Ferroportin (Fpn) is the only known iron exporter in vertebrate cells and plays a critical role in iron homeostasis regulating cytosolic iron levels and exporting iron to plasma. Ferroportin1 (FPN1) expression can be transcriptionally regulated by iron as well as other transition metals. Fpn can also be posttranslationally regulated by hepcidin-mediated internalization and degradation. We demonstrate that zinc and cadmium induce FPN1 transcription through the action of Metal Transcription Factor-1 (MTF-1). These transition metals induce MTF-1 translocation into the nucleus. Zinc leads to MTF-1 binding to the FPN1 promoter, while iron does not. Silencing of MTF-1 reduces FPN1 transcription in response to zinc but not in response to iron. The mouse FPN1 promoter contains 2 MTF-1 binding sites and mutation of those sites affects the zinc and cadmium-dependent expression of a FPN1 promoter reporter construct. We demonstrate that Fpn can transport zinc and can protect zinc sensitive cells from high zinc toxicity.
Introduction
Ferroportin (Fpn) is the only known mammalian iron exporter and plays an essential role in the entry of iron into plasma (for review, see Wessling-Resnick1 and Wrighting and Andrews2 ). Fpn is most highly expressed in cells that play a major role in iron acquisition: duodenal enterocytes, macrophages, hepatocytes and syncytial trophoblasts. Fpn, however, can be expressed on a wide variety of other cell types in response to heme3,4 or iron.5,–7 Iron can regulate Fpn expression through transcriptional and translational mechanisms. Ferroportin1 (FPN1), the gene that encodes Fpn, contains a 5′ iron-responsive element (IRE), which places its translation under the control of iron regulatory proteins. It is thought that the increased expression of Fpn by iron is a response to cellular iron load, as increased cytosolic iron would lead to increased iron export.
FPN1 mRNA levels are also increased when cells are exposed to transition metals such as zinc, manganese, cadmium, copper, and other transition metals.6 In both J774 cells and Caco cells, the increase in Fpn expression resulted in increased iron export. What is unclear, however, is the physiologic function of transition metal-induced expression of Fpn. Iron export in response to increased transition metals may protect cells from transition metal toxicity, perhaps by reducing the potential for iron-induced Fenton chemistry. Alternatively, Fpn might export other metals in addition to iron and thus increased expression of Fpn would directly protect cells from metal toxicity.
In this study, we identify at least 1 mechanism by which transition metals induce Fpn expression. We show that zinc and cadmium can activate FPN1 transcription through the Metal Transcription Factor-1 (MTF-1). We also demonstrate that Fpn transports zinc and can protect cells from zinc toxicity.
Methods
Vector
pcDNA3.1-mouse-MTF1–Flag was a gift from Dr G. K. Andrews.8 We produced a chimera pcDNA3.1-mouse/human-MTF1–Flag by switching the mouse fragment with the human corresponding fragment between KpnI/FseI sites of mouse MTF1-Flag. To express an shRNA against mouse MTF-1, we inserted a double strand oligonucleotide targeting the gtacttcgccaccgctgta sequence of mouse MTF-1 into BamHI and EcoRI sites of pSIREN-DNR-DSRed (Clontech) according to manufacturer's instructions. The mouse/human chimera MTF-1 is resistant to shRNA against mouse MTF-1. The pGL3-control vector (Promega) was modified to perform the luciferase assay. The SV40 promoter was removed by HindIII/NheI digestion and the mouse Fpn promoter (starting at −2378, −1154, and −623 from the TATAA box) was amplified by polymerase chain reaction (PCR) and inserted using same restriction sites. The 5′IRE in the FPN1 promoter was deleted by BamHI/SamI digestion as described.9 The putative metal-responsive elements (MREs) in the Fpn promoter in pGL3 were mutagenized by PCR to produce TCCAGCA*GA*AT*CT*CG and TGGAAGAA*TT*CG*AG (the consensus sequence is underlined, *mutagenized base). Fpn-green fluorescent protein (GFP) was carried by a peGFP-N1 vector (Clontech). All constructs have been sequence verified.
Cell culture
Mouse fibroblast NIH3T3 cells and human epithelial HeLa cells were maintained in Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS; ThermoFisher). Baby hamster kidney (BHK) clone 3286-8-8 cells (kindly given by Dr Richard Palmiter, University of Washington) were maintained in DMEM with pyruvate and 10% FBS. Primary cultured mouse bone marrow macrophages were grown in RPMI 1640 with 20% equine serum (Invitrogen) for 4 days, then in RPMI 1640 with 20% FBS and 30% L cell-conditioned medium.
Transfections
Mouse bone marrow macrophages were transfected with a pool of siRNA oligonucleotides targeting mouse MTF-1 (Dharmacon RNA Technologies) using Oligofectamine reagent following manufacturer' instructions (Invitrogen). NIH3T3 cells, BHK clone 3286-8-8 cells, and HeLa cells were transfected with constructs as mentioned above using Amaxa nucleofection according to manufacturer's directions (Lonza).
RNA extraction and reverse transcription PCR (RT-PCR) total RNA was isolated using RNeasy Mini kit (QIAGEN), purity assayed with a Nanodrop spectrophotometer and stored at −80°C. RT-PCR was performed in a single step reaction using Verso-1-Step–RT-PCR HotStart kit (Thermo Scientific) on 30 ng of total RNA using the following primer sequences: FPN1 5′-TTGCAGGAGTCATTGCTGCTA-3′ (forward) and 5′-TGGAGTTCTGCACACCATTGAT-3′ (reverse); MTF-1 5′-CAGGTTTGTGGATGACAACG-3′ (forward) and 5′-TCACCTTCCACCAGAAAAGG-3′ (reverse); MT-1 5′-GCTGTGCCTGATGTGACGAA-3′ (forward) and 5′-AGGAAGACGCTGGGTTGGT-3′ (reverse) as published.10 RT-PCR was performed for FPN1 by 30 cycles, MTF-1 by 25 cycles, the metallothionein 1 gene (MT-1) by 18 cycles, and actin, 25 cycles. PCR products were loaded on a 2% agarose gel and bands were analyzed using ImageJ image software Version 1.34S (National Institutes of Health). Alternatively, real-time RT-PCR was performed using same primers with Absolute SYBR Green ROX Mix (Thermo Scientific).
Immunofluorescence and microscopy
Cells expressing MTF-1-Flag were fixed for 20 minutes with 4% paraformaldehyde (PFA) in cacodylate buffer pH 7.3, permeabilized 3 minutes in phosphate-buffered saline (PBS) 0.1% Triton X-100, saturated 1 hour in PBS 3% BSA, incubated with mouse anti-Flag (1:1000, clone M2, Sigma-Aldrich) overnight at 4°C, and incubated with Alexa 594 conjugated goat anti–mouse antibody (1:1000; Invitrogen) for 1 hour at room temperature. BHK clone 3286-8-8 expressing Fpn-GFP or GFP-control vector were fixed in PFA, permeabilized, and mounted. Fixed cells were mounted in Vectashield mounting medium containing DAPI (4′,6-diamidino-2-phenylindole) and visualized using an epifluorescence microscope (Olympus). Images were capture using Pictureframer software Version 3.00.03 (Olympus).
Chromatin immunoprecipitation
NIH3T3 cells were seeded in 150-mm dishes. Cells were harvested in PBS. Protein-DNA complexes were cross-linked with 1% formaldehyde for 10 minutes. Cross-linking was quenched by adding 125mM glycine. Cells were washed with PBS, harvested, resuspended in lysis buffer (5mM PIPES [piperazine-N,N≪-bis(2-ethanesulfonic acid] pH 8.0, 85mM KCl, 0.5% NP-40 containing protease inhibitors [Roche]), harvested, resuspended in high salt lysis buffer (PBS, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, and Protease Inhibitor Cocktail) and sonicated 4× for 30 seconds each time. The soluble chromatin was collected by centrifugation, and an aliquot of the chromatin was put aside and represented the input fraction. The supernatant was precleared with 50 μL of protein A/G PLUS Agarose (Santa Cruz Biotechnology) for 30 minutes at 4°C, then the beads were removed. The supernatant was incubated with 0.5 μg of rabbit anti-Flag (Sigma-Aldrich) overnight at 4°C. Fifty microliters of protein A/G PLUS Agarose was added and incubated for 2 hours at 4°C. Beads were harvested by centrifugations and were washed twice in high salt lysis buffer, 4 times in wash buffer (100mM Tris (pH 8.0), 500mM LiCl, 1% NP-40, and 1% deoxycholate). Protein-DNA complexes were resuspended in 400 μL of elution buffer (1% sodium dodecyl sulfate, 0.1M NaHCO3) and the cross-links were reversed by overnight incubation at 67°C. The beads were removed after 2 hours. DNA was isolated in phenol/chloroform/isoamyl alcohol (25:24:1), and finally concentrated in 50 μL 10mM Tris, 1mM EDTA (ethylenediaminetetraacetic acid), pH 8.1. Chromatin immunoprecipitation DNA (5 μL) was amplified by PCR with primers 5′-AGACCTTTGGGGCTTCTGAT-3′ and 5′-TGCAGATAAAGCCCTTTTGG-3′ for the putative MRE 1 in the FPN1 promoter, 5′-ACCCCATTTTGCAACTTCAG-3′ and 5′-ATCAGAAGCCCCAAAGGTCT-3′ for the putative MRE 2 in FPN1, 5′-GATAGGCCG-TAATATCGGGGAAAGCAC-3′ and 5′-GAAGTACTCAGGACGTTGAAGTCGTGG-3′ for MRE in the MT-1 promoter as described.11
Intracellular zinc assay
Cells were loaded with 70μM zinc for 2 hours. After 3 washes, cells were incubated with 1μM FluoZin-3AM (Sigma-Aldrich) in medium without FBS for 30 minutes. Fluorescence was read at 485/520 nm (excitation/emission) in HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid)–buffered saline (10mM HEPES, 150mM NaCl, pH 7.4).
Other procedures
Luciferase assays were done with the Dual Luciferase Reporter (Promega) following manufacturer's directions. Data were normalized with Renilla luminescence. Ferritin enzyme-linked immunosorbent assay was carried out with the enzyme-linked immunosorbent assay test ferritin kit (Laguna Scientific) following manufacturer's directions. Data were normalized to total protein concentration. Zinc, copper, manganese, cobalt, and cadmium were prepared as salt sulfate as a 50-mM stock solution. Ferric ammonium citrate (J. T. Baker) was prepared as a 10mM (iron) stock solution. All solutions were filtered and stored away from light at 4°C. Western analysis of Fpn was performed as described.12
Results
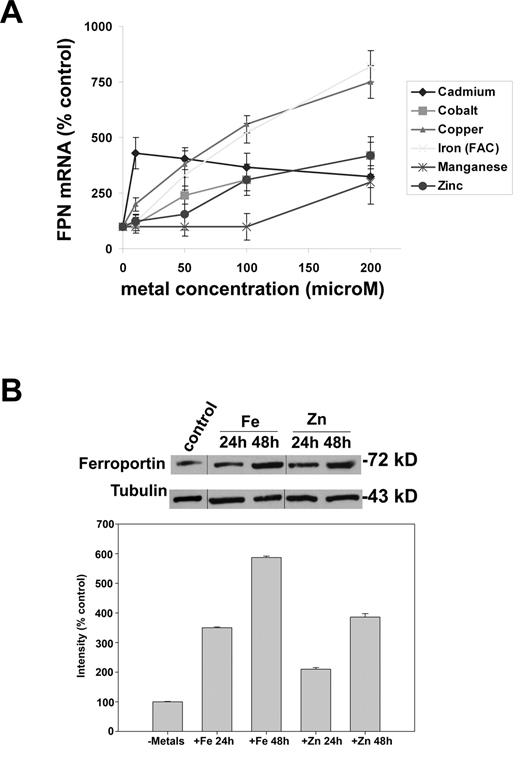
Differentiated mouse bone marrow macrophages were exposed to serial concentrations of cadmium, cobalt, copper, iron, manganese, and zinc for 16 hours. FPN1 mRNA levels were assayed by semiquantitative RT-PCR, and the data were normalized to actin mRNA. Cobalt, copper, iron, and zinc induced the expression of FPN1 mRNA in a dose-dependent manner (Figure 1A). Cadmium induced FPN1 mRNA at the 10μM concentration. Manganese had no significant effect on FPN1 mRNA at low concentrations, but increased FPN1 expression at 200μM.
FPN1 mRNA increases upon exposure of cells to transition metals. (A) Cultured mouse bone marrow macrophages were exposed to serial concentrations of transition metals ranging from 0 to 200μM for 16 hours, total RNA was harvested and semi quantitative RT-PCR were performed for FPN1 and actin mRNA. For this and all other experiments, FPN1 mRNA or other specified mRNA were normalized to actin mRNA. (B) Cells as in A were exposed to 10μM iron or zinc for 24-48 hours, cells lysed in 0.15M NaCl/10mM tris-HCl pH 7.2/0.5mM EDTA/1% Triton X-100 plus protease inhibitors and 50 μg of protein run on sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Samples were analyzed for Fpn by Western blot using a polyclonal rabbit antibody directed against Fpn followed by peroxidase conjugated goat anti–rabbit immunoglobulin G. To control for protein loading the blots were assayed for tubulin using a mouse anti-tubulin antibody followed by peroxidase conjugated goat anti–mouse immunoglobulin G. The righthand panel is a quantification of the Western blot. All experiments were performed a minimum of 3 times.
FPN1 mRNA increases upon exposure of cells to transition metals. (A) Cultured mouse bone marrow macrophages were exposed to serial concentrations of transition metals ranging from 0 to 200μM for 16 hours, total RNA was harvested and semi quantitative RT-PCR were performed for FPN1 and actin mRNA. For this and all other experiments, FPN1 mRNA or other specified mRNA were normalized to actin mRNA. (B) Cells as in A were exposed to 10μM iron or zinc for 24-48 hours, cells lysed in 0.15M NaCl/10mM tris-HCl pH 7.2/0.5mM EDTA/1% Triton X-100 plus protease inhibitors and 50 μg of protein run on sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Samples were analyzed for Fpn by Western blot using a polyclonal rabbit antibody directed against Fpn followed by peroxidase conjugated goat anti–rabbit immunoglobulin G. To control for protein loading the blots were assayed for tubulin using a mouse anti-tubulin antibody followed by peroxidase conjugated goat anti–mouse immunoglobulin G. The righthand panel is a quantification of the Western blot. All experiments were performed a minimum of 3 times.
We then examined if the increase in Fpn mRNA was followed by increased Fpn protein levels. We focused on iron and zinc, as iron is known to increase Fpn and acts as a positive control. Cells were incubated with 0-10μM iron or zinc for up to 48 hours. The amount of iron or was dropped to 10μM to maintain cell viability. The amount of Fpn was enhanced in presence of iron or zinc (Figure 1B). These data suggest that Fpn may play a role in zinc homeostasis.
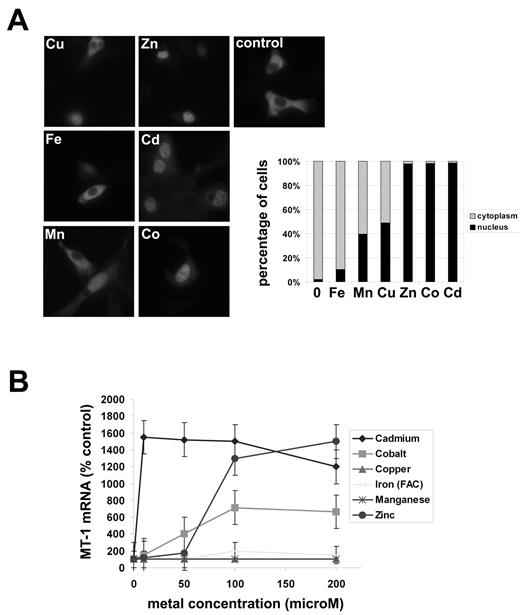
Activation of genes by transition metals, notably zinc, cobalt, and cadmium, depends on MTF-1, a unique zinc finger transcription factor conserved from insects to human (for review13 ). When cytosolic concentrations of transition metals are low, MTF-1 resides in the cytosol. Increased cytosolic levels of transition metals induce the translocation of MTF-1 into the nucleus where it binds to the promoters of target genes activating transcription. We examined the ability of different transition metals to induce the translocation of cytosolic MTF-1 into the nucleus. NIH3T3 cells were transfected with a construct expressing MTF-1-Flag and the subcellular distribution of the protein was examined by immunofluorescence. MTF-1-Flag showed a 100% cytoplasmic localization in control cells and cells exposed to 100μM iron, 40%-50% in the cytoplasm in cells exposed to 100μM manganese or copper for 2 hours (Figure 2A). In contrast, MTF-1-Flag was found in 100% of the nuclei of cells exposed to 100μM zinc, cadmium, and cobalt for 2 hours.
Zinc, cobalt, and cadmium induce MTF-1 translocation into the nucleus and increased MT-1 mRNA. (A) NIH3T3 cells were transfected with pcDNA-mouse-MTF-1-Flag. Two days later, the cells were exposed to the specified transition metals (100μM) for 2 hours. Cells were fixed and MTF1-Flag localization detected by indirect immunofluorescence as described in “Immunofluorescence and Microscopy.” The right hand panel is a quantification of the imaging where > 200 cells were examined for nuclear or cytosolic MTF-1-Flag. (B) Macrophages were exposed for 16 hours to serial concentrations of transition metals ranging from 0 to 200μM, total RNA was harvested and semiquantitative RT-PCR were performed for MT-1 and actin mRNA. All experiments were performed a minimum of 3 times.
Zinc, cobalt, and cadmium induce MTF-1 translocation into the nucleus and increased MT-1 mRNA. (A) NIH3T3 cells were transfected with pcDNA-mouse-MTF-1-Flag. Two days later, the cells were exposed to the specified transition metals (100μM) for 2 hours. Cells were fixed and MTF1-Flag localization detected by indirect immunofluorescence as described in “Immunofluorescence and Microscopy.” The right hand panel is a quantification of the imaging where > 200 cells were examined for nuclear or cytosolic MTF-1-Flag. (B) Macrophages were exposed for 16 hours to serial concentrations of transition metals ranging from 0 to 200μM, total RNA was harvested and semiquantitative RT-PCR were performed for MT-1 and actin mRNA. All experiments were performed a minimum of 3 times.
We then examined the effect of these metals on the expression of MT-1, a well-known target gene of MTF-1.14 Exposure of cells to different concentrations of zinc, cadmium, or cobalt for 16 hours led to increased levels of MT-1 mRNA (Figure 2B). In contrast, iron, manganese, or copper exposure had no effect on MT-1 expression.
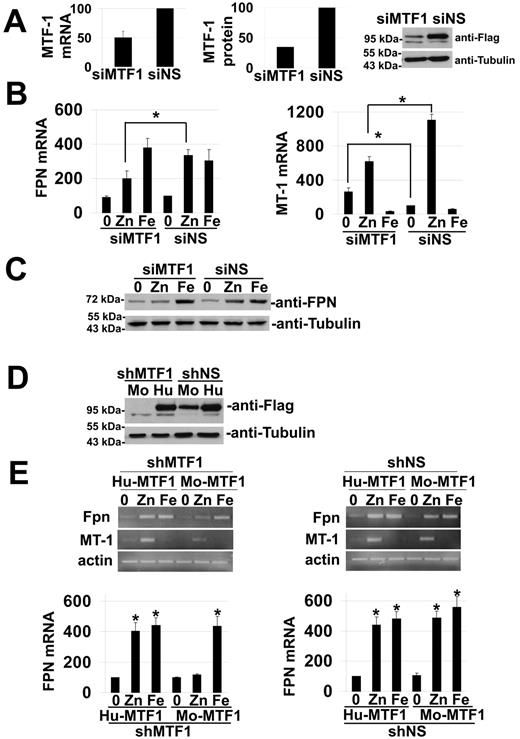
We focused our study on zinc regulation of FPN1 expression because MTF-1 serves as a potent zinc sensor that responds to changes in free cytosolic zinc concentrations.8,15 To confirm that MTF-1 acts as a transcription factor for FPN1 we studied transition metal-induced FPN1 mRNA expression in cells treated with siRNA oligonucleotide pool specific for MTF-1. The commercially available MTF-1 antibody did not detect mouse MTF-1; therefore, we obtained an MTF-1-Flag expression construct, transfected it or control vector into NIH3T3, followed by silencing with nonspecific or mouse specific siRNA oligonucleotide pools and examined the levels of MTF-1 mRNA or MTF-1-Flag using Western blot. MTF-1-Flag protein levels were decreased by approximately 65% (Figure 3A). Reduced levels of MTF-1-Flag resulted in decreased MT-1 and FPN1 mRNA steady state levels (45% and 35% respectively) in response to zinc compared with cells transfected with nonspecific RNAi pools (Figure 3B). We note that MT-1 mRNA levels were elevated in MTF-1 silenced cells. While induction of MT-1 transcription is Drosophila is dependent on MTF-1,16 a similar dependency has not been reported in mammalian cells. We examined the levels of Fpn in response to zinc or iron in cells treated with MTF-1 specific or nonspecific siRNA oligonucleotide pools. Fpn levels increased in response to iron or zinc (Figure 3C). In MTF-1 silenced cells, the zinc-induced increase in Fpn levels was lost suggesting that regulation of Fpn expression by zinc was dependent on MTF-1.
The transcription factor MTF-1 is required for zinc-induced increase in FPN1 transcription. (A) NIH3T3 cells transfected with pcDNA-mouse-MTF-1-Flag were incubated with nonspecific (siNS) or MTF-1 specific (si MTF1) oligonucleotides. Seventy-two hours after silencing, cells were lysed and MTF-1 mRNA analyzed (left panel) or MTF-1-Flag and tubulin levels determined by Western blot analysis (right panel). The middle panel is a quantification of the Western blot. (B) Macrophages incubated with nonspecific (siNS) or MTF-1 specific (siMTF1) oligonucleotides as in panel A for 48 hours were exposed to 100μM iron (FAC) or 50μM zinc overnight at 37°C. RNA was harvested and semi quantitative RT-PCR was performed for MTF-1, MT-1 and actin mRNA. The data were quantified and normalized to actin with 100% FPN1 or MT-1 mRNA representing the mRNA levels in the non specific (siNS) samples without iron or zinc. The data represent the average of 3 independent experiments. Asterisks in the figures with lines identify columns being compared and show a P value < .05 as determined by Student t test. (C) NIH3T3 cells treated as in panel B were lysed and Fpn and tubulin levels determined by Western blot analysis. (D) NIH3T3 cells expressing either mouse or human MTF1-Flag (Mo and Hu, respectively) were transfected with non specific shRNA (shNS) or sh-mouse MTF1 (shMTF1) containing vectors. Ninety-six hours after shRNA transfection cells were harvested, lysed as described in “Immunofluorescence and microscopy,” and the levels of MTF-1-Flag and tubulin determined by Western blot. (E) NIH3T3 were transfected with shMTF1 or shNS and 72 hours later with either pcDNA-human-MTF-1-Flag (Hu-MTF1) or pcDNA-mouse-MTF-1-Flag (Mo-MTF1). Cells were exposed to 100μM iron (FAC) or 50μM zinc overnight. RNA was harvested and semi quantitative RT-PCR were performed for FPN1, MT-1 and actin mRNA. Histograms show gel quantification of PCR band intensity for 3 independent experiments. Asterisks represent P values less than .05 as determined by Student t test.
The transcription factor MTF-1 is required for zinc-induced increase in FPN1 transcription. (A) NIH3T3 cells transfected with pcDNA-mouse-MTF-1-Flag were incubated with nonspecific (siNS) or MTF-1 specific (si MTF1) oligonucleotides. Seventy-two hours after silencing, cells were lysed and MTF-1 mRNA analyzed (left panel) or MTF-1-Flag and tubulin levels determined by Western blot analysis (right panel). The middle panel is a quantification of the Western blot. (B) Macrophages incubated with nonspecific (siNS) or MTF-1 specific (siMTF1) oligonucleotides as in panel A for 48 hours were exposed to 100μM iron (FAC) or 50μM zinc overnight at 37°C. RNA was harvested and semi quantitative RT-PCR was performed for MTF-1, MT-1 and actin mRNA. The data were quantified and normalized to actin with 100% FPN1 or MT-1 mRNA representing the mRNA levels in the non specific (siNS) samples without iron or zinc. The data represent the average of 3 independent experiments. Asterisks in the figures with lines identify columns being compared and show a P value < .05 as determined by Student t test. (C) NIH3T3 cells treated as in panel B were lysed and Fpn and tubulin levels determined by Western blot analysis. (D) NIH3T3 cells expressing either mouse or human MTF1-Flag (Mo and Hu, respectively) were transfected with non specific shRNA (shNS) or sh-mouse MTF1 (shMTF1) containing vectors. Ninety-six hours after shRNA transfection cells were harvested, lysed as described in “Immunofluorescence and microscopy,” and the levels of MTF-1-Flag and tubulin determined by Western blot. (E) NIH3T3 were transfected with shMTF1 or shNS and 72 hours later with either pcDNA-human-MTF-1-Flag (Hu-MTF1) or pcDNA-mouse-MTF-1-Flag (Mo-MTF1). Cells were exposed to 100μM iron (FAC) or 50μM zinc overnight. RNA was harvested and semi quantitative RT-PCR were performed for FPN1, MT-1 and actin mRNA. Histograms show gel quantification of PCR band intensity for 3 independent experiments. Asterisks represent P values less than .05 as determined by Student t test.
To ascertain the role of MTF-1 in FPN1 mRNA expression we rescued the knockdown phenotype using an RNAi resistant MTF-1. The MTF-1 siRNA oligonucleotide pool targeted both human and mouse MTF-1 (data not shown); therefore, we designed an shRNA specifically targeting mouse MTF-1 and rescued silenced cells by transforming them with a human MTF-1-Flag resistant to this shRNA. Western blot analysis of MTF-1-Flag showed that mouse MTF-1-Flag was sensitive to the silencing shRNA whereas human MTF-1-Flag was resistant to the silencing construct (Figure 3D). Silencing of MTF-1 led to reduced MT-1 induction, showing the efficacy of the silencing procedure. Cells expressing a nonspecific shRNA and either mouse or human MTF-1-Flag showed an up-regulation of FPN1 mRNA after incubation with zinc or iron (Figure 3E). In cells expressing the specific shRNA against mouse MTF-1, we observed that FPN1 mRNA levels were still increased upon exposure to iron but were not increased upon exposure to zinc. Transformation of cells with the human silencing resistant construct of MTF-1 restored zinc regulation of FPN1 transcription. These results show that MTF-1 is required for zinc induction of FPN1 mRNA but is not required for iron induction of FPN1 mRNA.
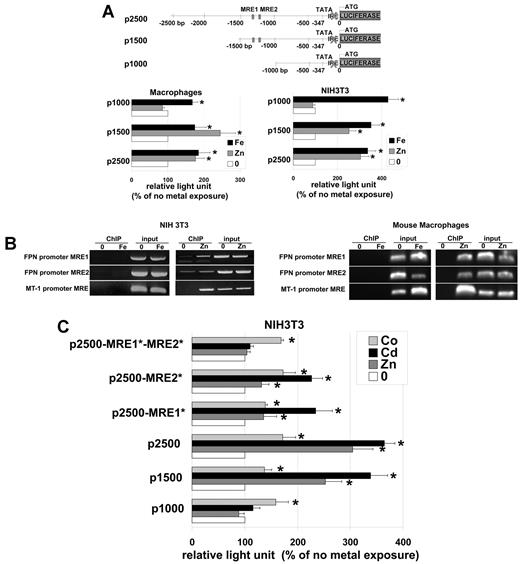
MTF-1 targets gene expression by binding to MREs.17,18 The minimal MRE consensus sequence 5′-TGCRCnCGGCCC-3′ is highly conserved. Using in silico analysis, we determined that the human, rat and mouse FPN1 promoters have 6, 4, and 2 putative MREs, respectively. In the mouse FPN1 promoter the 2 putative MREs are located 120 bp apart at the positions −990 (tgcaccc) and −879 (tgcactc on the reverse strand) from the TATAA box (supplemental Figure 1A, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). MREs are also found in the promoter regions of human and rat FPN. Mice have 2 MREs, whereas there are 5 to 6 in the human promoter and 4 in the rat promoter (supplemental Figure 1B). We cloned the mouse FPN1 promoter lacking the IRE in the 5′ untranslated region into a luciferase reporter vector and generated 3 FPN1 promoter constructs containing approximately 2500, 1500, or 1000 bp upstream of the start (Figure 4A). These constructs were transformed into mouse bone marrow macrophages or NIH3T3 cells and the effects of metals on reporter expression were determined. Luciferase assays on macrophages and NIH3T3 cells showed that addition of iron activated luciferase synthesis from all 3 constructs. In contrast, zinc increased luciferase production in cells transformed with the 2 longest promoters (2500 and 1500 bp) but not in cells transformed with the 1000-bp construct. The 1000-bp construct lacks the putative MREs (Figure 4A), suggesting they are required for zinc-mediated FPN1 transcriptional activation.
The role of MREs in the mouse FPN1 promoter. (A) Diagram of the mouse FPN1 promoter shows 2 putative MREs at positions −990 (tgcaccc) and −879 (tgcactc on the reverse strand) from the TATAA box. Plasmids were constructed containing portions of FPN1 promoter region in front of a Firefly luciferase reporter. The areas of the FPN1 promoter contained regions −2378, −1154, or −623 to the TATAA box lacking the IRE region (called p2500, p1500, p1000). Macrophages or NIH3T3 cells were transfected with p2500, p1500, and p1000 plasmids and subsequently treated with 100μM zinc or 100μM FAC. Cells were also transfected with a Renilla luciferase containing plasmid to control for transfection efficiency. Sixteen hours later, cells were assayed for luciferase activity. Data presented are expressed as percentage of Firefly luciferase light units that have been normalized using the Renilla luciferase. (B) Chromatin immunoprecipitations were performed on MTF-1-Flag expressing NIH3T3 cells exposed to zinc, iron or no metal using antibodies against Flag as described in “Chromatin immunoprecipitation.” (C) The putative MREs in the p2500 plasmid were mutagenized, the plasmid transfected into cells, and cells incubated with cobalt or cadmium and luciferase assayed after 16 hours. All experiments were performed a minimum of 3 times.
The role of MREs in the mouse FPN1 promoter. (A) Diagram of the mouse FPN1 promoter shows 2 putative MREs at positions −990 (tgcaccc) and −879 (tgcactc on the reverse strand) from the TATAA box. Plasmids were constructed containing portions of FPN1 promoter region in front of a Firefly luciferase reporter. The areas of the FPN1 promoter contained regions −2378, −1154, or −623 to the TATAA box lacking the IRE region (called p2500, p1500, p1000). Macrophages or NIH3T3 cells were transfected with p2500, p1500, and p1000 plasmids and subsequently treated with 100μM zinc or 100μM FAC. Cells were also transfected with a Renilla luciferase containing plasmid to control for transfection efficiency. Sixteen hours later, cells were assayed for luciferase activity. Data presented are expressed as percentage of Firefly luciferase light units that have been normalized using the Renilla luciferase. (B) Chromatin immunoprecipitations were performed on MTF-1-Flag expressing NIH3T3 cells exposed to zinc, iron or no metal using antibodies against Flag as described in “Chromatin immunoprecipitation.” (C) The putative MREs in the p2500 plasmid were mutagenized, the plasmid transfected into cells, and cells incubated with cobalt or cadmium and luciferase assayed after 16 hours. All experiments were performed a minimum of 3 times.
To ascertain the binding of MTF-1 on both MREs, we did chromatin immunoprecipitation using MTF-1-Flag as bait. Addition of iron did not lead to binding of MTF-1 to either of the MREs on the FPN1 promoter or to the MRE in the MT-1 promoter in NIH3T3 cells and in bone marrow-derived macrophages (Figure 4B). In contrast, addition of zinc to cells resulted in the binding of MTF-1 to both of the MREs in FPN1 and to the MRE in the MT-1 promoter. In these experiments we have consistently seen binding of MTF-1 to one of the MREs in the NIH3T3 FPN promoter in the absence of zinc (n = 3). In contrast, we have not seen a similar binding of MTF-1 to the MRE of the macrophage FPN promoter in the absence of zinc. Currently, we have no explanation for this difference.
To examine the contribution of each of the FPN1 MREs we generated reporter plasmids in which each of the MRE were separately mutagenized and performed a luciferase reporter assay after stimulation by zinc, cobalt, or cadmium (Figure 4C). Mutations of each MRE in the FPN1 promoter diminished the metal-induced luciferase activity and simultaneous mutations in both MREs completely abolished zinc and cadmium-induced luciferase activity but not cobalt. These data show that both MREs are required for complete activation of FPN1 transcription in response to zinc and cadmium.
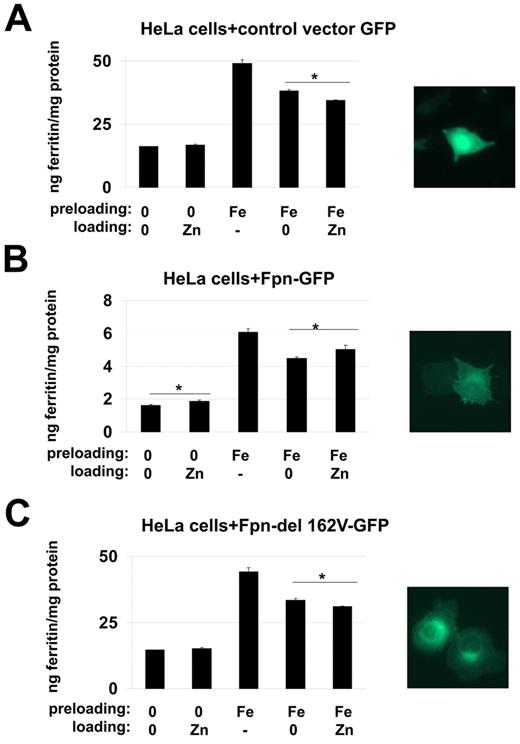
A critical question is why FPN1 transcription responds to zinc. Fpn is a well-characterized iron exporter, though it has been published that Fpn orthologues in Arabidopsis Thaliana can also transport cobalt.19 We explored the possibility that mammalian Fpn could export zinc. Cells were exposed to iron for 16 hours, the medium was removed, and the cells were extensively washed. Cells were incubated in the presence or absence of zinc and the level of ferritin determined. Expression of Fpn results in iron export and decreased cytosolic iron, which is assayed by the reduction of ferritin.20 If zinc competes with iron for Fpn-mediated transport, then cells exposed to zinc will have higher levels of cytosolic iron and higher levels of ferritin than cells not incubated with zinc. In cells transformed with a vector control, iron-loaded cells expressed less ferritin in the presence of zinc than with a zinc-free medium (Figure 5A). When cells expressed a cytomegalovirus (CMV)–regulated Fpn, which is not regulated by iron or zinc, the basal ferritin concentration was 10-fold lower than with a control vector. Addition of iron increased the amount of ferritin 3-fold. Addition of zinc led to an increase in the amount of ferritin compared with iron-loaded cells not exposed to added zinc. In contrast, there was no increase in ferritin when cells were transfected with a mutant Fpn [Fpn(V162-GFP)], which is unable to export metals because it is not targeted to the cell surface.21 Our explanation for the decrease in ferritin levels in cells not expressing wild-type Fpn is that zinc induces endogenous Fpn expression resulting in iron export and concomitantly lowering ferritin. In cells transfected with wild-type Fpn, the level of expressed Fpn is so high that zinc-induced endogenous Fpn has little effect over the CMV-expressed Fpn. Zinc, by competing with iron for transport, reduces iron transport and thus leads to a relative increase (or retention) in ferritin.
Fpn transports zinc. HeLa cells were transfected with (A) pCMVeGFP, (B) pCMVFpn-GFP or (C) pCMVFpn162V-GFP. Twenty-four hours after transfection, cells were incubated (preloading) without or with 100μM iron (FAC) for 16 hours (-). The cells were extensively washed and incubated (loading) without (0) or with 100μM zinc at 37°C for 6 hours. Cells were washed, lysed as described in “Immunofluorescence and microscopy,” and ferritin levels determined by ELISA. The asterisks represent P values less than .05 as determined by Student t test.
Fpn transports zinc. HeLa cells were transfected with (A) pCMVeGFP, (B) pCMVFpn-GFP or (C) pCMVFpn162V-GFP. Twenty-four hours after transfection, cells were incubated (preloading) without or with 100μM iron (FAC) for 16 hours (-). The cells were extensively washed and incubated (loading) without (0) or with 100μM zinc at 37°C for 6 hours. Cells were washed, lysed as described in “Immunofluorescence and microscopy,” and ferritin levels determined by ELISA. The asterisks represent P values less than .05 as determined by Student t test.
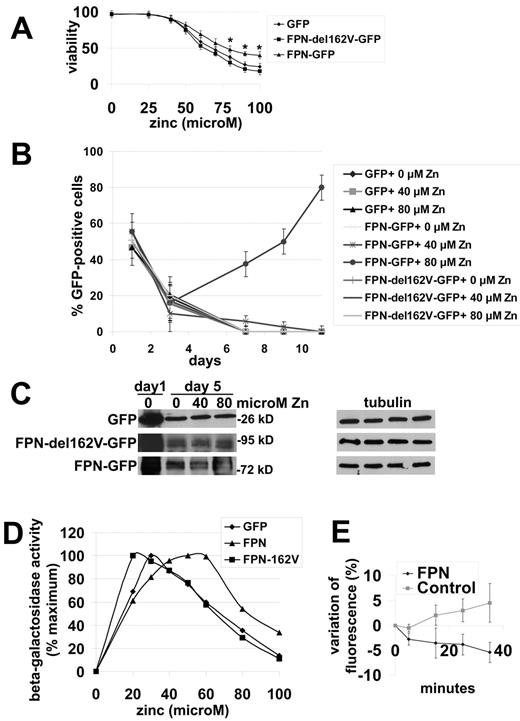
One interpretation of this result is that zinc by competing with iron for Fpn-mediated export gives rise to increased cytosolic iron and therefore increased ferritin synthesis. To further examine this hypothesis, we took advantage of a BHK cell line that is highly sensitive to zinc toxicity. This cell line (clone 3286-8-8) lacks ZnT-1, the zinc exporter. Incubation of cells with 80μM zinc leads to complete cell death.22 To determine whether overexpression of Fpn would suppress zinc toxicity, we transfected these cells with a GFP expressing vector, Fpn-GFP vector or a Fpn(162V)-GFP vector, and incubated these cells in different concentrations of zinc for 11 days. Cells expressing Fpn-GFP showed greater resistance to zinc as ascertained by trypan blue staining (Figure 6A). Transfection efficiency was approximately 50%, based on the number of GFP-positive cells 1 day after transfection. After 3 days, 15% of control GFP transformed cells and Fpn-GFP transformed cells were GFP-positive regardless of zinc concentration (Figure 6B). In cells incubated with 0μM or 40μM zinc for 7 days, there were no detectable GFP positive cells suggesting that there was no selective pressure to maintain the GFP or Fpn-GFP plasmids. In cells incubated with 80μM zinc there was an increase in the percentage of Fpn-GFP expressing cells. At day 11, 80% of still viable cells showed Fpn-GFP expression. A plausible explanation for these results is that at higher zinc concentration (80μM zinc) Fpn-GFP expressing cells can export enough zinc to survive, whereas, the untransfected or GFP expressing cells die due to zinc toxicity. An alternative explanation is that cells do not express the different constructs at equal levels. To determine whether expression was equivalent we performed Western blot analysis on transfected cells. As shown in Figure 6C, similar levels of Fpn-GFP were found, indicating that alterations in transfection frequency or Fpn expression levels cannot account for increased resistance due to Fpn.
Fpn-GFP expression protects cells against zinc toxicity. (A) BHK 3286-8-8 cells were transfected with a plasmid expressing GFP, Fpn-GFP, or Fpn162V-GFP under the control of the CMV promoter. The day after transfection, cells were exposed to different concentrations of zinc for 24 hours and cell viability assessed by trypan blue exclusion. The data are expressed as percent viable. (B) Cells as in panel A were exposed to 0, 40, and 80μM zinc and the percentage of GFP positive cells were determined over time. The data are expressed as the percentage of GFP-positive cells. (C) Cells as in panel A were harvested at days 1 and 5, cells lysed, and GFP and tubulin levels assessed by Western blot analysis. (D) Cells as in panel A were exposed to different concentrations of zinc to induced MRE-b-galactosidase expression, cells harvested, and b-galactosidase activity measured. The data are expressed as the percentage maximum. (E) BHK 3286-8-8 cells were transfected with pCMV-Fpn-Flag or empty vector, exposed to 70μM zinc for 2 hours, extensively washed, incubated with 1μM FluoZin-3AM for 30 minutes, and extensively washed. FluoZin-3AM fluorescence was detected with a BioTek plate reader fluorimeter at 485/520 nm (excitation/emission) and the data are expressed as the percentage of variation in FluoZin fluorescence (F) over time with the initial FluoZin fluorescence in control and Fpn-Flag expressing cells at zero time (F0). All experiments were performed a minimum of 3 times.
Fpn-GFP expression protects cells against zinc toxicity. (A) BHK 3286-8-8 cells were transfected with a plasmid expressing GFP, Fpn-GFP, or Fpn162V-GFP under the control of the CMV promoter. The day after transfection, cells were exposed to different concentrations of zinc for 24 hours and cell viability assessed by trypan blue exclusion. The data are expressed as percent viable. (B) Cells as in panel A were exposed to 0, 40, and 80μM zinc and the percentage of GFP positive cells were determined over time. The data are expressed as the percentage of GFP-positive cells. (C) Cells as in panel A were harvested at days 1 and 5, cells lysed, and GFP and tubulin levels assessed by Western blot analysis. (D) Cells as in panel A were exposed to different concentrations of zinc to induced MRE-b-galactosidase expression, cells harvested, and b-galactosidase activity measured. The data are expressed as the percentage maximum. (E) BHK 3286-8-8 cells were transfected with pCMV-Fpn-Flag or empty vector, exposed to 70μM zinc for 2 hours, extensively washed, incubated with 1μM FluoZin-3AM for 30 minutes, and extensively washed. FluoZin-3AM fluorescence was detected with a BioTek plate reader fluorimeter at 485/520 nm (excitation/emission) and the data are expressed as the percentage of variation in FluoZin fluorescence (F) over time with the initial FluoZin fluorescence in control and Fpn-Flag expressing cells at zero time (F0). All experiments were performed a minimum of 3 times.
The BHK cell line clone 3286-8-8 contains a stably integrated MRE-βGeo reporter that responds to zinc leading to increased β-galactosidase activity. To determine whether Fpn can export zinc we transfected cells with a control vector (GFP), Fpn-GFP or Fpn(162V)-GFP, and measured β-galactosidase activity. Addition of zinc resulted in increased β-galactosidase activity and cells expressing either GFP or Fpn(162V)-GFP showed similar zinc dependency curves (Figure 6D). In contrast, higher zinc concentrations were required to induce maximal activity in cells expressing Fpn-GFP. At higher zinc concentrations, β-galactosidase activity decreases reflecting cell mortality.22 In cells expressing Fpn-GFP the decrease in β-galactosidase occurred at higher concentrations of zinc than in control cells, supporting the hypothesis that Fpn protects cells from zinc toxicity by lowering intracellular zinc. We confirmed that Fpn decreased intracellular zinc using FluoZin-3AM. Within cells the reagent is cleaved by nonspecific esterases leading to the accumulation of FluoZin which by binding zinc fluoresces. Cells incubated in the presence of 70μM zinc showed an increase in zinc-associated fluorescence. In the absence of external zinc there is a continued increase in FluoZin fluorescence that we believe is the result of the movement of stored zinc into the cytosol. In contrast, cells expressing Fpn-Flag showed a decrease in FluoZin fluorescence, which is consistent with a reduction in intracellular zinc (Figure 6E). This result demonstrates that Fpn is able to transport zinc.
Discussion
Regulation of Fpn at the translational and posttranslational level is well described,23 but little is known about transcriptional regulation of Fpn. Previous studies have shown that FPN1 mRNA is increased in vivo in response to iron deficiency anemia and hereditary hemochromatosis24,25 but is unaffected by dietary iron loading.24 Studies determined that other transition metals could induce increased FPN1 transcription. There appears to be cell type variability in the induction of FPN1 transcription in response to specific metals. FPN1 mRNA is increased in J774 cells in response to copper but not zinc or manganese.26 In Caco-2 cells and as shown here in cultured mouse bone marrow macrophages, FPN1 mRNA levels increase in response to a wide variety of transition metals including zinc, copper, manganese, cobalt, and cadmium.
Cells change FPN1 mRNA levels in response to these metals, however, there is a clear difference in the mechanisms that underlie this response. Our data indicate that zinc and cadmium induce FPN1 transcription through the action of MFT-1. MTF-1 targets genes coping with heavy metal loading such as MT-1 or the zinc efflux transporter Znt1 but also the hypoxic/anoxic induction of the gene for placental growth factor27 and may be involved in tumor development.28 MTF-1 can also mediate the induction of MT genes in response to other stress situations, such as oxidative stress and hypoxia. We have determined that MTF-1 is important in the zinc-mediated induction of FPN1 mRNA. We have shown that MTF-1 binds to the FPN1 promoter in the presence of zinc and that mutagenesis of the 2 MREs in the FPN1 promoter abolishes the MTF-1 zinc responsiveness. We further demonstrated that cadmium induced FPN1 transcription in an MRE-dependent manner. Recently, it was shown that cadmium induction of FPN1 transcription was dependent on the generation of oxidative radicals.29 This observation is consistent with our data and with the established role of cadmium in induction of MTF-1 responsive genes. Cadmium can directly displace zinc from metallothionein or damage metallothionein by generating oxidative radicals that would act on metallothionein and release zinc.30 Released zinc then binds to MTF-1 inducing gene transcription. Our data rule out a role for MTF-1 in iron or copper-mediated FPN1 transcription, suggesting the existence of other yet to be identified transcription factors.
We examined the function of zinc induction of Fpn. Many studies show a connection between zinc and iron metabolism. In various systems, zinc is able to antagonize the catalytic properties of the redox-active transition metals iron and copper.10,31 A diet deficient in zinc can result in the accumulation of iron in numerous tissues, including testes, liver, kidney, and spleen, as well as in fetuses of zinc-deficient dams.32,–34 High concentrations of iron can inhibit zinc absorption35 and likewise high zinc content can reduce iron absorption although one study ascribed decreased iron transport to changes in hephaestin.36 In contrast, addition of large amounts of zinc to the diet of rats can retard growth, lower their hemoglobin levels and reduce storage of iron.37 Zinc supplementation in hepatoma cells can alter expression of the iron transporter divalent metal transporter 1 (DMT1) and thereby decrease iron absorption.38 The interaction between zinc and iron can be explained by the broad specificity of metal transporters. Most Fe(II) transporters also transport other transition metals. Notably, DMT1, identified as an intestinal iron transporter, as well as DMT1 family members in other species (eg, SMF1 in yeast), can transport iron, manganese, zinc, cobalt, and copper.39,40 The Fet4 transporter in yeast,41 IRT1 in plants,42 as well as ZIP14 in mammals43 transports other transition metals in addition to iron.
A recent study has shown that Fpn can transport manganese, suggesting that Fpn may accept other metals as substrates.44 Our data show that Fpn has modest zinc transport activity. Zinc transport activity by Fpn may provide an explanation for hypozincemia seen in inflammation. Decreased plasma zinc during inflammation has been ascribed to induction of the zinc and iron transporter Zip14 by cytokines.45 Inflammation leads to increased hepcidin, which by down-regulating Fpn may prevent zinc as well as iron export to plasma. The finding that Fpn can transport other metals provides a rationale for metal induction of FPN1 mRNA, as this induction provides a mechanism to protect cells from broad metal toxicity. Induction of Fpn by transition metals would lead to their export from the cytosol. The induction of a transporter that then regulates the concentration of the inducing agent is a classic example of a negative feedback system.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Richard Palmiter (University of Washington) for the BHK cell line 3286-8-8, Dr G. K. Andrews for the pcDNA mouse-MTF1-Flag vector, and members of the Kaplan laboratory for critically reading the manuscript.
This work was supported by National Institutes of Health grant DK070947 to J.K.
National Institutes of Health
Authorship
Contribution: M.B.T designed and performed research and wrote the paper; D.M.W. analyzed the data and wrote the paper; E.L. performed research; J.K. analyzed the data and wrote the paper; and I.D.D. performed research, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ivana De Domenico, Department of Internal Medicine, School of Medicine, 50 N Medical Dr, University of Utah, Salt Lake City, UT 84132; e-mail: ivana.dedomenico@path.utah.edu; or Jerry Kaplan, Department of Pathology, School of Medicine, 50 N Medical Dr, University of Utah, Salt Lake City, UT 84132; e-mail: jerry.kaplan@path.utah.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal