Abstract
The Ataxia Telangiectasia Mutated (ATM) gene is frequently inactivated in lymphoid malignancies such as chronic lymphocytic leukemia (CLL), T-prolymphocytic leukemia (T-PLL), and mantle cell lymphoma (MCL) and is associated with defective apoptosis in response to alkylating agents and purine analogues. ATM mutant cells exhibit impaired DNA double strand break repair. Poly (ADP-ribose) polymerase (PARP) inhibition that imposes the requirement for DNA double strand break repair should selectively sensitize ATM-deficient tumor cells to killing. We investigated in vitro sensitivity to the poly (ADP-ribose) polymerase inhibitor olaparib (AZD2281) of 5 ATM mutant lymphoblastoid cell lines (LCL), an ATM mutant MCL cell line, an ATM knockdown PGA CLL cell line, and 9 ATM-deficient primary CLLs induced to cycle and observed differential killing compared with ATM wildtype counterparts. Pharmacologic inhibition of ATM and ATM knockdown confirmed the effect was ATM-dependent and mediated through mitotic catastrophe independently of apoptosis. A nonobese diabetic/severe combined immunodeficient (NOD/SCID) murine xenograft model of an ATM mutant MCL cell line demonstrated significantly reduced tumor load and an increased survival of animals after olaparib treatment in vivo. Addition of olaparib sensitized ATM null tumor cells to DNA-damaging agents. We suggest that olaparib would be an appropriate agent for treating refractory ATM mutant lymphoid tumors.
Introduction
The Ataxia Telangiectasia Mutated (ATM) tumor suppressor gene encodes a principal DNA damage–signaling protein, and cells with ATM dysfunction exhibit increased radiosensitivity, loss of cell-cycle checkpoints, and p53 dysfunction.1–5 In addition to the impaired apoptotic response, the cellular phenotype of these cells can be attributed to subtle but significant defects in both major types of DNA double strand break (DSB) repair: error-prone non-homologous end joining (NHEJ), preferentially employed in the gap 1 (G1) phase of the cell cycle, and error-free homologous recombination (HR) repair, restricted to synthesis/gap 2/mitosis (S/G2/M) phases of the cell cycle.6–11 After DNA damage, ATM mutant cells consequently exhibit prolonged DNA DSBs and abnormal retention of DNA proteins at the sites of DNA DSB observed as intra-nuclear foci.6–11
Inactivation of ATM is a frequent event in lymphoid malignancies such as B-cell chronic lymphocytic leukemia (CLL),12–14 T-prolymphocytic leukemia (T-PLL)15,16 and mantle cell leukemia (MCL).17 CLL is the most common leukemia in western countries. While many patients do not require therapeutic intervention, those with progressive CLL have a poor overall outcome, and survival is greatly impaired by the presence of genetic abnormalities such as 11q deletions/ATM mutations and 17p deletions/TP53 mutations.18–23 The less-frequent malignancies, T-PLL and MCL, also commonly harbor 11q deletions and ATM mutations,15–17 which may contribute to their dismal clinical responses. Progressive lymphoid malignancies are currently treated with combinations of nucleoside analogues and alkylating agents, which typically exert their effect through the generation of DNA damage and subsequent induction of an ATM/p53-dependent apoptotic pathway.24 Consistent with this mechanism, ATM mutant CLL cells exhibit resistance to fludarabine-induced apoptosis in vitro.21 The adverse impact of ATM inactivation on clinical responses of this subtype to current therapies may be evident at several levels: The presence of pathogenic ATM mutations causes rapid clonal expansion of 11qdel subclones21 and significantly reduces overall survival21 as well as progression-free survival in patients treated in the United Kingdom CLL4 trial (A.S., V.W., D.O., G.P., A.M.R.T., T.S., manuscript in preparation). Recent developments in front-line regimens by the addition of rituximab to fludarabine and cyclophosphamide (FCR) and second-line alemtuzumab and flavopiridol22–27 has resulted in some clinical benefits for 11q del CLL24 as well as other progressive lymphomas.26,27 Despite these improvements, immunosuppression and toxicity associated with these new first- and second-line agents emphasizes the requirement for the identification of novel targeted drugs for the treatment of chemoresistant ATM mutant lymphoid tumors.
A favorable approach to designing targeted therapy is either to undermine redundant pathways or enhance deficiencies, already present in the cell, that are potentially lethal for the tumor. For example, a DNA repair inhibitor can facilitate conversion of one form of DNA damage into another form, which, in a cell with a particular mutated gene, cannot be repaired and leads to cell death.28 This is the concept of synthetic lethality: when inactivation of either of 2 genes alone allows cell viability but simultaneous inactivation of both genes causes cell death.28 Using this mechanism, poly (ADP-ribose) polymerase (PARP) inhibitors were recently shown to selectively target DNA DSB repair–deficient BRCA1/2 null cells for killing.29,30 After inhibition of PARP1, a component of the DNA single strand break (SSB) repair machinery,29–31 unrepaired SSB lesions are converted into DNA DSBs during DNA replication and require activation of HR repair proteins (eg, BRCA1/2) for their resolution. Thus, BRCA1/2 functionally null tumor cells treated with PARP inhibitor accumulate extensive DNA DSBs and undergo cellular death.29–31 HR-impaired cells are, therefore, sensitive to PARP inhibition.32
ATM null cells show some deficiency in HR repair. Indeed, it has been shown in 2 independent studies that cells with ATM knockdown, like BRCA1/2 null cells, also exhibit selective sensitivity to PARP inhibition.33,34 In the present study we investigated whether the synthetic lethality resulting from PARP inhibitor treatment of ATM null cells33,34 would also be applicable to ATM mutant lymphoid tumors and result in their specific killing. We have demonstrated a differential in vitro and in vivo sensitivity of primary and transformed ATM mutant CLL and MCL tumor cells to a new clinically tested PARP inhibitor, olaparib.35,36
Methods
Patients
CLL samples were obtained from Birmingham and Bournemouth Hospitals. Ethical approval was obtained from the South Birmingham Ethics Committee. A total of 17 CLLs with wildtype ATM and 14 CLL tumors with ATM dysfunction were used in the study, and their cellular features are given in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Cell lines, retroviral ATM knockdown, and pharmacologic ATM inhibition
Lymphoblastoid cell lines (LCLs) generated from 5 healthy donors and 5 individuals with ataxia-telangiectasia (A-T), MCL cell lines with either a confirmed ATM defect (Granta-519) or functional ATM (JVM-2; supplemental Figure 1), and the CLL cell line, PGA37 were employed. Stable knockdown in PGA cells was performed using RNA oligonucleotides (Dharmacon) targeting either green fluorescent protein (GFP) as a negative control (PGA-GFPsh) or ATM (PGA-ATMsh) as previously described.38 Knockdown was shown to be stable by assessment of ATM activity through ionizing radiation (IR)–induced phosphorylation of the known ATM targets: ATM itself, structural maintenance of chromosomes 1 (SMC1), and Nijmegen breakage syndrome 1 (Nbs1; supplemental Figure 2). Pharmacologic inhibition of ATM was performed using the specific ATM inhibitor, KU-55 933 (AstraZeneca), at the dose of 5μM, shown to fully inhibit ATM kinase activity.39
Induction of primary CLL cell proliferation using CD40L/IL-4
Primary CLL leukemia cells obtained from the peripheral blood were typically arrested at gap 1/gap 0 (G1/G0) of the cell cycle. To stimulate and sustain proliferation of these cells, we compared 5 different mitogenic stimuli (see supplemental Figure 3) and found the CD40L/IL-4 culture system the most effective and reproducible. Briefly, after adherence of irradiated (50 Gy) CD40L-expressing murine fibroblasts at 3 × 105 cells/well, 1-1.5 × 106 primary CLL cells were seeded into each well with 10 ng/mL IL-4 (R&D Systems) in a total volume of 2 mL RPMI containing 10% fetal calf serum (Sigma-Aldrich/PAA Laboratories) and incubated at 37°C for 3-4 days. At this point, survival assays were initiated. As bromodeoxyuridine (BrdU) staining revealed that CLL proliferation could only be sustained for 7-11 days in culture (supplemental Figure 3), primary CLL cells initiated to cycle over 3-4 days were then treated with 0-10μM olaparib (AstraZeneca) for an additional 7 days. For consistency, therefore, in all survival assays, all cell types were exposed to olaparib for only 7 days.
Isolation and culture of normal B and T cells
B and T cells were isolated from the blood of healthy donors using RosetteSep Human B-cell and T-cell Enrichment Cocktails (StemCell Technologies) according to the manufacturer's instructions. Cycling of normal B cells at a concentration of 1 × 106 cells/mL was induced as for primary CLL cells. T cells were cultured at a concentration of 1 × 106 cells/mL in RPMI containing 10% fetal calf serum with 100 IU/mL interleukin 2 (IL-2; R&D Systems).
Cell survival assays
Suspensions of lymphoid cells were exposed to increasing concentrations of olaparib for up to 7 days in triplicate experiments and counted 3 times using a hemocytometer; the surviving fraction was then calculated. In experiments using a single olaparib dose, 3μM was used irrespective of cell type, as this produced a survival response on the second part of the curve beyond the initial sharp reduction and ensured a maximal differ-ential between normal and ATM-deficient cells. It also reflected the maximum clinically achievable dose, making the cellular effects at this dose clinically important.
Western blotting
Western blotting was performed as described12 with the following antibodies: rabbit anti-poly(ADP-ribose) (pADPr; Calbiochem); mouse anti-PARP1 (Enzo Life Sciences); rabbit anti–phospho-ATM S1981 (Rockland Immunochemicals); mouse 11G12 anti-ATM;12 rabbit anti–phospho-SMC1 S966 and anti-SMC1, both from Bethyl Laboratories; rabbit anti–phospho-Nbs1 S343 and anti-Nbs1, both from Abcam; mouse anti–β-actin (Sigma-Aldrich); sheep anti-p53 (D. Lane, University of Dundee, Scotland); rabbit anti-p21 (Santa Cruz Biotechnology); and rabbit anti–caspase 7 and anti–caspase 3 from Cell Signaling Technology.
BrdU staining of proliferating cells and FACS analysis
Cells were treated with 100μM BrdU (Sigma-Aldrich) for 24 hours, fixed in ethanol, treated with 2M HCl/0.1 mg/mL pepsin (VWR/Sigma-Aldrich), labeled with anti-BrdU monoclonal antibody and resuspended in 25 μg/mL propidium iodide containing 0.1 mg/mL RNase A (Sigma-Aldrich) before analysis using a Coulter Epics XL-MCL flow cytometer (Beckman Coulter).
Annexin V apoptosis assay
Apoptosis was assayed using an annexin V apoptosis kit (Becton Dickinson) according to the manufacturer's instructions and analyzed using a Coulter Epics XL-MCL flow cytometer. At least 40 000 events were scored for each condition.
Immunofluorescence
Cells were fixed to poly-L lysine–coated slides, treated with methanol (Sigma-Aldrich), and stained with the following antibodies at room temperature: For DNA damage and repair analyses, they were stained with mouse monoclonal anti-γH2AX (Millipore) and rabbit anti-RAD51 (Santa Cruz Biotechnology); for mitotic catastrophe analysis, they were stained with goat anti–Lamin B (Santa Cruz Biotechnology) and rabbit anti–phospho-Histone H3 serine-10 (Cell Signaling). After staining, they were treated with secondary antibodies anti–mouse immunoglobulin G (IgG [Alexa Fluor 594]) and anti–rabbit (Alexa Fluor 488; Invitrogen). Slides were mounted in Vectorshield with DAPI (4,6 diamidino-2-phenylindole; Vector Laboratories). Analysis was performed using a Nikon Eclipse E600 fluorescence microscope and Velocity Version 4.1.0 software (Improvision).
Murine xenograft model
Animals were treated in accordance with United Kingdom Home Office guidelines, Schedule 1. For all experiments, tumor cell engraftment in the bone marrow and spleen before initiation of olaparib treatment was confirmed both by FACS analysis of human anti-CD45 (eBioscience)– and murine anti-CD45 (BD Pharmingen)–stained cells and by immunohistochemistry using anti–human CD5 (Leica Microsystems), anti–human Pax5 (Thermo Scientific), and anti–human Ki-67 (Dako) antibodies.
For assessment of MCL tumor load in murine primary lymphoid organs, sublethally irradiated nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice (aged 5 weeks) were intravenously injected with 3 × 106 Granta-519 cells. Fourteen days after injection, animals received either 50 mg/kg/d olaparib (n = 12) or vehicle, 10% (wt/vol) 2-hydroxy-propyl-β-cyclodextrin (Sigma-Aldrich; n = 11), via intraperitoneal injection, daily for 14 days. Mice were culled at day 30-36, and tumor load in the bone marrow and spleen was assessed by FACS analysis of human CD45+ cells.
To compare tumor size and survival between treated and untreated animals, subcutaneous tumors were initiated by injection of 3 × 106 Granta-519 cells. Tumors were allowed to grow for 5 days before initiation of treatment with 100 mg/kg/d olaparib (tumor size, n = 15; survival, n = 10) or vehicle alone (tumor size, n = 20; survival, n = 10) via oral gavage for no more than 28 days. Tumor volume was measured manually using a calliper 3× per week. Mice were killed upon signs of illness or when tumors reached > 1250 mm.3
Any mice that died early in the experiments due to graft-unrelated causes were excluded from the experiments.
Combination olaparib/cytotoxic treatment
Granta-519 cells seeded in triplicate at 1 × 105 cells/mL in a 200μL volume were pretreated with olaparib (dose range 0-10μM) for 2 days. Subsequently, increasing doses of 4-hydroxycyclophosphamide (4HC; 0-0.25μM; NIOMECH), fludarabine (0-0.5μM), valproic acid (VPA; 0-10mM), bendamustine (0-12.5μM; Sigma-Aldrich), and IR (0-5 Gy) were added to the olaparib-containing culture for an additional 5 days. This time frame enabled consistency in the duration of olaparib treatment (7 days total) and allowed sufficient time for the cytotoxic effects of the conventional agents to occur before calculation of the surviving fraction of cells. Cell viability was measured using the CellTiter-Glo luminescent cell viability assay (Promega) according to the manufacturer's instructions. Luminescence was quantified using a Wallac Victor2 1420 multilabel counter (Perkin Elmer). Synergism was determined using Calcusyn Version 2.1 for Windows software (Biosoft).
Statistical analysis
In vitro data were analyzed using 2-tailed t tests, in vivo tumor load data by the nonparametric Mann-Whitney U test, tumor size data by 2-way analysis of variance, and xenograft survival data by the log rank (Mantel-Cox) test. Data are presented as mean ± SEM.
Results
Olaparib selectively targets ATM mutant lymphoid cells, including proliferating primary CLL cells
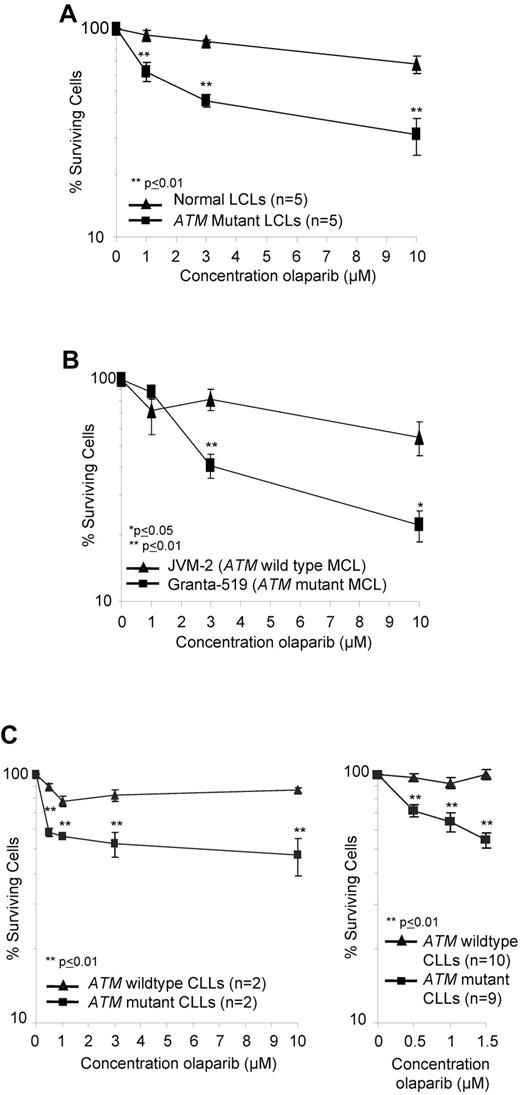
PARP1 activity is associated with the synthesis of poly(ADP-ribose) (pADPr), which modifies a number of proteins including PARP1 itself.32 We used immunoblotting to assess the impact of olaparib on pADPr levels as an indicator of inhibition of PARP activity. The analysis indicated comparable dose-dependent inhibition of PARP activity by olaparib in both ATM-wildtype and ATM-null LCLs and primary CLL cells with significant inhibition consistently achieved from 0.5μM olaparib (supplemental Figure 4A-B). Inhibition continued at a lesser rate with increasing doses of olaparib. We then tested the sensitivity of a range of lymphoid cells with and without ATM inactivation to the PARP inhibitor, olaparib, at the concentrations capable of inactivating PARP activity in lymphoid cells. These included 10 LCLs (5 control and 5 derived from ataxia telangiectasia patients; Figure 1A) and 2 MCL cells lines (ATM mutant Granta-519 and ATM wildtype JVM-2; Figure 1B). In addition, we tested 19 primary CLLs (10 ATM mutant and 9 ATM wild type), which were first induced to proliferate in vitro by coculture with irradiated CD40L–expressing murine fibroblasts and IL-4 for 4 days before exposure to the drug (Figure 1C). In all lymphoid cell types tested, olaparib exposure induced greater dose-dependent reductions in the number of cells with ATM inactivation compared with those harboring functional ATM (Figure 1A-C). LCLs with no functional ATM revealed significant differential sensitivity to 1μM olaparib, the lowest tested dose associated with inhibition of PARP1 activity in this cell type, compared with control LCLs (Figure 1A; supplemental Figure 4A). The ATM mutant Granta-519 MCL cells were significantly more sensitive to olaparib at 3μM compared with JVM-2 MCL cells with ATM function (Figure 1B). Furthermore, ATM deficient–cycling CLL tumor cells, irrespective of the type of ATM mutation and mode of ATM inactivation, revealed a highly significant differential sensitivity to olaparib even at submicromolar doses of 0.5μM olaparib compared with ATM wild type primary tumor cells (Figure 1C). In ATM null CLL cells, the most prominent killing occurred between 0 and 0.5μM olaparib, consistent with the major inhibition of pADP ribose formation at 0.5μM observed by Western blot (supplemental Figure 4B). Normal proliferating B and T cells were not sensitive to olaparib at the same doses (supplemental Figure 5).
ATM mutant lymphoid cells are sensitive to olaparib. Effect of increasing doses of olaparib after 7 days exposure on the percentage of surviving cells (logarithmic scale) in ATM null and ATM wild-type LCLs (A), MCL cell lines (B), and proliferating primary CLL cells (C) over a broad range of doses (left) and at lower doses in an expanded cohort (right).
ATM mutant lymphoid cells are sensitive to olaparib. Effect of increasing doses of olaparib after 7 days exposure on the percentage of surviving cells (logarithmic scale) in ATM null and ATM wild-type LCLs (A), MCL cell lines (B), and proliferating primary CLL cells (C) over a broad range of doses (left) and at lower doses in an expanded cohort (right).
Sensitivity to olaparib is mediated by absence of ATM activity and by cell proliferation
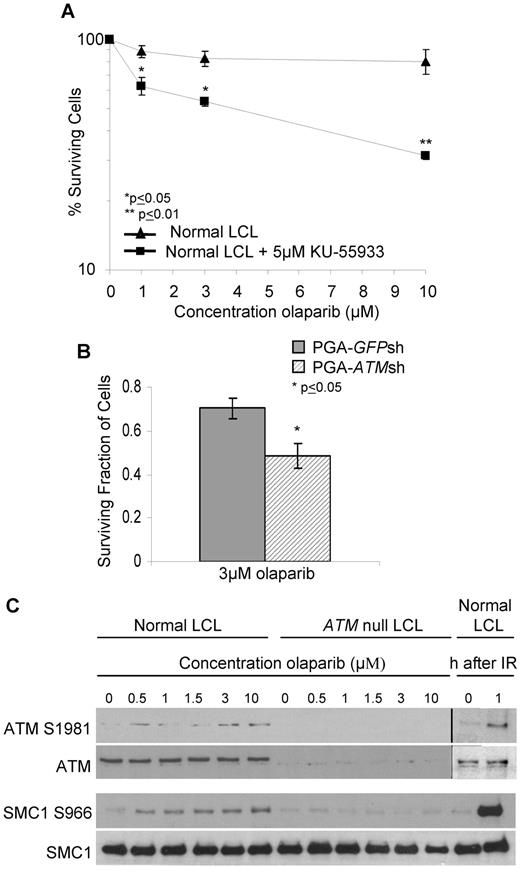
To determine whether sensitivity to olaparib was a consequence of specific ATM inactivation, we first used the small molecule ATM inhibitor, KU-55 933, to inhibit ATM activity in a normal LCL. KU-55 933 (5μM) caused complete inhibition of ATM activity (not shown) and significant sensitization of the LCL cells at a dose of 1μM olaparib (Figure 2A). Next, we down-regulated ATM gene expression by shRNA in the CLL cell line, PGA. ATM knockdown at the time of experiment was confirmed by immunoblot assessment of ATM-dependent DNA damage responses (supplemental Figure 2). We then compared the sensitivity of the isogenic cell lines pairs (PGA-ATMsh and PGA-GFPsh) to 3μM olaparib and observed significant differential sensitivity of cells with ATM knockdown (Figure 2B). The effects of both chemical ATM inhibition and ATM knockdown in sensitizing cells to 3μM olaparib measured over a relatively short period of time were significant. This most likely relates to the biologic function of ATM, which unlike BRCA2, is not a principle component of HR repair. Nonetheless, immunoblot analysis revealed that in ATM wildtype LCLs, but not ATM null LCLs, phosphorylation of the ATM-dependent targets ATM S1981 and SMC1 S966 was induced in a dose-dependent manner by olaparib (Figure 2C). This was consistent with PARP inhibition causing accumulation of DNA DSB, as well as the cellular response being impaired in the absence of functional ATM. Collectively, these data confirm that ATM is required for the normal cellular response to PARP inhibition and that this response is defective in lymphoid cells with ATM deficiency, resulting in sensitivity to olaparib.
The cytotoxic response to olaparib is ATM-specific. Effect of increasing doses of olaparib after 7 days exposure on survival of control LCL cells treated with 5μM of the ATM inhibitor, KU-55 933 (A) and PGA cells without (GFPsh) and (B) with (ATMsh) ATM knockdown. (C) Immunoblotting shows olaparib-induced dose-dependent phosphorylation of ATM-dependent targets in normal cells. IR provides a positive control.
The cytotoxic response to olaparib is ATM-specific. Effect of increasing doses of olaparib after 7 days exposure on survival of control LCL cells treated with 5μM of the ATM inhibitor, KU-55 933 (A) and PGA cells without (GFPsh) and (B) with (ATMsh) ATM knockdown. (C) Immunoblotting shows olaparib-induced dose-dependent phosphorylation of ATM-dependent targets in normal cells. IR provides a positive control.
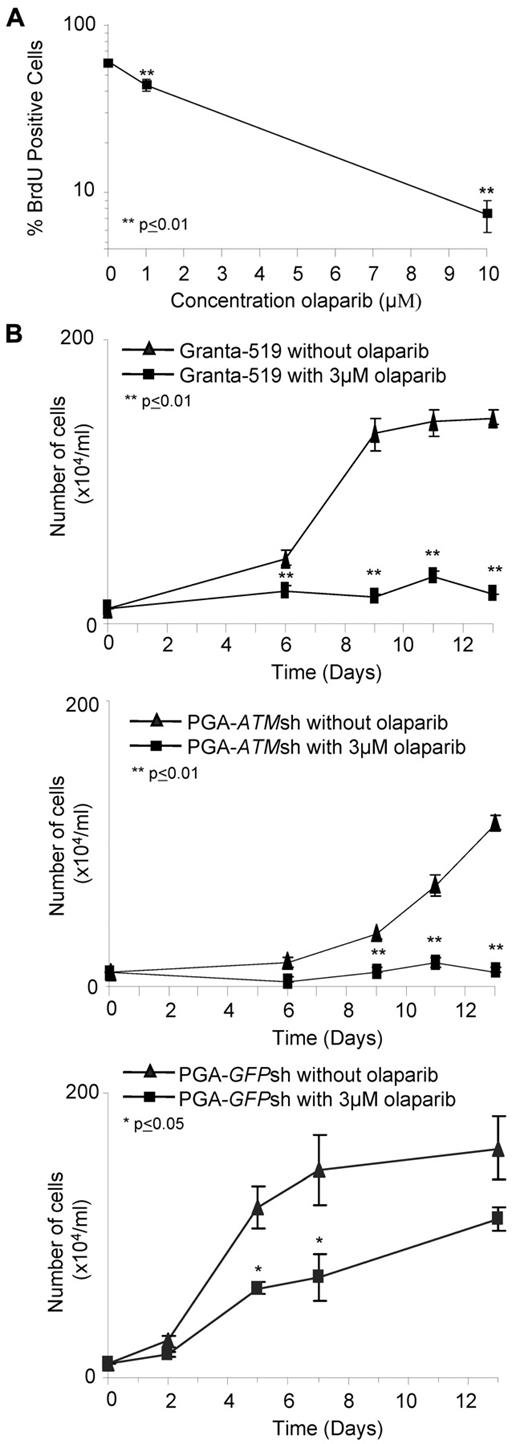
Interestingly, we did not observe any differential effect of olaparib on primary ATM mutant CLL cells that had not been stimulated to proliferate in vitro (supplemental Figure 6), leading us to assume that olaparib preferentially targeted cells that were cycling. This was consistent with the requirement for replication during the conversion of SSBs to DSBs in the absence of PARP activity. Consequently, we assessed the sensitivity of ATM mutant Granta-519 cells staining positive for BrdU, which incorporates during DNA replication, after 7 days exposure to olaparib. We observed a dose-dependent decrease in the fraction of BrdU-stained cells after treatment with olaparib (Figure 3A). Furthermore, comparing 2 representative proliferating primary CLL tumors, we observed a similar trend in the percentage of BrdU-stained cells in the ATM-mutant but not ATM-wildtype CLL cells after olaparib treatment (not shown). Our findings indicate that the PARP inhibitor, olaparib, specifically targets cells that are actively cycling and exhibiting ATM dysfunction.
Olaparib targets proliferating lymphoid cells. (A) Percentage of BrdU-positive cells significantly decreases 7 days after exposure to increasing doses of olaparib in Granta-519 cells. (B) Effect of continued olaparib exposure on proliferation of ATM null cells. After 7 days exposure to 3μM olaparib, Granta519 (top), PGA-ATMsh (middle), and PGA-GFPsh (bottom), cells were reseeded at a low number and then continuously exposed to either 0μM or 3μM olaparib for an additional 2 weeks.
Olaparib targets proliferating lymphoid cells. (A) Percentage of BrdU-positive cells significantly decreases 7 days after exposure to increasing doses of olaparib in Granta-519 cells. (B) Effect of continued olaparib exposure on proliferation of ATM null cells. After 7 days exposure to 3μM olaparib, Granta519 (top), PGA-ATMsh (middle), and PGA-GFPsh (bottom), cells were reseeded at a low number and then continuously exposed to either 0μM or 3μM olaparib for an additional 2 weeks.
Given the targeting by olaparib of proliferating cells with ATM inactivation, we reasoned that the impact of the drug would be cumulative over longer exposure periods as a result of more cells going through 1 or more cell cycles. After treatment with olaparib for 7 days in vitro, Granta-519, PGA-ATMsh, and PGA-GFPsh cells were reseeded at a low number into new cultures with or without olaparib. Notably, the ATM null Granta-519 and PGA-ATMsh cells continuously treated with olaparib maintained complete suppression of proliferation over the additional 2 weeks of exposure, whereas PGA-GFPsh cells showed no such response (Figure 3B).
Olaparib sensitivity of ATM dysfunctional cells is related to the accumulation of DNA damage and is apoptosis independent
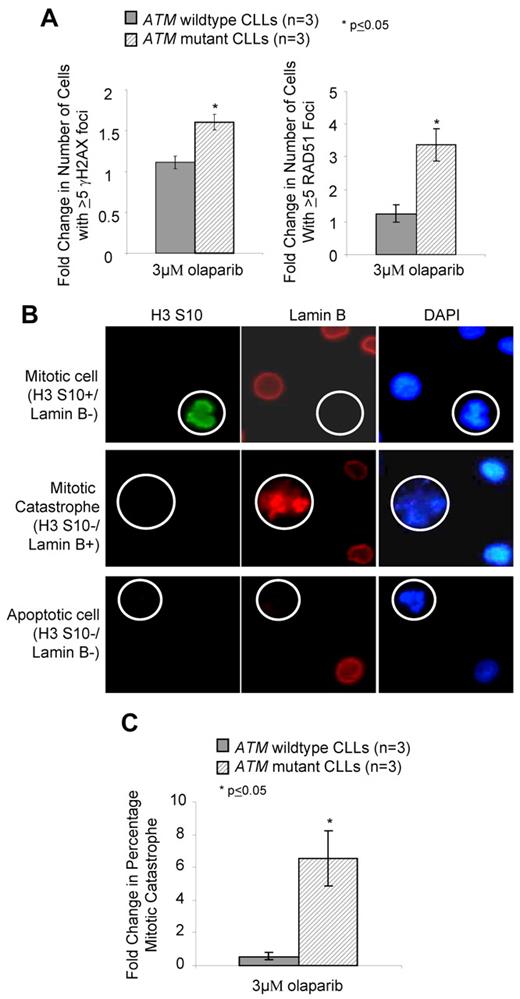
The molecular basis for olaparib activity in ATM deficient cells was presumed to occur by exacerbating the existing DNA DSB repair defect. We used immunofluorescence to assess olaparib-mediated DNA damage measured by γH2AX foci, a marker of DNA DSBs, and by activation of HR repair (Rad51 foci). Although rapid damage-induced formation of γH2AX foci is compromised in ATM null cells,3 their persistence during prolonged PARP inhibition reflects unrepaired DSBs. We found that compared with proliferating primary CLL cells with wildtype ATM, those harboring ATM mutations exhibited significantly elevated γH2AX and RAD51 foci after 7 days exposure to 3μM olaparib (Figure 4A). This demonstrated that olaparib and ATM dysfunction cooperate in compromising DNA DSB repair leading to persistence of DNA damage and retention of repair foci. Sensitivity of ATM-deficient cells to olaparib could not be attributed to apoptosis-mediated cell death, as FACS analysis of annexin V/PI staining revealed an absence of apoptosis in CLL cells undergoing olaparib-induced killing (supplemental Figure 7A). This observation was corroborated, first, by the absence of stabilization of the p53 protein, its downstream target, the p21 protein, and absence of cleavage and activation of the effector caspase 3 (supplemental Figure 7B) and, second, by failure of the pan-caspase inhibitor Z-VAD-fmk to alter olaparib sensitivity (supplemental Figure 7C).
Olaparib induces DNA damage and mitotic catastrophe in ATM mutant lymphoid cells. (A) Induction of γH2AX (left) and RAD51 (right) foci after 7 days exposure to 3μM olaparib measured by immunofluorescence. (B) Differential immunofluorescent staining of cells undergoing normal mitosis (positive for H3 ser-10 phosphorylation and negative for Lamin B), mitotic catastrophe (nega-tive for H3 serine-10 and positive for Lamin B), and apoptosis (negative for both H3 serine-10 and Lamin B), shown at original magnification ×500. (C) Induction of mitotic catastrophe determined by immunofluorescence-based morphologic analysis after 72 hours exposure to 3μM olaparib.
Olaparib induces DNA damage and mitotic catastrophe in ATM mutant lymphoid cells. (A) Induction of γH2AX (left) and RAD51 (right) foci after 7 days exposure to 3μM olaparib measured by immunofluorescence. (B) Differential immunofluorescent staining of cells undergoing normal mitosis (positive for H3 ser-10 phosphorylation and negative for Lamin B), mitotic catastrophe (nega-tive for H3 serine-10 and positive for Lamin B), and apoptosis (negative for both H3 serine-10 and Lamin B), shown at original magnification ×500. (C) Induction of mitotic catastrophe determined by immunofluorescence-based morphologic analysis after 72 hours exposure to 3μM olaparib.
In the absence of apoptosis after olaparib treatment, we investigated the possibility of another mechanism of cell death, mitotic catastrophe, using immunofluorescence-based morphologic analysis. Mitotic catastrophe occurs when cells undergo aberrant mitosis in the presence of unrepaired DNA damage, resulting in the formation of multimicronucleated cells featuring damaged chromosomes.40,41 Cells undergoing normal mitosis stain positive for H3 ser-10 phosphorylation yet lose integrity of the nuclear envelope, indicated by absence of lamin B1 staining. In contrast, cells undergoing mitotic catastrophe do not stain for H3 ser-10 phosphorylation but retain the nuclear envelope (and lamin B staining) due to failed cytokinesis (Figure 4B). These cells contain multiple micronuclei and are distinguishable from the nuclear blebbing of apoptotic cells, which lose integrity of the nuclear envelope. After olaparib exposure, we observed a significant elevation in the number of ATM mutant cells undergoing mitotic catastrophe compared with ATM wild-type, proliferating primary CLL cells (Figure 4C).
ATM-mutant lymphoid tumor cells are sensitive to olaparib in vivo
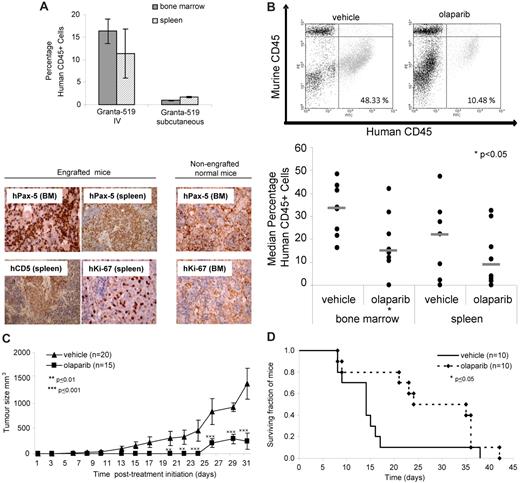
To investigate the in vivo impact of olaparib, we generated murine xenograft models of the ATM mutant MCL cell line, Granta-519. To determine whether infiltration and engraftment of the tumor cell line had already taken place before initiation of olaparib treatment in the different animal cohorts, 3 representative mice from each cohort were analyzed on the day that treatment was to begin (14 days after intravenous or 5 days after subcutaneous injection of cells). The presence of tumor cells at the level of at least 1% of all cells, which is considered to be engraftment, was observed by FACS analysis in the bone marrow and spleen both at 5 days (subcutaneous) and 14 days (intravenous) after injection (Figure 5A). Furthermore, using immunocytochemistry and anti–human CD5, Pax-5, and Ki-67 antibodies, we confirmed significant infiltration of proliferating human B-lymphoid tumor cells in both the spleen and bone marrow at both time points before treatment initiation (Figure 5A).
Olaparib impedes growth of ATM mutant tumor cells in vivo and lengthens survival in a Granta-519 murine xenograft model. (A) Engraftment of a human tumor cell line in bone marrow and spleen of representative mice before treatment. Engraftment was demonstrated 14 days after intravenous injection or 5 days after subcutaneous injection of 3 × 106 Granta-519 cells/animal by FACS assessment of the percentage of human CD45+cells (top) and by immunohistochemistry using anti–human antibodies for B-cell lineage (CD5 and Pax5) and proliferating cells (Ki-67; bottom) in the spleen and bone marrow of mice (shown either at original magnification ×20 or ×40). Brown color indicates positive immunostaining. Organs from non-engrafted mice did not show nuclear staining for either hKi-67 or hPax5. (B) Effect of olaparib exposure on tumor burden in Granta-519–engrafted NOD/SCID mice. Representative FACS dot plots showing percentage of human CD45+ cells in murine bone marrow after treatment with olaparib or vehicle alone (top) and median percentage of human CD45+ cells in the bone marrow and spleen of mice intravenously injected with Granta-519 cells after 5 weeks treatment with olaparib (n = 8) or vehicle alone (n = 8; bottom). (C) Effect of olaparib treatment on size of subcutaneous tumors generated by localized Granta-519 injection (n = 15) compared with vehicle alone (n = 20). (D) Impact of olaparib treatment on overall survival of mice engrafted with Granta-519 cells (n = 10) compared with vehicle alone (n = 10).
Olaparib impedes growth of ATM mutant tumor cells in vivo and lengthens survival in a Granta-519 murine xenograft model. (A) Engraftment of a human tumor cell line in bone marrow and spleen of representative mice before treatment. Engraftment was demonstrated 14 days after intravenous injection or 5 days after subcutaneous injection of 3 × 106 Granta-519 cells/animal by FACS assessment of the percentage of human CD45+cells (top) and by immunohistochemistry using anti–human antibodies for B-cell lineage (CD5 and Pax5) and proliferating cells (Ki-67; bottom) in the spleen and bone marrow of mice (shown either at original magnification ×20 or ×40). Brown color indicates positive immunostaining. Organs from non-engrafted mice did not show nuclear staining for either hKi-67 or hPax5. (B) Effect of olaparib exposure on tumor burden in Granta-519–engrafted NOD/SCID mice. Representative FACS dot plots showing percentage of human CD45+ cells in murine bone marrow after treatment with olaparib or vehicle alone (top) and median percentage of human CD45+ cells in the bone marrow and spleen of mice intravenously injected with Granta-519 cells after 5 weeks treatment with olaparib (n = 8) or vehicle alone (n = 8; bottom). (C) Effect of olaparib treatment on size of subcutaneous tumors generated by localized Granta-519 injection (n = 15) compared with vehicle alone (n = 20). (D) Impact of olaparib treatment on overall survival of mice engrafted with Granta-519 cells (n = 10) compared with vehicle alone (n = 10).
Subsequently, the degree of tumor load was compared in the lymphoid organs of 23 NOD/SCID Granta-519 cell–injected mice 5 weeks after intravenous injection of cells and 14 days after treatment with olaparib. However, early in the experiment, 7 mice died of graft-unrelated causes, leaving 16 mice, which were treated with either olaparib (n = 8) or vehicle alone (n = 8). Analysis of the percentage of human CD45 staining by FACS analysis (Figure 5B) revealed a significant reduction in the percentage of Granta-519 cells in the bone marrow and a trend toward reduced tumor cell load in the spleen of mice treated with olaparib compared with those receiving vehicle alone (Figure 5B).
We next assessed the effect of olaparib on the growth of subcutaneous tumors generated by the localized injection of ATM mutant Granta-519 cells into mice and found a significant positive correlation between olaparib treatment and reduced tumor size (Figure 5C). Finally, the overall survival of mice engrafted with Granta-519 cells was significantly increased by olaparib treatment compared with vehicle alone (Figure 5D).
Collectively, these data provide convincing evidence that 4 weeks of continuous intraperitoneal or oral treatment with 50 or 100 mg/kg olaparib significantly impedes the growth of ATM mutant tumor cells and consequently lengthens the survival of animals harboring these tumors.
We conclude that ATM mutant malignant lymphoid cells show differential sensitivity to PARP inhibition both in vitro and in vivo.
Olaparib sensitizes ATM mutant cells to conventional cytotoxic agents
Finally, to address the ability of PARP inhibition to increase the effect of standard CLL treatments as well as other chemotherapy agents, we tested the ability of olaparib to sensitize ATM mutant cells to the purine analogue fludarabine, the alkylating agents 4HC and bendamustine, the histone deacetylase inhibitor VPA, and IR (supplemental Table 2; Figure 6A). When treated with the agents alone, Granta-519 cells were resistant to bendamustine and 4HC, yet were sensitive to VPA. Olaparib pretreatment was able to significantly sensitize cells to all the agents tested (Figure 6A). The greatest synergism was observed between olaparib and VPA, whereas olaparib and IR revealed only moderate synergism (supplemental Table 2). The effect of 4HC and olaparib was largely additive (supplemental Table 2), although moderate synergism was observed at the 4HC dose of 0.1μM (Figure 6A). Finally, the impact of olaparib on fludarabine and bendamustine cytotoxicity was generally additive, although synergistic activity could be detected at some doses of fludarabine (supplemental Table 2). Western blot analysis demonstrated enhanced cleavage of PARP1 and caspases 3 and 7 when Granta-519 cells were exposed to either 4HC, VPA, or IR in combination with 1μM olaparib (Figure 6B), indicating that drug-induced apoptosis was increased in the presence of olaparib.
Pretreatment with olaparib sensitizes ATM null cells to cytotoxic agents. (A) Effect of 1μM olaparib on killing by 12.5μM bendamustine, 0.1μM fludarabine, 0.1μM 4HC, 5mM VPA, and 1 Gy IR in ATM mutant Granta-519 cells. Synergistic effects (see supplemental Table 2) for the given doses are shown below the graph (+++ synergism; ++ moderate synergism; + slight synergism; ± nearly additive). (B) Western blot analysis of Granta-519 cells showing an increased cleavage of PARP1, caspase 7, and caspase 3 after combined olaparib plus 4HC, VPA, or IR treatment compared with treatment with 4HC, VPA, or IR alone. Actin shows equal loading.
Pretreatment with olaparib sensitizes ATM null cells to cytotoxic agents. (A) Effect of 1μM olaparib on killing by 12.5μM bendamustine, 0.1μM fludarabine, 0.1μM 4HC, 5mM VPA, and 1 Gy IR in ATM mutant Granta-519 cells. Synergistic effects (see supplemental Table 2) for the given doses are shown below the graph (+++ synergism; ++ moderate synergism; + slight synergism; ± nearly additive). (B) Western blot analysis of Granta-519 cells showing an increased cleavage of PARP1, caspase 7, and caspase 3 after combined olaparib plus 4HC, VPA, or IR treatment compared with treatment with 4HC, VPA, or IR alone. Actin shows equal loading.
Overall, these data indicate that olaparib is able to sensitize ATM null cells in vitro to conventional chemotherapy agents used for the treatment of leukemias.
Discussion
ATM mutation represents the single most frequent genetic alteration in CLL, as well as in the more rare, aggressive lymphoproliferative disorders T-PLL and MCL.12–17 The nature of chemoresistant ATM mutant leukemia20,21 is reflected at the biologic level by an apoptotic defect in response to DNA damaging agents.42,43 Importantly, and distinct from TP53 mutant cells, which are also apoptosis resistant, the DNA damage response defect of ATM mutant lymphoid cells extends to multiple pathways, demonstrated by impaired phosphorylation of downstream protein targets, such as SMC1 and Nbs1, and DNA repair.20,42,43 Thus, manipulating the DNA damage response may enable sensitization of ATM mutant lymphoid cells to DNA damage despite the presence of an apoptotic defect.
In the current study, we have demonstrated that the PARP inhibitor olaparib is able to enhance a pre-existing DNA repair defect in ATM mutant lymphoid tumor cells, leading to the accumulation of unrepaired DNA DSBs and apoptotic-independent cell death, which involved the process of mitotic catastrophe. The growth of ATM mutant Granta-519 tumor cells in a NOD/SCID xenograft model was significantly impaired in the presence of olaparib, both in primary lymphoid organs as well as in subcutaneous tumors. Furthermore, the overall survival of the Granta-519–engrafted NOS/SCID mice was significantly increased by olaparib treatment compared with mice receiving vehicle alone. Overall, our findings indicate that olaparib impedes the growth of ATM mutant cells in vitro and in vivo by instigating the accumulation of intolerable levels of DNA damage in cycling cells.
The observed effect of olaparib in ATM mutant lymphoid cells as a result of off-target effects has to be formally considered. Off-target effects are unlikely, as, first, we observed a tumor-specific effect in ATM null lymphoid tumor cells with olaparib, and, second, olaparib treatment produced a cytotoxic effect, which led to both the accumulation of DNA damage and inhibition of PARP activity in primary CLL cells at similar submicromolar/micromolar doses. Thus, the activity of olaparib was clearly dependent on ATM dysfunction and represents a novel targeted therapy for ATM mutant lymphoid tumors.
These observations suggest that primary ATM mutant leukemias may be predicted to show a differential sensitivity to PARP inhibition–mediated killing in vivo in a manner dependent on their highly proliferative status and, importantly, independent of p53-mediated apoptosis. Olaparib, here, targets only proliferating cells with ATM dysfunction, consistent with a cytotoxic mechanism involving the conversion of SSBs into DSBs during DNA replication that cannot be repaired efficiently in cells with a HR repair defect. PARP inhibition did not lead to the same degree of cytotoxicity of ATM deficient tumor cells as BRCA mutant cells,29,30 because the major role of ATM is in sensing the damage that is subsequently repaired by HR repair in which Rad51, BRCA2 and BRCA1 proteins play a major role. A mechanism that is independent of an HR defect and allows some ATM mutant cells to survive in the presence of olaparib could potentially account for the lesser effect. However, this possibility is less likely given that several other proteins in the HR pathway, when inactivated, also result in cellular sensitivity to olaparib comparable in scale with ATM null cells.34 The response of ATM mutant lymphoid tumor cells to PARP inhibition is, therefore, comparable to, although not the same as, the scenario previously described for BRCA1/2 mutant breast carcinoma cells,29,30 which has resulted in Phase I and ongoing Phase II clinical trials with orally administrated olaparib, providing evidence that this agent is well tolerated and exhibits clinical potency.36,44 Thus, the clear differential sensitivity of ATM mutant lymphoid cells to submicromolar concentrations of olaparib and the necessity to improve treatment for chemoresistant ATM mutant lymphoid tumors makes olaparib a compelling candidate for trials in these malignancies. Indeed, progressive tumors with especially active proliferation centers45,46 may provide the ideal cellular scenario for targeting by olaparib with the aim of at least delaying disease progression.
There is a possibility that different ATM mutant lymphoid tumors will exhibit differential sensitivity to olaparib, depending on the type of ATM mutation(s) and, consequently, the degree of residual ATM activity. In the current study, we did not observe a difference in response to olaparib between primary CLL tumors with either a single or 2 identified ATM mutations. This may not be surprising, given that ATM mutant tumors were initially selected on the basis of their defective response to DNA damage and therefore loss of ATM kinase activity. It is also possible that some single mutant alleles might act in a dominant negative manner. As we have shown in the context of CLL, loss of a single ATM allele by 11q deletion does not affect ATM function,21 and it is therefore conceivable that only 11q-deleted tumors that exhibit mutation in the remaining ATM allele and consequently lose ATM function will respond differentially to treatment with olaparib. Although accurate measurement of the in vivo proliferative rate of ATM-deficient CLL tumors is not yet available, there is data to suggest that cases of progressive disease, which are often associated with ATM mutations, are generally associated with increased cell turnover.45,46 Consistent with this notion, 11qdel CLL subclones expand more rapidly in vivo after acquisition of homozygous ATM gene inactivation.21 While olaparib monotherapy is an attractive proposition for treating these challenging tumors, there is also the possibility of combining olaparib with chemotherapy agents. Indeed, we have shown that addition of olaparib acts in a cooperative manner with several conventional chemotherapy agents and increases the sensitivity of ATM null lymphoid tumor cells to alkylating agents and purine anologs. Interestingly, clear synergistic activity of olaparib was observed with the histone deacetylase inhibitor, VPA. Thus, the combination of VPA and olaparib may prove to be a useful and novel clinical approach to treating chemoresistant ATM null CLLs. Finally, it remains to be determined whether, in addition to ATM inactivation, refractory CLL tumors with chromosomal instability47 exhibit other types of HR repair defects that might render them sensitive to PARP inhibition.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Claire Baker, Clemency Hawksley, the BMSU staff, and Oliver G. Best for technical support; Professor John Gordon for CD40L-expressing fibroblasts; and Anders Rosen for CLL cell lines.
This work was supported by Leukaemia Lymphoma Research UK, Cancer Research UK, and AstraZeneca.
Authorship
Contribution: V.J.W. and C.E.O. designed the work, performed experiments, interpreted the data, and wrote the manuscript; A.S. performed experiments; D.G.O., G.P., and M.J.S.D. wrote the manuscript; G.S. designed the work and wrote the manuscript; J.E.P. analyzed the data; Z.R. performed experiments; P.K. designed the work; P.A.H.M. wrote the manuscript; A.M.R.T. designed the work, interpreted the data, and wrote the manuscript; and T.S. designed the work, interpreted the data, and wrote the manuscript.
Conflict-of-interest disclosure: C.E.O. has been supported by AstraZeneca, and G.S. is an employee of AstraZeneca. The remaining authors declare no competing financial interests.
Correspondence: Tatjana Stankovic, School of Cancer Sciences, Birmingham University, Vincent Dr, Edgbaston, Birmingham, B15 2TT, United Kingdom; e-mail: t.stankovic@bham.ac.uk.
References
Author notes
V.J.W. and C.E.O. contributed equally to the manuscript.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal