Abstract
HGAL is a germinal center (GC)–specific gene that negatively regulates lymphocyte motility and whose expression predicts improved survival of patients with diffuse large B-cell lymphoma (DLBCL) and classical Hodgkin lymphoma (cHL). We demonstrate that HGAL serves as a regulator of the RhoA signaling pathway. HGAL enhances activation of RhoA and its down-stream effectors by a novel mechanism – direct binding to the catalytic DH-domain of the RhoA-specific guanine nucleotide exchange factors (RhoGEFs) PDZ-RhoGEF and LARG that stimulate the GDP-GTP exchange rate of RhoA. We delineate the structural domain of HGAL that mediates its interaction with the PDZ-RhoGEF protein. These observations reveal a novel molecular mechanism underlying the inhibitory effects of GC-specific HGAL protein on the motility of GC-derived lymphoma cells. This mechanism may underlie the limited dissemination and better outcome of patients with HGAL-expressing DLBCL and cHL.
Introduction
HGAL (Human Germinal center Associated Lymphoma, also named germinal center-expressed transcript 2 (GCET2), is a novel germinal center (GC)–specific gene identified by gene expression profiling.1,2 HGAL and its murine homolog, M17 protein, are specifically expressed in GC B-lymphocytes.1,3,4 HGAL is also expressed in GC-derived lymphomas and distinguishes biologically distinct subgroups of diffuse large B-cell lymphomas (DLBCL) as well as classic Hodgkin lymphoma (cHL) associated with improved survival.1,5,6 The HGAL gene is located on chromosome 3q13 and encodes a 178-amino acid (aa) protein with 51% identity and 62% similarity to the murine M17 protein. Studies in M17 knockout mice revealed that this protein is dispensable for GC formation, immunoglobulin somatic hypermutation, class-switch recombination, and for mounting of T cell–dependent antibody responses.7 However, in contrast to their wild-type littermates, M17-deficient mice exhibited reduced-sized Peyer patches.7
HGAL is a cytoplasmatic protein that may also associate with cell membrane.1,3,8 Protein sequence analysis of HGAL and M17 demonstrates presence of an immunoreceptor tyrosine-based activation motif (ITAM), usually implicated in signal transduction in B lymphocytes, suggesting that these proteins have a specific signaling function. We have demonstrated that IL-6 induces phosphorylation of the C-terminal tyrosine residue of the HGAL protein via the Lyn kinase and promotes its relocalization from the cytoplasm to filopodia and podosome-like structures.9 We have reported that HGAL interacts with actin and myosin proteins and inhibits migration of GC B-cells and HGAL-expressing lymphoma cells, thus potentially constraining lymphocytes to the GC9 and inhibiting lymphoma dissemination. However, the molecular mechanism underlying HGAL effects on lymphocyte motility is unknown.
The specific proteins and signaling pathways regulating the shape and motility of GC lymphocytes and lymphoma cells are presently unknown. In the GC, B-lymphocytes are functionally and spatially segregated from extra-GC compartments and also between the light and dark zones of the GC due to limited inter-zonal and inter-compartmental lymphocyte movement.10 Stationary B cells may be observed throughout the light and dark zones, and GC lymphocytes frequently exhibit irregular contours with shifting prominent cytoplasmic processes resulting in polarized shapes, which are usually not observed in largely spherical naive and memory B cells.10-13 This restricted motility of GC lymphocytes as well as alterations of their cellular contours may be necessary for successful completion of the GC reaction.
Cell shape and migration are controlled by dynamic remodeling of the actin cytoskeleton. Reorganization of the actin cytoskeleton is temporally and spatially regulated by Rho family small GTPases.14,15 Rho-family GTPases function as bi-molecular switches by adopting different conformational states in response to binding GTP or GDP. The best-studied members of the family are Rac1, Cdc42 and RhoA, which regulate the formation of focal adhesions and complexes and control formation of filopodia, lamellipodia and membrane ruffling as well as stress fiber formation, respectively.16
GTP-bound RhoA activates several effectors including Rho kinase (ROCK) and citron kinase.17,18 Both kinases induce direct phosphorylation of myosin regulatory light chain (MRLC) at Ser19/Thr1819 that regulates actin-activated Mg-ATPase activity of myosin II. The major phosphorylation site is Ser19, which promotes the interaction of myosin II with actin, assembly of the actomyosin complex and the initiation of contraction. Phosphorylation at both Ser19 and Thr18 further promotes filament assembly. In addition, ROCK induces inhibitory phosphorylation of myosin phosphatase (myosin PPTase) subunit MYPT1 at Thr696 and Thr853,20,21 inhibiting MRLC dephosphorylation and contributing to myosin activation.22,23 These downstream effects of RhoA regulate actomyosin contractility. ROCK also stimulates LIM kinase (LIMK) to phosphorylate cofilin (P-cofilin), thereby inactivating its function.24,25 Activated cofilin severs actin filaments to produce free barbed ends leading to the elongation of newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp 2/3 complex and G-actin resulting from the depolymerization of pointed ends produced by the same severing reaction, thus reorganizing the cytoskeleton and contributing to cell motility. The complex interrelations between the RhoA effectors control actomyosin cytoskeleton and cell motility. Furthermore, RhoA is reported to regulate the transcriptional activation by serum response factor (SRF) and is implicated in oncogenesis and cellular transformation.26,27
Here we demonstrate that HGAL plays an essential function in the physiological activation of the RhoA signaling pathway. HGAL-induced activation of RhoA and its downstream effectors results in inhibition of lymphoma cell motility and induction of transcriptional activation by serum response factor. HGAL-induced activation of RhoA may also regulate normal GC lymphocyte motility. HGAL's effect on RhoA is mediated by its direct interaction with RhoA-specific guanine nucleotide exchange factors (RhoGEFs) PDZ-RhoGEF and LARG that stimulate the GDP-GTP exchange rate. These observations reveal a novel molecular mechanism underlying the inhibitory effects of HGAL on the motility of GC-derived lymphoma cells and may contribute to the favorable outcome of DLBCL and cHL patients whose tumors express high levels of HGAL protein.
Methods
Reagents and antibodies
Mouse monoclonal anti-HGAL antibody was generated in our laboratory, as reported previously.3 Rabbit polyclonal anti-LARG (H-70), rabbit polyclonal anti–PDZ-RhoGEF (H-300), rabbit polyclonal anti-paxillin (H-114), rabbit polyclonal anti-CDC42 (P1), rabbit polyclonal anti-Rac1 (C-14), rabbit polyclonal anti-RhoA (119), mouse monoclonal anti-RhoA (26C4), rabbit polyclonal anti-MYPT1 (H-130), rabbit polyclonal anti–p-MYPT1 (Thr-696), rabbit polyclonal anti–p-MYPT1 (Thr-853), rabbit polyclonal anti–p-cofilin (hSer3), rabbit polyclonal anti-phosphorylated myosin regulatory light chain (p-MRLC; Thr18) antibodies were from Santa Cruz Biotechnology Inc. Rabbit polyclonal anti-MRLC, rabbit polyclonal anti–p-MRLC (Ser19), rabbit polyclonal anti–p-MRLC (Thr18/Ser19), rabbit polyclonal anti-cofilin antibodies were from Cell Signaling Technology. Monoclonal anti-V5, Alexa Fluor 488 goat anti–mouse IgG, Alexa Fluor 555 goat anti–mouse IgG, Alexa Fluor 647 goat anti–mouse IgG, Alexa Fluor 488 goat anti–rabbit IgG, Alexa Fluor 555 goat anti–rabbit IgG, Alexa Fluor 647 goat anti–rabbit IgG antibodies were from Invitrogen. Mouse monoclonal anti-phosphotyrosine (PY20), mouse monoclonal anti–β-actin antibodies were from Sigma-Aldrich; rabbit polyclonal anti–myosin IIa and IIb antibodies were from CovΛnce; anti–human IgM antibody was from Biosource. Rhodamine labeled phalloidin and DAPI were from Molecular Probes (Invitrogen). Recombinant human IL-6 was purchased from R&D Systems. G418 was purchased from GIBCO (Invitrogen-GIBCO). Human fibronectin was purchased from BD Biosciences; Latrunculin B, Y-27 632 and sodium lysophosphatidic acid were from BIOMOL Research Laboratories Inc. ON-TARGETplus SMARTpool siRNA for HGAL and PDZ-RhoGEF, as well as ON-TARGETplus Non-Targeting pool siRNA were purchased from Dharmacon RNA Technologies. A different siRNA for PDZ-RhoGEF and siRNA for LARG was from Santa Cruz Biotechnology Inc; siRNA for RhoA was from Santa Cruz Biotechnology Inc.
Plasmids and plasmid constructs
The full-length wild-type human HGAL cDNA expression plasmid (pcDNA3.1-HGAL) and the HGAL truncated mutant pcDNA3.1-(1-118)HGAL encoding the HGAL cDNA amino acids 1-118 were previously reported.9 The pCMV6-XL5-RhoA (Human cDNA clone, accession No:NM_001664.2) and pCMV6-PDZ-RhoGEF-Myc/DDK (Human cDNA clone, accession No:NM_198236.1) were purchased from OriGene Technologies Inc; pT-ADV-LARG plasmid was a generous gift of Dr Alexander Belyavsky (Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, Moscow, Russia). These plasmids were used as templates for amplification and cloning, using standard techniques, of the full-length HGAL cDNAs tagged with FLAG and cMyc (HGAL-FLAG-cMyc), HGAL PDZ binding motif truncated mutant (HGALΔPDZ), pTRX-HGAL, full-length PDZ-RhoGEF and its truncated mutants (128-1562)PDZ-RhoGEF, (489-1562)PDZ-RhoGEF, (597-1562)PDZ-RhoGEF, (955-1562)PDZ-RhoGEF, (1-488)PDZ-RhoGEF), (738-953)PDZ-RhoGEF, pGEX-4T-2-(597-1080)PDZ-RhoGEF, full-length LARG and full-length RhoA. The RhoAQ63L vector was generated using the QuickChange Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer's instructions. Primers used for amplification of templates are shown in supplemental Table 1 (available on the Blood .Web site; see the Supplemental Materials link at the top of the online article).
Cells and cell culture
Human non-Hodgkin lymphoma (NHL) cell lines VAL, Raji, SUDHL-6, MC116 and cervical cancer cell line HeLa were cultured, as previously reported9 and described in supplemental Methods. For fibronectin (FN) stimulation, plates (Corning Incorporated), were precoated overnight at 4°C with a 50 μg/mL solution of FN in phosphate-buffered saline (PBS, Ca2+, Mg2+ free, 1 mL/35 mm dish). For sodium lysophosphatidic acid (LPA) treatment, HeLa, VAL or Raji cells grown in serum-free culture medium for 8 hour were treated with LPA (1μg/mL) for 3 minutes.
Peripheral blood B lymphocytes and human tonsil CD77+ GC centroblasts and centrocytes (referred as GC B cells) were isolated, as previously reported.28 Informed consent was obtained in accordance with the Declaration of Helsinki from all patients, and tissue collection was approved by the University of Miami institutional review committee.
Cell transfection and gene silencing
Western blotting, immunoprecipitation and immunofluorescence microscopy
Western blotting and immunoprecipitation were performed as previously reported29 and described in supplemental Methods. The subcellular localization of HGAL, PDZ-RhoGEF, LARG, pTyr, Paxillin, and actin was assessed by confocal immunofluorescence and interference reflection contrast microscopy, as reported previously29 and described in supplemental Methods.
F-actin, RhoA, CDC42 and Rac1 activation, Rho-kinase, RhoA guanidine nucleotide exchange and chemotaxis assays
F-actin assay in lymphoma cell was performed as described by Vicente-Manzanares et al.30 Details are described in supplemental Methods. RhoA activation was measured by pull down assay (Cytoskeleton Inc) or G-LISA RhoA Activation Assay Biochem kit (Cytoskeleton Inc). CDC42 and Rac1 pull down assays (EZ-Detect CDC42 and Rac1 activation kits; Pierce Biotechnology) were performed according to the manufactures' instructions using 1000 μg of fresh cell lysate per assay. Rho-kinase assay was performed in triplicates according to the manufacture's protocol (CyeLex Co Ltd). Rho-kinase inhibitor Y-27 632 at final concentration of .1mM was used as a control. RhoA N-methylanthraniloyl (mant-GTP) exchange assay was performed according to the manufacturer's instructions (Cytoskeleton Inc).
Chemotaxis assays were performed in 24-well plates (Nalge Nunc International) containing 5.0 μm porous polycarbonate membranes inserts (Corning Incorporated Life Sciences) as was previously reported9 and described in supplemental Methods.
Luciferase reporter assays
HeLa cell, VAL and Raji cells were co-transfected in triplicate with RhoA modulated serum response factor (SRF)–driven luciferase reporter construct pSRE-Luc (4 μg; Stratagene), the constitutively active Renilla reniformis luciferase-producing vector pRL-TK (40 ng; Promega), and one of the expression plasmids pcDNA3.1-HGAL, pcDNA3.1-HGALΔPDZ or pcDNA3.1 control (4 μg each) or ON-TARGETplus SMARTpool HGAL siRNA or ON-TARGETplus Non-Targeting pool siRNA (2 μg each) using the Amaxa Nucleofector Kit. Firefly (Photinus pyralis) and R reniformis luciferase activities were detected with the Dual Luciferase assay kit according to manufacturer's instructions (Promega). Data are presented as average plus or minus SEM.
Transformations and focus-formation assay
Three days after transfection, the NIH3T3 cells were split into 6 × 100-mm plates, 3 plates containing Dulbecco modified Eagle medium (DMEM) with 5% FCS and 3 plates containing DMEM medium with 10%FCS supplemented with 1.2 mg/mL G418. The cell cultures were fed every 3 days with fresh medium. After 3 weeks, the cells cultured in the DMEM medium with 5% FCS were stained with 0.5% crystal violet in 25% methanol for 10 minutes and then washed with methanol. The foci images were taken on FluorChem FC2 (Alpha Innotech Corporation) and foci number was counted with AlphaView V.1.3.0 software (Alpha Innotech). The cells cultured in the DMEM medium with 10% FCS supplemented with G418 were used to assess transfection efficiency. Because all the transfected plasmids yielded comparable numbers of G418-resistant colonies, differences in the focus-forming ability were attributed to the biological effects of specific plasmids expressing distinct proteins.
Statistical analysis
A 2-tailed Student t test was used and a P value less than .05 was considered statistically significant.
Results
HGAL activates RhoA but not CDC42 and Rac1
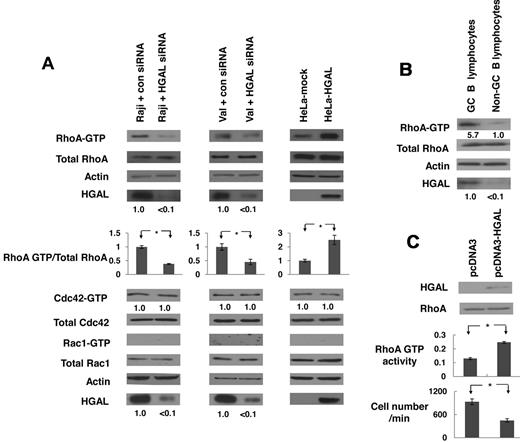
Rho-family members RhoA, Cdc42 and Rac1 are key regulators of cell shape, adhesion, migration and polarization.16 Because GC lymphocytes exhibit restricted cell motility and irregular cell contours, and because GC-specific protein HGAL was demonstrated to inhibit lymphocyte migration and to induce bleb-like membrane protrusions,9,10 we investigated whether RhoA, Cdc42 and Rac1 are affected by the presence or absence of HGAL. GTP-bound forms of RhoA, Cdc42 and Rac1 were observed in cells exposed to serum, but not in growth factors-deprived (starved) cells (data not shown). Starved HGAL-expressing Raji and VAL lymphoma cells, transfected with either HGAL siRNA or control siRNA, were plated on FN (Figure 1) or stimulated with LPA (supplemental Figure 1). In both Raji and VAL cells, knockdown of HGAL expression markedly decreased levels of active GTP-bound RhoA, but not of GTP-bound Cdc42 or Rac1. Ectopic expression of HGAL in HeLa cells markedly increased levels of GTP-bound RhoA upon exposure to FN (Figure 1A) and LPA (supplemental Figure 1), while not affecting levels of GTP-bound Cdc42 and Rac1. These results indicate that HGAL specifically increases levels of active GTP-bound RhoA upon exposure to FN or LPA stimulation. Concordantly, markedly higher levels of GTP-bound RhoA were observed in HGAL expressing CD77+ GC B cells enriched from normal tonsils compared with CD77– cells expressing low levels of HGAL (Figure 1B). Ectopic expression of HGAL in normal human peripheral blood B-lymphocytes stimulated with LPA enhanced levels of GTP-bound RhoA (Figure 1C) and inhibited chemotaxis in response to IL-6 (Figure 1C) and SDF1 (supplemental Figure 2).
HGAL induces RhoA activation. (A) Raji and VAL lymphoma cells, transfected with siRNA for HGAL or scrambled control siRNA 48 hours before the experiments, and HeLa cells stably transfected with pcDNA3.1-HGAL or pcDNA3.1-mock plasmids, were starved for 8 hours and then seeded on fibronectin for 60 minutes. Cellular extracts were prepared and RhoA, CDC42 and Rac1 pull down assays were performed. HGAL knockdown and equal loading were confirmed by immunoblotting with HGAL and actin antibodies. Results are representative of 3 independent experiments. Densitometry analysis of normalized RhoA-GTP to total RhoA is presented. The values in the control samples were arbitrarily defined as 1. Error bars represent SD; * indicates a statistically significant difference (P < .05) between experimental conditions. Normalized densitometry measurements for CDC42 and HGAL are also shown below the corresponding blots. (B) RhoA pull down assays in CD77+ GC B cells and CD77– cells enriched from normal tonsils. (C) B lymphocytes, enriched from peripheral blood of healthy volunteers, were transfected with control or HGAL-encoding plasmids. 48 hours later the cells were stimulated with LPA and RhoA activity was assessed in triplicates by G-Lisa RhoA Activation Assay kit (Cytoskeleton Inc). * indicates a statistically significant difference (P = 3.3 × 10−5). Fraction of the same lymphocytes was used in IL-6 chemotaxis assay, demonstrating significant (P = 6.6 × 10−4) inhibition in chemotaxis of HGAL-expressing normal lymphocytes.
HGAL induces RhoA activation. (A) Raji and VAL lymphoma cells, transfected with siRNA for HGAL or scrambled control siRNA 48 hours before the experiments, and HeLa cells stably transfected with pcDNA3.1-HGAL or pcDNA3.1-mock plasmids, were starved for 8 hours and then seeded on fibronectin for 60 minutes. Cellular extracts were prepared and RhoA, CDC42 and Rac1 pull down assays were performed. HGAL knockdown and equal loading were confirmed by immunoblotting with HGAL and actin antibodies. Results are representative of 3 independent experiments. Densitometry analysis of normalized RhoA-GTP to total RhoA is presented. The values in the control samples were arbitrarily defined as 1. Error bars represent SD; * indicates a statistically significant difference (P < .05) between experimental conditions. Normalized densitometry measurements for CDC42 and HGAL are also shown below the corresponding blots. (B) RhoA pull down assays in CD77+ GC B cells and CD77– cells enriched from normal tonsils. (C) B lymphocytes, enriched from peripheral blood of healthy volunteers, were transfected with control or HGAL-encoding plasmids. 48 hours later the cells were stimulated with LPA and RhoA activity was assessed in triplicates by G-Lisa RhoA Activation Assay kit (Cytoskeleton Inc). * indicates a statistically significant difference (P = 3.3 × 10−5). Fraction of the same lymphocytes was used in IL-6 chemotaxis assay, demonstrating significant (P = 6.6 × 10−4) inhibition in chemotaxis of HGAL-expressing normal lymphocytes.
HGAL induces activation of RhoA downstream effectors regulating actin cytoskeleton
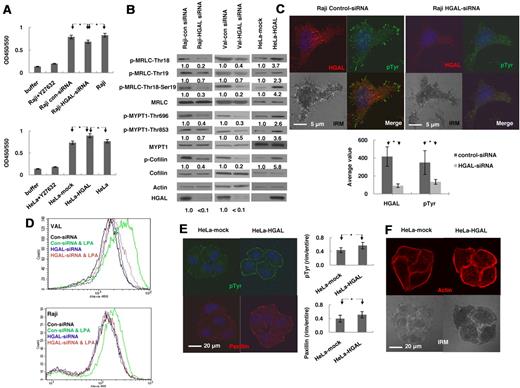
GTP-bound RhoA interacts with a large number of downstream effectors that regulate a variety of cellular processes, including actin-myosin cytoskeleton remodeling.17,18 Rho kinase (ROCK) is one of the key downstream effectors activated by RhoA and leading to cytoskeleton remodeling.21 To determine whether HGAL regulates ROCK activity, we transiently knocked down the expression of HGAL in Raji cells using siRNAs. FN stimulation of these cells resulted in significantly decreased ROCK activity (P = .003) compared with nontransfected cells as well as control siRNA transfected cells (Figure 2A). HGAL expression in FN stimulated HeLa cells resulted in significantly increased ROCK activity (P = 1.5 × 10−5; Figure 2A). ROCK activity was similar in control transfected and nontransfected HeLa cells (Figure 2A).
HGAL induces activation of RhoA downstream effectors regulating actin cytoskeleton. (A-B) Raji and VAL lymphoma cells, transfected with siRNA for HGAL or control siRNA, and HeLa cells stably transfected with pcDNA3.1-HGAL or pcDNA3.1-plasmids, were starved for 8 hours and then seeded on fibronectin for 60 minutes. (A) ROCK enzymatic activity was measured in triplicates in the indicated cells. Data are presented as mean ± SD of the mean. (B) Cellular lysates were immunoblotted for indicated proteins. Densitometry measurements of respective phosphorylated to nonphosphorylated proteins are presented. The values in control specimens were arbitrarily defined as 1. (C) Raji lymphoma cells, transfected with siRNA for HGAL or control siRNA were starved for 8 hours and then seeded on fibronectin for 90 minutes. The cells were stained with anti-HGAL (red), anti-pTyr (green) and DAPI (blue). Interference reflection contrast (IRM) images were obtained and p-Tyr staining intensity was measured and depicted as mean ± SD of the mean. (D) VAL or Raji cells transfected with siRNA for HGAL or control siRNA were starved for 8 hours and then left unstimulated or treated with LPA (1.0 μg/mL) for 45 seconds followed by staining with Alexa-488 phalloidin and analyzed by flow cytomety. (E-F) Serum-starved HeLa cells stably transfected with pcDNA3.1-HGAL or pcDNA3.1-plasmids were seeded on FN-coated slides for 90 minutes. (E) The cells were stained with anti-pTyr (green), anti-paxillin (red) and DAPI (blue); and (F) rhodamine-labeled phalloidin (red); IRM images were obtained and visualized on a Carl Zeiss LSM510/UV confocal microscope. The intensity of each staining was calculated in 25 cell tetrads as described in supplemental Mehtods and depicted as mean ± SD of the mean for p-Tyr and paxillin (E). Results in panels A through E are representative of 3 independent experiments. * indicate statistically significant difference (all below P < .01).
HGAL induces activation of RhoA downstream effectors regulating actin cytoskeleton. (A-B) Raji and VAL lymphoma cells, transfected with siRNA for HGAL or control siRNA, and HeLa cells stably transfected with pcDNA3.1-HGAL or pcDNA3.1-plasmids, were starved for 8 hours and then seeded on fibronectin for 60 minutes. (A) ROCK enzymatic activity was measured in triplicates in the indicated cells. Data are presented as mean ± SD of the mean. (B) Cellular lysates were immunoblotted for indicated proteins. Densitometry measurements of respective phosphorylated to nonphosphorylated proteins are presented. The values in control specimens were arbitrarily defined as 1. (C) Raji lymphoma cells, transfected with siRNA for HGAL or control siRNA were starved for 8 hours and then seeded on fibronectin for 90 minutes. The cells were stained with anti-HGAL (red), anti-pTyr (green) and DAPI (blue). Interference reflection contrast (IRM) images were obtained and p-Tyr staining intensity was measured and depicted as mean ± SD of the mean. (D) VAL or Raji cells transfected with siRNA for HGAL or control siRNA were starved for 8 hours and then left unstimulated or treated with LPA (1.0 μg/mL) for 45 seconds followed by staining with Alexa-488 phalloidin and analyzed by flow cytomety. (E-F) Serum-starved HeLa cells stably transfected with pcDNA3.1-HGAL or pcDNA3.1-plasmids were seeded on FN-coated slides for 90 minutes. (E) The cells were stained with anti-pTyr (green), anti-paxillin (red) and DAPI (blue); and (F) rhodamine-labeled phalloidin (red); IRM images were obtained and visualized on a Carl Zeiss LSM510/UV confocal microscope. The intensity of each staining was calculated in 25 cell tetrads as described in supplemental Mehtods and depicted as mean ± SD of the mean for p-Tyr and paxillin (E). Results in panels A through E are representative of 3 independent experiments. * indicate statistically significant difference (all below P < .01).
ROCK induces phosphorylation of MRLC at Ser19/Thr18 and myosin phosphatase subunit MYPT1, inhibiting MRLC dephosphorylation and also stimulates LIMK to phosphorylate cofilin (P-cofilin), thereby inactivating its function.24,25 We next examined the effects of HGAL on these ROCK targets. siRNA-induced knockdown of HGAL in Raji and VAL cells stimulated with FN decreased phosphorylation of cofilin, of MRLC at Ser19/Thr18 and of MYPT1 at Thr696 and Thr853 compared with cells transfected with control siRNA (Figure 2B). HGAL expression in the FN stimulated HeLa cells also increased phosphorylation of these proteins compared with vector-control expressing cells (Figure 2B).
Phosphorylation of the downstream effectors of RhoA regulates the function of actin-myosin cytoskeleton whose reorganization results in the formation of focal adhesions and stress fibers in adherent cells or actin polymerization in nonadherent cells.31 These changes control cell migration and motility. We examined by immunofluorescence and interference reflection contrast microscopy whether HGAL expression affects the formation of focal adhesions and stress fibers. A significant decrease (P = 1.7 × 10−10) in the number of focal adhesions, visualized with anti-phosphotyrosine (pTyr) staining, was observed in the FN-adherent Raji cells after siRNA-induced knockdown of HGAL protein (Figure 2C). Flow cytometry analysis showed marked decrease in LPA-induced phalloidin-stained polymerized F-actin in both Raji and VAL lymphoma cells transfected with HGAL siRNA compared with cells transfected with control siRNA (Figure 2D). In HGAL-transfected HeLa cells plated on FN, there was a statistically significant increase in the number of focal adhesions visualized by staining with anti-pTyr (P = 2.1 × 10−5) and anti-paxillin (P = 9.8 × 10−5) antibodies (Figure 2E). Stress fibers, visualized by staining with rhodamine-labeled phalloidin, were also significantly increased (P = .014) compared with vector-control stably transfected HeLa cells (Figure 2F). Taken together, these results indicate that HGAL enhances the activation of RhoA and its downstream effectors, regulating actin cytoskeleton remodeling.
HGAL augments transcriptional effects and transformation potential of RhoA
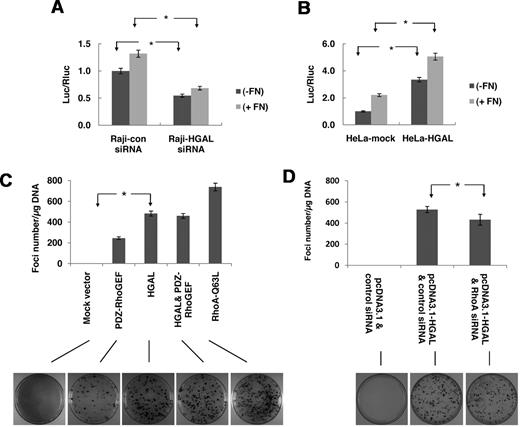
Our results demonstrate that HGAL expression enhances the activation of RhoA and its downstream effectors, known to regulate actin cytoskeleton and cell motility. However RhoA is also implicated in regulation of transcriptional activity by inducing expression from serum responsive element (SRE) through the transcriptional activation of the serum response factor (SRF).27 To analyze HGAL effects on the RhoA-SRF-dependent signaling, we measured c-fos SRE transcriptional activity, assessed by a luciferase reporter assay, after transient siRNA-induced knockdown of HGAL expression in Raji cells. Knockdown of HGAL expression led to a significant decrease in both baseline, FN-induced (P = 7.9 × 10−5; Figure 3A) and LPA-induced (P = 1.1 × 10−4; supplemental Figure 3A) luciferase expression from the SRE reporter plasmid. HGAL expression in HeLa cells markedly increased baseline, FN-induced (Figure 3B) and LPA-induced (supplemental Figure 3B) luciferase expression from the SRE reporter plasmid.
HGAL augments transcriptional effects and transformation potential of RhoA. (A) Raji lymphoma cells were transfected with SRF-driven luciferase reporter construct pSRE-Luc and pRL-TK plasmids and with either siRNA for HGAL or scrambled control siRNA. Forty-eight hours after transfection the cells were starved for 8 hours and then either plated on FN-coated or noncoated plates for 60 minutes followed by determination of luciferase activity. Numbers refer to luciferase activities representing means + SD of the mean of 3 independent experiments, each performed in triplicate. * indicate statistically significant (all below P < .01) difference. (B) HeLa cells stably transfected with pcDNA3.1-HGAL or pcDNA3.1-mock plasmids were transiently transfected with pSRE-Luc and pRL-TK. Forty-eight hours after transfection the cells were starved for 8 hours and then either plated on FN-coated or noncoated plates for 60 minutes and luciferase activity was determined as described in panel A. (C-D) NIH 3T3 cells were transfected with pcDNA3.1-mock, or pcDNA3.1-RhoAQ63L, pcDNA3.1-HGAL and pcDNA3.1-PDZ-RhoGEF alone or together with pcDNA3.1-HGAL (C) or pcDNA3.1-HGAL with control or RhoA siRNA (D). Cells were stained 3 weeks after transfection. Representative plates are depicted. Foci of transformation were counted in triplicate plates in independent transfections and the number of foci per μg of transfected DNA for each condition is shown as mean ± SD of the mean. * indicate statistically significant difference (all below P < .001 in panel C and P = .016 in panel D).
HGAL augments transcriptional effects and transformation potential of RhoA. (A) Raji lymphoma cells were transfected with SRF-driven luciferase reporter construct pSRE-Luc and pRL-TK plasmids and with either siRNA for HGAL or scrambled control siRNA. Forty-eight hours after transfection the cells were starved for 8 hours and then either plated on FN-coated or noncoated plates for 60 minutes followed by determination of luciferase activity. Numbers refer to luciferase activities representing means + SD of the mean of 3 independent experiments, each performed in triplicate. * indicate statistically significant (all below P < .01) difference. (B) HeLa cells stably transfected with pcDNA3.1-HGAL or pcDNA3.1-mock plasmids were transiently transfected with pSRE-Luc and pRL-TK. Forty-eight hours after transfection the cells were starved for 8 hours and then either plated on FN-coated or noncoated plates for 60 minutes and luciferase activity was determined as described in panel A. (C-D) NIH 3T3 cells were transfected with pcDNA3.1-mock, or pcDNA3.1-RhoAQ63L, pcDNA3.1-HGAL and pcDNA3.1-PDZ-RhoGEF alone or together with pcDNA3.1-HGAL (C) or pcDNA3.1-HGAL with control or RhoA siRNA (D). Cells were stained 3 weeks after transfection. Representative plates are depicted. Foci of transformation were counted in triplicate plates in independent transfections and the number of foci per μg of transfected DNA for each condition is shown as mean ± SD of the mean. * indicate statistically significant difference (all below P < .001 in panel C and P = .016 in panel D).
Activated forms of RhoA protein have been shown to facilitate the appearance of transformed foci in NIH 3T3 cells, suggesting a potential role in oncogenesis.26,32 Consequently we have examined whether HGAL may affect formation of fibroblast transformed foci. NIH3T3 cells were transfected with control plasmid or plasmids encoding HGAL, activated form of RhoA (RhoAQ63L) and PDZ-RhoGEF—a guanine nucleoside exchange factor that is known to increase activity of RhoA and to induce formation of fibroblast transformed foci.33 Both RhoAQ63L and PDZ-RhoGEF induced the appearance of transformed foci, with RhoAQ63L inducing a larger number of transformed foci compared with PDZ-RhoGEF, as previously reported33 (Figure 3C). HGAL expression also induced formation of transformed foci of similar morphology. Furthermore, the number of formed foci was increased up to 2-fold in cells transfected with HGAL compared with cells transfected with PDZ-RhoGEF (Figure 3C). Cotransfection of HGAL and PDZ-RhoGEF did not further increase the number of transformed foci, suggesting that HGAL and PDZ-RhoGEF share a common mechanism of RhoA activation. Transient siRNA-induced knockdown of RhoA decreased the number of HGAL-induced foci (P = .016; Figure 3D). Overall, these observations suggest that HGAL-enhanced activation RhoA launches multiple downstream signaling cascades that may affect various cellular processes regulated by RhoA protein.
HGAL interacts with PDZ-RhoGEF and LARG and stimulates guanidine exchange activity
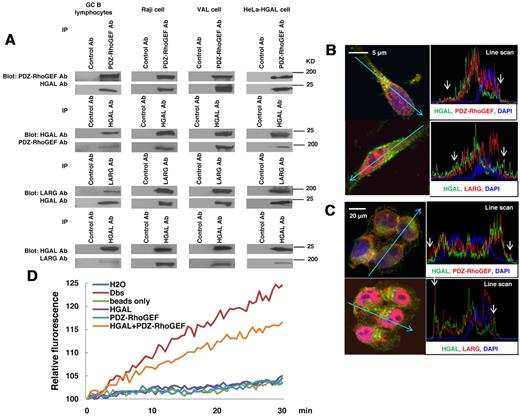
HGAL-mediated enhancement of active RhoA levels might be attributable to a direct effect of HGAL on RhoA or to indirect HGAL effects on the upstream regulators of RhoA. To evaluate the possibility of a direct effect, HGAL-RhoA coimmunoprecipitation experiments were performed (supplemental Figure 4). No binding between HGAL and RhoA, as well as Cdc42 or Rac1 were detected. We next hypothesized that HGAL may enhance RhoA activation indirectly by binding to Rho-specific guanine nucleotide exchange factors (RhoGEFs) that activate RhoA by stimulating its GTP-GDP exchange rate. Absence of an additive effect on the formation of transformed foci in NIH 3T3 cells upon cotransfection of HGAL with PDZ-RhoGEF (Figure 3C) suggested that HGAL-enhanced activation of RhoA may be at least partially mediated by PDZ-RhoGEF, a member of the regulators of G protein signaling-like (RGS)–domain containing RhoGEF family that also includes LARG and p115-RhoGEF.34 PDZ-RhoGEF and LARG proteins specifically activate RhoA and contain a PDZ domain, missing in the p115-RhoGEF, while HGAL protein contains a C-terminal PDZ binding motif. To assess whether HGAL protein may bind to PDZ-RhoGEF, LARG and/or p115-RhoGEF proteins, coimmunoprecipitation experiments were performed. Endogenous HGAL was detected in immunoprecipitates of endogenous PDZ-RhoGEF and LARG proteins and, vice versa, endogenous PDZ-RhoGEF and LARG were detected in the immunoprecipitates of endogenous HGAL from GC B lymphocytes, Raji and VAL lymphoma cells (Figure 4A), as well SUDHL6 (supplemental Figure 5A) and MC116 (not shown) lymphoma cell lines. Similarly, PDZ-RhoGEF and LARG proteins coimmunoprecipitated with cellular HGAL protein expressed in HeLa cells (Figure 4A). In contrast, HGAL protein did not coimmunoprecipitate with p115-RhoGEF protein (supplemental Figure 5C). Confocal microscopy studies of FN-adherent Raji (Figure 4B) and HGAL-expressing HeLa (Figure 4C) cells confirmed colocalization of HGAL protein with both PDZ-RhoGEF and LARG proteins in cell membrane and cytoplasm. Enlarged images of individual cells are shown in supplemental Figure 6A through H.
HGAL interacts with PDZ-RhoGEF and LARG and stimulates guanidine exchange activity. (A) Cellular lysates were extracted from unmanipulated isolated GC B lymphocytes and HeLa cells stably transfected with pcDNA3.1-HGAL, Raji and VAL cells seeded on fibronectin and subjected to immunoprecipitation with anti–PDZ-RhoGEF and LARG or HGAL antibodies, as well as control antibodies, followed by anti-HGAL and anti–PDZ-RhoGEF or anti-LARG Western immunoblotting, respectively. (B-C) Raji (B) and HeLa cells stably transfected with pcDNA3.1-HGAL (C) were seeded on fibronectin for 90 minutes and stained with DAPI (blue nuclear staining), antibodies to HGAL (green) and PDZ-RhoGEF or LARG (red). Slides were viewed on a Carl Zeiss LSM510/UV confocal microscope with 63×/1.4 NA plan apochromat objective lens and 2× zoom and processed with Zeiss LSM510 AIM 3.2 SP2 confocal microscope software. Z stack linescan images demonstrating proteins colocalization at the cell membrane (arrow) are shown; higher resolution images are shown in supplemental Figure 6. (D) PDZ-RhoGEF was immunoprecipitated from Raji cells and used in the RhoA N-methylanthraniloyl exchange assay, either alone or with purified HGAL protein. Purified Dbs protein served as a positive control, while immunoprecipitates with beads only, purified HGAL alone and water were used as negative controls. Results are representative of 3 independent experiments.
HGAL interacts with PDZ-RhoGEF and LARG and stimulates guanidine exchange activity. (A) Cellular lysates were extracted from unmanipulated isolated GC B lymphocytes and HeLa cells stably transfected with pcDNA3.1-HGAL, Raji and VAL cells seeded on fibronectin and subjected to immunoprecipitation with anti–PDZ-RhoGEF and LARG or HGAL antibodies, as well as control antibodies, followed by anti-HGAL and anti–PDZ-RhoGEF or anti-LARG Western immunoblotting, respectively. (B-C) Raji (B) and HeLa cells stably transfected with pcDNA3.1-HGAL (C) were seeded on fibronectin for 90 minutes and stained with DAPI (blue nuclear staining), antibodies to HGAL (green) and PDZ-RhoGEF or LARG (red). Slides were viewed on a Carl Zeiss LSM510/UV confocal microscope with 63×/1.4 NA plan apochromat objective lens and 2× zoom and processed with Zeiss LSM510 AIM 3.2 SP2 confocal microscope software. Z stack linescan images demonstrating proteins colocalization at the cell membrane (arrow) are shown; higher resolution images are shown in supplemental Figure 6. (D) PDZ-RhoGEF was immunoprecipitated from Raji cells and used in the RhoA N-methylanthraniloyl exchange assay, either alone or with purified HGAL protein. Purified Dbs protein served as a positive control, while immunoprecipitates with beads only, purified HGAL alone and water were used as negative controls. Results are representative of 3 independent experiments.
To examine whether HGAL protein may stimulate the guanidine nucleotide exchange activity, PDZ-RhoGEF protein was immunoprecipitated from starved Raji cells and used in the RhoA N-methylanthraniloyl exchange (mant-GTP) assay, either alone or with purified HGAL protein (Figure 4D). In comparison to PDZ-RhoGEF protein alone, addition of HGAL protein to PDZ-RhoGEF markedly increased the RhoA guanidine nucleotide exchange, as reflected by increased fluorescence of the RhoA-bound mant-GTP. These results suggest that binding of HGAL protein to PDZ-RhoGEF and probably LARG proteins stimulates release of GDP from the RhoA leading to its replacement with GTP.
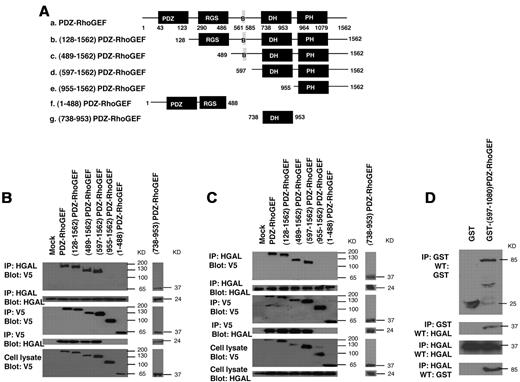
PDZ-RhoGEF and LARG proteins exhibit similar molecular structure and domain composition containing PDZ, regulators of G protein signaling (RGS), Dbl homology (DH) and pleckstrin homology (PH) domains (Figure 5A). To determine which of these domains bind HGAL protein, we have focused on the PDZ-RhoGEF protein. 7 V5-tagged plasmid constructs of PDZ-RhoGEF were generated (Figure 5A): (128-1562)PDZ-RhoGEF construct in which the N terminal PDZ domain is deleted; (489-1562)PDZ-RhoGEF construct in which both PDZ and RGS domains are deleted; (597-1562)PDZ-RhoGEF construct in which PDZ and RGS domains as well as an additional sequence (B) involved in the interaction with actin35 are deleted; (955-1562)PDZ-RhoGEF construct encoding the PH domain and the C terminal sequence of the protein; (1-488)PDZ-RhoGEF construct encoding the N terminal PDZ and RGS domains; and (738-953)PDZ-RhoGEF construct encoding only the DH domain. These mutants could be expressed and were detected by the anti-V5 tag antibody.
PDZ-RhoGEF interacts with HGAL through DH domain. (A) Structure of wild-type and 6 different V5 epitope tagged truncated mutants of PDZ-RhoGEF that were used to transfect Raji lymphoma cells expressing endogenous HGAL (B) and HEK-293T cells cotransfected with pcDNA3.1-HGAL vector (C). PDZ: PDZ domain; RGS: regulators of G protein signaling domain, B: protein sequence binding to actin, DH: Dbl-homology domain, PH: pleckstrin-homology domain. (B-C) Total cellular extracts prepared from the transfected Raji (B) and HEK-293T (C) cells were immunoprecipitated with anti-HGAL and anti-V5 antibodies and analyzed by Western blotting with anti-V5 and anti-HGAL antibodies, as indicated. The corresponding total cellular lysates were subjected to Western blot analysis using anti-HGAL and anti-V5 antibodies. (D) The purified GST-(597-1080)PDZ-RhoGEF or GST proteins were incubated with purified TRX-HGAL or TRX proteins for 12 hours. The co-precipitated HGAL and (597-1080)PDZ-RhoGEF were detected by immunoprecipitation with anti-HGAL and anti-GST antibodies followed by Western blotting with anti-GST and anti-HGAL antibodies, as indicated. Results are representative of 3 independent experiments.
PDZ-RhoGEF interacts with HGAL through DH domain. (A) Structure of wild-type and 6 different V5 epitope tagged truncated mutants of PDZ-RhoGEF that were used to transfect Raji lymphoma cells expressing endogenous HGAL (B) and HEK-293T cells cotransfected with pcDNA3.1-HGAL vector (C). PDZ: PDZ domain; RGS: regulators of G protein signaling domain, B: protein sequence binding to actin, DH: Dbl-homology domain, PH: pleckstrin-homology domain. (B-C) Total cellular extracts prepared from the transfected Raji (B) and HEK-293T (C) cells were immunoprecipitated with anti-HGAL and anti-V5 antibodies and analyzed by Western blotting with anti-V5 and anti-HGAL antibodies, as indicated. The corresponding total cellular lysates were subjected to Western blot analysis using anti-HGAL and anti-V5 antibodies. (D) The purified GST-(597-1080)PDZ-RhoGEF or GST proteins were incubated with purified TRX-HGAL or TRX proteins for 12 hours. The co-precipitated HGAL and (597-1080)PDZ-RhoGEF were detected by immunoprecipitation with anti-HGAL and anti-GST antibodies followed by Western blotting with anti-GST and anti-HGAL antibodies, as indicated. Results are representative of 3 independent experiments.
Raji cells were transiently transfected with plasmids encoding the wild-type PDZ-RhoGEF protein or the 6 truncated mutants. Endogenous HGAL protein was immunoprecipitated with HGAL antibody and immunoblotted with anti-V5 tag antibody or, vice versa, PDZ-RhoGEF protein and its mutants were immunoprecipitated with anti-V5 tag antibody and immunoblotted with HGAL antibody (Figure 5B). HGAL protein interacted with the catalytic DH domain of the PDZ-RhoGEF protein. Similar results were observed in 293T (Figure 5C) and HeLa (not shown) cells transiently transfected with both HGAL and PDZ-RhoGEF or its mutants. Purified bacterially expressed GST and GST-(597-1080)PDZ-RhoGEF protein segment containing the DH and PH domains was able to pull-down purified bacterially expressed TRX-HGAL and, vice versa, TRX-HGAL protein pulled-down the (597-1080)PDZ-RhoGEF protein segment (Figure 5D). However, HGAL was unable to complex with GST protein. Taken together, these results indicate that the interaction between PDZ-RhoGEF and HGAL is direct and is mediated by the catalytic DH domain of the PDZ-RhoGEF protein.
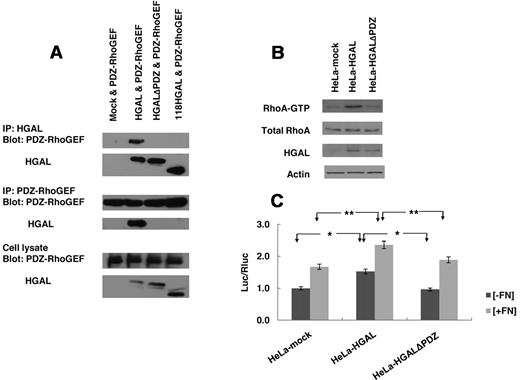
To determine the sequence of HGAL protein necessary for interaction with PDZ-RhoGEF protein, we generated 2 HGAL truncated mutants, one encoding an N terminal 118 amino acids (1-118)HGAL segment and a second mutant in which the HGAL C-terminus PDZ domain binding motif (174QFSHL) was deleted (HGALΔPDZ). Immunoprecipitates of the wild-type, full-length HGAL, but none of its mutants, contained the PDZ-RhoGEF protein (Figure 6A). Similarly, immunoprecipitates of the PDZ-RhoGEF protein contained the wild-type, full-length HGAL protein but none of its deletion mutants. These findings suggest that the C-terminal PDZ domain interacting sequence of the HGAL protein is required for complex formation with PDZ-RhoGEF protein.
HGAL interacts with PDZ-RhoGEF through its C-terminal PDZ binding motif. (A) HeLa cells were co-transfected with pcDNA3.1-PDZ-RhoGEF and one of the following plasmids: pcDNA3.1, pcDNA3.1-HGAL, pcDNA3.1-HGALΔPDZ and pcDNA3.1-(1-118)HGAL. Total cellular lysates were prepared and immunoprecipitated with anti-HGAL and anti–PDZ-RhoGEF antibodies and analyzed by Western blotting with anti–PDZ-RhoGEF and anti-HGAL antibodies, as indicated. The corresponding total cellular lysates were subjected to Western blot analysis using anti-HGAL and anti–PDZ-RhoGEF antibodies. (B-C) HeLa cells transfected with pcDNA3.1, pcDNA3.1-HGAL or pcDNA3.1-HGALΔPDZ were used for RhoA pull-down (B) and analysis of luciferase activity from the SRF-driven luciferase reporter construct pSRE-Luc as described in “Luciferase reporter assays” and shown in Figures 1B and 3A. Numbers refer to luciferase activities representing means + SD of the mean of 3 independent experiments, each performed in triplicate. * indicate statistically significant difference (all below P < .01). Results in panels A and B are also representative of 3 independent experiments.
HGAL interacts with PDZ-RhoGEF through its C-terminal PDZ binding motif. (A) HeLa cells were co-transfected with pcDNA3.1-PDZ-RhoGEF and one of the following plasmids: pcDNA3.1, pcDNA3.1-HGAL, pcDNA3.1-HGALΔPDZ and pcDNA3.1-(1-118)HGAL. Total cellular lysates were prepared and immunoprecipitated with anti-HGAL and anti–PDZ-RhoGEF antibodies and analyzed by Western blotting with anti–PDZ-RhoGEF and anti-HGAL antibodies, as indicated. The corresponding total cellular lysates were subjected to Western blot analysis using anti-HGAL and anti–PDZ-RhoGEF antibodies. (B-C) HeLa cells transfected with pcDNA3.1, pcDNA3.1-HGAL or pcDNA3.1-HGALΔPDZ were used for RhoA pull-down (B) and analysis of luciferase activity from the SRF-driven luciferase reporter construct pSRE-Luc as described in “Luciferase reporter assays” and shown in Figures 1B and 3A. Numbers refer to luciferase activities representing means + SD of the mean of 3 independent experiments, each performed in triplicate. * indicate statistically significant difference (all below P < .01). Results in panels A and B are also representative of 3 independent experiments.
To further confirm that the C-terminal PDZ domain interacting sequence of the HGAL protein is necessary for binding to RhoGEFs, we assessed the levels of the GTP-bound activated RhoA and the c-fos SRE transcriptional activity measured by the luciferase reporter assay in the HGAL wild-type, HGALΔPDZ deletion mutant, and control vector-expressing HeLa cells plated on FN or stimulated with LPA. In contrast to the wild-type HGAL protein, the mutant missing the PDZ domain interacting sequence neither enhanced the amount of GTP-bound RhoA (Figure 6B and supplemental Figure 7A) nor increased the baseline and the FN or LPA–induced luciferase expression from the SRE reporter plasmid (Figure 6C and supplemental Figure 7B). These findings indicate that disruption of the binding between HGAL and PDZ-RhoGEF leads to loss of activation of RhoA and its downstream effectors.
Relative contribution of PDZ-RhoGEF and LARG to HGAL-induced activation of RhoA
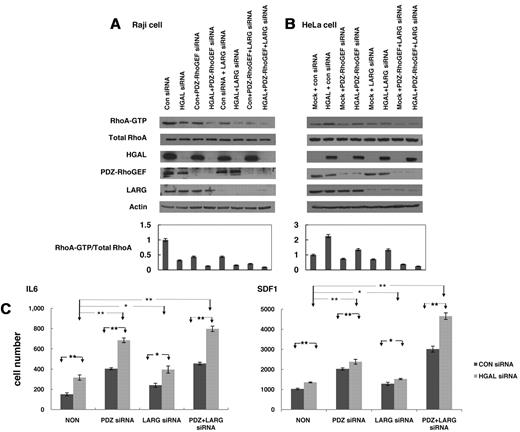
Our results suggest that HGAL-enhanced activation of the RhoA is mediated by HGAL binding to PDZ-RhoGEF and LARG proteins. To assess the relative contributions of these 2 proteins, we measured GTP-bound RhoA after individual or concomitant siRNA-mediated knockdown of HGAL, PDZ-RhoGEF and LARG proteins in Raj cell stimulated with FN (Figure 7A) and LPA (supplemental Figure 8). Knockdown of HGAL protein led to a more pronounced decrease in the levels of GTP-bound RhoA than individual knockdown of PDZ-RhoGEF or LARG proteins. Simultaneous knockdown of HGAL and PDZ-RhoGEF proteins or HGAL and LARG proteins resulted in even further decrease in the levels of activated RhoA compared with knockdown of HGAL alone. Furthermore, knockdown of all 3 proteins led to almost complete absence GTP-bound RhoA. In HeLa-pcDNA3 cells stimulated with FN (Figure 7B) and LPA (supplemental Figure 8), individual knockdown of PDZ-RhoGEF or LARG proteins markedly decreased levels of GTP-bound RhoA. Similar effects were observed after PDZ-RhoGEF knockdown with different siRNAs (not shown). In contrast, in HeLa HGAL cells that harbor higher levels of activated RhoA compared with HeLa cells transfected with control vector, individual knockdown of PDZ-RhoGEF or LARG proteins only mildly decreased the levels of the GTP-bound RhoA, while their concomitant knockdown led to almost complete absence of activated RhoA. Taken together, these results suggest that HGAL protein enhances activation of RhoA by binding to both PDZ-RhoGEF and LARG proteins.
Cooperative effects of HGAL, PDZ-RhoGEF and LARG on RhoA activation and lymphocyte motility. (A) Raji cells transfected with either control siRNA or siRNA for HGAL alone or in combination with siRNAs for PDZ-RhoGEF, LARG or both were seeded on fibronectin. Cellular lysates were prepared and used for RhoA pull-down assay and Western blotting with anti-HGAL, PDZ-RhoGEF, LARG and actin antibodies. Densitometry analysis of normalized RhoA-GTP to total RhoA from 3 independent Western blot experiments is presented. The values in the control specimens were arbitrarily defined as 1. Error bars represent SD. (B) HeLa cells stably transfected with pcDNA3.1 or pcDNA3.1-HGAL were transfected with siRNAs for PDZ-RhoGEF, LARG or both and were seeded on fibronectin. Cellular lysates were prepared and used for RhoA pull-down assay and Western blotting with anti-HGAL, PDZ-RhoGEF, LARG and actin antibodies. Densitometry analysis of normalized RhoA-GTP to total RhoA from 3 independent Western blot experiments is presented. The values in the control specimens were arbitrarily defined as 1. Error bars represent SD. (C) VAL lymphoma cells were transfected with control siRNA or siRNA for HGAL and siRNAs for PDZ-RhoGEF, LARG or both. 48 hours after siRNA transfection, the cells were used for IL-6 and SDF1 chemotaxis assay performed in triplicate, as described in the “Chemotaxis assays.” Means + SD of the mean of 3 independent experiments are demonstrated. Statistically significant differences with *P < .05 and **P < .01.
Cooperative effects of HGAL, PDZ-RhoGEF and LARG on RhoA activation and lymphocyte motility. (A) Raji cells transfected with either control siRNA or siRNA for HGAL alone or in combination with siRNAs for PDZ-RhoGEF, LARG or both were seeded on fibronectin. Cellular lysates were prepared and used for RhoA pull-down assay and Western blotting with anti-HGAL, PDZ-RhoGEF, LARG and actin antibodies. Densitometry analysis of normalized RhoA-GTP to total RhoA from 3 independent Western blot experiments is presented. The values in the control specimens were arbitrarily defined as 1. Error bars represent SD. (B) HeLa cells stably transfected with pcDNA3.1 or pcDNA3.1-HGAL were transfected with siRNAs for PDZ-RhoGEF, LARG or both and were seeded on fibronectin. Cellular lysates were prepared and used for RhoA pull-down assay and Western blotting with anti-HGAL, PDZ-RhoGEF, LARG and actin antibodies. Densitometry analysis of normalized RhoA-GTP to total RhoA from 3 independent Western blot experiments is presented. The values in the control specimens were arbitrarily defined as 1. Error bars represent SD. (C) VAL lymphoma cells were transfected with control siRNA or siRNA for HGAL and siRNAs for PDZ-RhoGEF, LARG or both. 48 hours after siRNA transfection, the cells were used for IL-6 and SDF1 chemotaxis assay performed in triplicate, as described in the “Chemotaxis assays.” Means + SD of the mean of 3 independent experiments are demonstrated. Statistically significant differences with *P < .05 and **P < .01.
PDZ-RhoGEF and LARG proteins mediate HGAL-induced inhibition of cell motility
We hypothesized that HGAL-induced activation of RhoA may contribute to the inhibition of lymphocytes and lymphoma cells cell motility reported by us.9 To elucidate the relative role of PDZ-RhoGEF and LARG proteins in mediating HGAL effects on cell motility, we examined the effects of knockdown PDZ-RhoGEF and LARG proteins on HGAL-induced inhibition of chemotaxis in response to IL-6 and SDF1 in VAL (Figure 7C) and Raji lymphoma cells (not shown). As expected, SDF1 induced more pronounced chemotaxis of lymphoma cells compared with the IL-6 (note different y-axis scale for IL-6 and SDF1-induced chemotaxis in Figure 7C). Knockdown of HGAL markedly increased lymphoma cell chemotaxis in response to IL-6 (P = .001) and SDF1 (P = .004). Knockdown of LARG protein moderately increased IL-6 and SDF1-induced chemotaxis of VAL cells, in which HGAL protein was expressed (P = .04 and P = .03, respectively) or knocked-down (P = .16 and P = .03, respectively). In contrast, knockdown of PDZ-RhoGEF protein had a more pronounced effect, significantly increasing IL-6 and SDF1-induced chemotaxis of VAL cells, in which HGAL protein was expressed (P = .001 and P = 8.3 × 10−4, respectively) or knocked-down (P = .005 and P = .004, respectively). Although further slight increase in the IL-6 and SDF1 induced chemotaxis was observed upon concomitant knockdown of both PDZ-RhoGEF and LARG proteins, the effect was not additive. Examination of spontaneous motility of Raji cells using timelapse images also demonstrated increased cell migration upon concomitant knockdown of HGAL and PDZ-RhoGEF compared with cells transfected with control siRNA or after individual knockdown of HGAL and PDZ-RhoGEF (supplemental Figure 9 and supplemental Videos). These findings suggest that HGAL inhibits cell motility and IL-6– and SDF1-induced chemotaxis by activating RhoA. Furthermore, they suggest that PDZ-RhoGEF is the major mediator of HGAL effects on chemotaxis and cell motility.
Discussion
HGAL is a novel GC-specific gene whose expression is correlated with better survival of patients with DLBCL and cHL.1,5 Recently we have demonstrated that HGAL is involved in negative regulation of lymphocyte motility,9 thus potentially constraining lymphocytes to the GC microenvironment. The mechanism of HGAL-mediated inhibition of lymphocyte motility is unknown. Herein, we demonstrate that HGAL serves as a regulator of the RhoA signaling pathway in B-cell lymphoma cells and may have a similar role in GC lymphocytes. HGAL-mediated enhancement of RhoA activation results in inhibition of lymphocyte motility and chemotaxis by activation of its downstream effector ROCK, leading to phosphorylation of MRLC, myosin PPTase subunit MYPT1 and cofilin, which mediate cytoskeletal reorganization, as reflected by increased formation of focal adhesions and stress fibers. Increased RhoA activity has been previously shown to decrease motility of macrophages and fibroblasts,36 and thus may contribute to the inhibitory effects of HGAL on lymphocyte motility. The latter may be attributed to firmer attachment to the extracellular matrix due to increased focal adhesions formation. Alternatively, it is possible that HGAL-mediated RhoA-induced phosphorylation of MRLC and myosin II activation may result in cell motility inhibition, because recent studies demonstrated that myosin II ablation leads to increased cell migration while myosin activation decreased cell motility.37
HGAL enhances activation of RhoA indirectly by binding and stimulating the guanidine nucleotide exchange activity of the RhoA-specific RhoGEFs, PDZ-RhoGEF and LARG, leading to inhibition of lymphoma cell motility. Knockdown of PDZ-RhoGEF and LARG increase lymphoma cell chemotaxis, similarly to previously reported migration increase of marginal zone B-lymphocytes in p115-RhoGEF−/− mice.38 Despite identification of more than 70 distinct RhoGEFs, the mechanisms by which these proteins are activated are still not fully understood.39 Several nonreceptor-type kinases have been shown to activate specific RhoGEFs. For example, FAK was reported to phosphorylate PDZ-RhoGEF and LARG in response to agonist stimulating G proteins.40 These RhoGEFs can also associate through their PDZ domain to Plexin B41 and insulin-like growth factor receptor,42 leading to increased guanidine nucleotide exchange activity, possibly attributable to alleviation of the N-terminus autoinhibition on the DH-domain. We demonstrate that HGAL activates PDZ-RhoGEF by a novel mechanism—direct binding to the catalytic DH-domain. This binding may lead to conformational changes in the PDZ-RhoGEF protein that either enhances the intrinsic guanidine nucleotide exchange activity of the DH-domain or alleviates autoinhibitory effects of other protein domains.
The specific expression of HGAL exclusively in the GC lymphocytes and GC-derived lymphomas may lead to particular effects on these cells. GC lymphocytes demonstrate limited inter-zonal and inter-compartmental lymphocyte movement, leading to their confinement to the GC and segregation from other lymph node areas.10 Stationary lymphocytes with irregular contours are frequently observed in the GC.10 HGAL may at least partially contribute to these phenotypic changes and motility restrictions. The function of the HGAL protein in the malignant counterparts of GC lymphocytes is still unclear. Expression of HGAL was reported to be associated with improved survival of DLBCL patients.1 Similarly, expression of fibronectin, another protein implicated in the activation of RhoA signaling, was also reported to be associated with improved survival of these patients.43 Our observations reveal a novel molecular mechanism underlying the inhibitory effects of HGAL on the motility of GC-derived lymphoma cells that may contribute to the favorable outcome of DLBCL and cHL patients whose tumors express high levels of HGAL protein. Inhibition of lymphoma cell motility may explain the correlation between HGAL expression and limited DLBCL stage.44 However, it is possible that HGAL may have additional functions in these tumors. Our observations that HGAL modulates RhoA-mediated transcriptional activation by serum response factor and RhoA transforming potential suggest that HGAL may indirectly regulate gene expression and may contribute to B cell transformation. Furthermore, it is possible that persistent HGAL expression may predispose to lymphomagenesis by confining cells to the GC microenvironment and thus exposing them to high proliferation rates and active mutational machinery. Further studies addressing function of HGAL in lymphomagenesis using transgenic mice are presently underway in our laboratory.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
I.S.L. is supported by National Institutes of Health (NIH) grants NIH CA109335 and NIH CA122105, and the Dwoskin Family, Bankhead-Coley, and Fidelity Foundations. D.M.H. is supported by the WCU Program through the National Research Foundation of Korea funded by the Ministry of Education Science and Technology (R31-2008-000-10 071-0). E.C. is supported by the Fellowship of Fundacion Alfonso Martin Escudero
National Institutes of Health
Authorship
Contribution: X.J. performed experiments, analyzed the data, and wrote the paper; X. Lu, G.M., E.C., and K.A.S. performed experiments and analyzed the data; X. Liu performed experiments; I.S.-G. analyzed the data; D.M.H. supervised the experiments, analyzed the data, and wrote the paper; I.S.L. conceptualized the idea of the study, supervised the experiments, analyzed the data, and wrote the paper; and all authors approved the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Izidore Lossos, MD, Professor of Medicine, Sylvester Comprehensive Cancer Center, University of Miami, 1475 NW 12th Ave (D8-4), Miami, FL 33136; e-mail: ILossos@med.miami.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal