miR551a downregulated) was seen in our model when AML cells became invasive and exited the niche at 4 months.
1) Altered miRNA expression profile of pediatric AML cells is observed after niche exposure (1Mo) compared to baseline AML cells without niche exposure (d0).
2) Altered miRNA expression profile is found when pediatric AML cells spread via the bloodstream (4Mo). In this model, miRNA expression can be correlated to biological behavior of leukemia cells at different time points.
3) Pediatric AML cells that escape from the leukemic stem cell niche and become invasive have the same miRNA expression pattern as published CNS-relapsed ALL cells.
4) The leukemic stem cell niche model can help identify novel treatment targets for pediatric leukemia patients.
(1)Zhang et al. 2009, PloSOne, 4, p1
(2)Raaijmakers et al. 2010, Nature, 464, p852
(3)Vaiselbuh et al, 2010, Tissue Eng Part C Methods, June 29 (ePub)
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal