Abstract
EZH2 is the catalytic subunit of the PRC2 Polycomb complex and mediates transcriptional repression through its histone methyltransferase activity. EZH2 is up-regulated in normal germinal center (GC) B cells and is implicated in lymphomagenesis. To explore the transcriptional programs controlled by EZH2, we performed chromatin immunoprecipitation (ChIP-on-chip) in GC cells and found that it binds approximately 1800 promoters, often associated with DNA sequences similar to Droso-phila Polycomb response elements. While EZH2 targets overlapped extensively between GC B cells and embryonic stem cells, we also observed a large GC-specific EZH2 regulatory program. These genes are preferentially histone 3 lysine 27–trimethylated and repressed in GC B cells and include several key cell cycle–related tumor suppressor genes. Accordingly, siRNA-mediated down-regulation of EZH2 in diffuse large B-cell lymphoma (DLBCL) cells resulted in acute cell cycle arrest at the G1/S transition and up-regulation of its tumor suppressor target genes. At the DNA level, EZH2-bound promoters are hypomethylated in GC B cells, but many of them are aberrantly hypermethylated in DLBCL, suggesting disruption of normal epigenetic processes in these cells. EZH2 is thus involved in regulating a specific epigenetic program in normal GCs, including silencing of antiproliferative genes, which may contribute to the malignant transformation of GC B cells into DLBCLs.
Introduction
Polycomb proteins (PcG) are chromatin regulators with a crucial role in establishing and maintaining epigenetic memory during development and cellular differentiation. PcG is organized into 2 main sets of protein complexes: PRC1 and PRC2. EZH2 is a subunit of PRC2,1 and its SET domain catalyzes trimethylation of H3K27,1-3 a histone modification associated with transcriptional silencing. H3K27me3 helps recruit PRC1 to chromatin; it is thought that PRC1 is the effector of PcG-mediated silencing and long-term epigenetic memory.4-6 It has been observed that H3K27me3 and DNA methylation, a distinct epigenetic mark, are associated with different sets of genes in murine and human embryonic stem cells (hESCs);7,8 moreover DNA methylation and H3K27me3 are mutually exclusive at the imprinted Rasgrf1 locus.9 However, this pattern of mutual exclusion between the 2 epigenetic mechanisms appears to be disrupted in cancer cells, where many hypermethylated promoters have been shown to also be H3K27-trimethylated.10 From a functional point of view, mice deficient in PcG complexes show developmental abnormalities and embryonic lethality.11
Within the B-cell lineage, it was shown that EZH2 is highly expressed in lymphoid progenitors, and EZH2 deficiency induces defects in early lymphopoiesis.12 EZH2 declines in resting B cells but is then massively up-regulated when activated B cells form germinal centers (GCs), wherein they undergo rapid proliferation and immunoglobulin affinity maturation.13 The latter observations suggest an important role for EZH2 in GC B-cell proliferation and a possible contribution to diffuse large B-cell lymphomas (DLBCLs), which are derived from GC B cells. The potential importance of EZH2 in lymphomagenesis is further supported by the discovery of a missense mutation in the EZH2 SET domain in a sizeable fraction of DLBCLs, especially those featuring the GC B-cell gene expression signature.14 More generally, EZH2 is overexpressed in several other types of cancer (eg, in metastatic prostate cancer,15 breast cancer,16 and mantle cell lymphoma17 ).
The mechanisms by which EZH2-mediated transcriptional repression confers a growth advantage to cells remain unclear. The genomic determinants of PcG binding are also unclear, although recent chromatin immunoprecipitation (ChIP-chip) studies in Drosophila and mammals have started shedding some light on these sequences.18,19 More specifically, how EZH2 contributes to the GC phenotype and whether it targets GC B cell–specific genes and pathways are also unknown. We reasoned that mapping the EZH2 regulatory network and characterizing its target genes would help explain its function in normal and malignant B cells. Therefore, in this study we used ChIP coupled with microarrays to identify promoters bound by EZH2 in GC B cells. We characterized these promoters and genes using a combination of computational analyses and functional assays. Our results indicate a significant role for EZH2 in regulating gene expression and epigenetic patterning in normal and malignant B cells.
Methods
Cell isolation
Tonsil mononuclear cells were affinity-purified using magnetic beads to specifically enrich for naive B cells (NBCs), centroblasts, and centrocytes. Naive and centroblast cells were purified by staining primary tonsil mononuclear cells with anti-immunoglobulin D (IgD) or anti-CD77 antibodies, respectively. CD77+ selection accurately defines pure populations of rapidly dividing centroblast lymphocytes, while IgD+ selection defines highly enriched (≥ 75%) populations of NBCs. Centrocytes were purified as CD10+ cells following a negative selection for centroblast (CD77+) lymphocytes.
ChIP
We performed ChIP with anti-EZH2 (07-689; Upstate) and anti-H3K27m3 (07-449; Upstate) antibodies. For the anti-EZH2 ChIP, we performed a sequential 2-step cross-linking (2mM disuccinimidyl glutarate in 10% dimethyl sulfoxide for 1 hour, followed by 1% formaldehyde for 15 minutes). For the genome-wide localization of EZH2 targets and H3K27m3 marks, we used NimbleGen genomic microarray covering approximately 1.5 kb promoter sequence from 24 000 genes, where each promoter is represented by 15 50-mer oligos. We performed rigorous quality control of all ChIP datasets for hybridization artifacts. EZH2 ChIP-chip experiments in centroblasts were performed in quadruplicate (biologic repeats). H3K27m3 ChIP-chip in NBCs and centroblasts were performed in duplicate.
ChIP-chip peak discovery
We used the following computational analysis for peak discovery. We first smoothed ChIP over input signal intensity log-ratios for probes covering each promoter, by averaging log-ratios across a sliding window of 3 consecutive probes. We then determined the maximum log-ratio (maximum peak height) for each promoter. To estimate the statistical significance of these maximum peak heights (ie, determine how likely such peak heights can be obtained by chance), we compared them to a null distribution of maximum peak heights. To build this distribution, we constructed 1000 artificial probe sets by randomly selecting probes from the original dataset and performed smoothing and maximum peak height calculation as described above. The distribution of random maximum peak heights closely resembles an extreme value distribution. We thus fitted an extreme value distribution to the random maximum peak heights, using the R software (http://www.r-project.org). For a given promoter, we then calculate the probability of observing a peak of the same or higher height by chance using this distribution. These probabilities (P values) are adjusted for multiple testing using the Benjamini-Hochberg procedure.20 We reasoned that even peaks of moderately low height represent true peaks if they can be replicated in more than one ChIP-chip experiment. Thus, we first select peaks with a relatively low stringency cutoff (false discovery rate [FDR] = 10%), but require peaks to be called in more than half of the replicate experiments.
Pathway and motif analyses
Pathway analyses were performed using the iPAGE program.21 CpG islands were detected using the newcpgreport EMBOSS program (http://emboss.sourceforge.net/), with default parameters. FIRE22 was used for ab initio motif discovery, using −minr = 2 as the only nondefault parameter. Promoter sequences used in FIRE were those covered by probes in the promoter array. FIRE motifs were compared with binding sites in JASPAR and TRANSFAC using the CompareACE approach.23
EZH2 and H3K27me3 binding in hESCs
EZH2 and H3K27me3 ChIP-seq data in hESCs was obtained from a published study.18 ChIP-seq peaks were detected using ChIPseeqer (manuscript in preparation) with significance threshold for peaks set using −t = 5. To compare ChIP-chip and ChIP-seq data, the approximately 1.5-kb regions covered for each promoter in the promoter array were first extended by 1 kb on each side; we then asked whether each of these genomic regions overlapped with the peaks detected by ChIPseeqer using the CompareIntervals tool in the ChIPseeqer framework.
Microarray data
Microarray experiments of normal and malignant B cells24,25 were downloaded from GEO. Interarray quantile normalization was performed on these arrays, irrespective of the summarization and normalization procedures used originally by the authors (eg, RMA, MAS5). Differential expression between NBCs (5 arrays) and centroblasts (5 arrays)25 was calculated using 2-tailed t tests; P values were adjusted for multiple testing using the Benjamini-Hochberg procedure and a FDR of 5% was used to select differentially expressed genes.
EZH2 siRNA-mediated knockdown and Q-PCR
Transfection of SUDHL4 cells treated with 100pM siRNA was performed by electroporation using Nucleofector reagents (Amaxa). Quantitative polymerase chain reaction (Q-PCR) was performed 48 hours after transfection, using primers described in Table 1.
Primers used in this study
| Gene . | RefSeq . | Forward . | Reverse . |
|---|---|---|---|
| EZH2 | NM_ 004456NM_152998 | 5′-GGTTCAGACGAGCTGATGAAG-3′ | 5′-CGCTGTTTCCATTCTTGGTT-3′ |
| CDKN1A/p21 | NM_078467 | 5′-GGCAGACCAGCATGACAGATTTC-3′ | 5′-CGGATTAGGGCTTCCTCTTGG-3′ |
| CDKN2A/p16 | NM_000077 | 5′-GGGTCGGGTAGAGGAGGTG-3′ | 5′-TGCCCATCATCATGACCTG-3′ |
| CDKN1B/p27 | NM_004064 | 5′-AATGCGCAGGAATAAGGAAG-3′ | 5′-TTGGGGAACCGTCTGAAAC-3′ |
| BCL2 | NM_000633 | 5′-TGTGTGTGGAGAGCGTCAA-3′ | 5′-ACAGTTCCACAAAGGCATCC-3′ |
| GAPDH | NM_002046 | 5′-CGACCACTTTGTCAAGCTCA-3′ | 5′-CCCTGTTGCTGTAGCCAAAT-3′ |
| HPRT1 | NM_000194 | 5′-AAAGGACCCCACGAAGTGTT-3′ | 5′-TCAAGGGCATATCCTACAACAA-3′ |
| Gene . | RefSeq . | Forward . | Reverse . |
|---|---|---|---|
| EZH2 | NM_ 004456NM_152998 | 5′-GGTTCAGACGAGCTGATGAAG-3′ | 5′-CGCTGTTTCCATTCTTGGTT-3′ |
| CDKN1A/p21 | NM_078467 | 5′-GGCAGACCAGCATGACAGATTTC-3′ | 5′-CGGATTAGGGCTTCCTCTTGG-3′ |
| CDKN2A/p16 | NM_000077 | 5′-GGGTCGGGTAGAGGAGGTG-3′ | 5′-TGCCCATCATCATGACCTG-3′ |
| CDKN1B/p27 | NM_004064 | 5′-AATGCGCAGGAATAAGGAAG-3′ | 5′-TTGGGGAACCGTCTGAAAC-3′ |
| BCL2 | NM_000633 | 5′-TGTGTGTGGAGAGCGTCAA-3′ | 5′-ACAGTTCCACAAAGGCATCC-3′ |
| GAPDH | NM_002046 | 5′-CGACCACTTTGTCAAGCTCA-3′ | 5′-CCCTGTTGCTGTAGCCAAAT-3′ |
| HPRT1 | NM_000194 | 5′-AAAGGACCCCACGAAGTGTT-3′ | 5′-TCAAGGGCATATCCTACAACAA-3′ |
Cell synchronization and cell-cycle analysis
SUDHL4 cells were incubated for 24 hours in complete RPMI medium, followed by a 24-hour incubation in serum-free medium. Starved cells were released into complete medium, and cell-cycle progression was measured in 24 hours. S-phase arrest was measured by fluorescence-activated cell sorting (FACS) using propidium podide staining (50 μg/mL at 4°C for 3 hours in the dark) of ethanol-fixed (75% ethanol, ice-cold) RNase A-pretreated (0.5 mg/mL for 30 minutes) cells. For cell-cycle progression, SUDHL4 cells were synchronized by a reversible double thymidine block. Briefly, exponentially growing cells (∼ 1.5 × 106/mL) were incubated in complete RPMI medium supplemented with 2.0mM thymidine for 19 hours at 37°C. After the first thymidine block, cells were collected, washed twice with thymidine-free RPMI medium, and resuspended in fresh thymidine-free RPMI medium. After 9 hours incubation, 2mM thymidine were added again, and incubation continued for an extra 16 hours (a second thymidine block). Next, cell culture was washed twice with RPMI medium and released into fresh thymidine-free RPMI medium at concentration approximately 1.0 × 106/mL. Cell-cycle progression was monitored by FACS analysis of collected samples every 4-5 hours (for 33 hours total). Collected samples were also analyzed by Western blotting.
DNA methylation profiling using HELP
DLBCL primary cell isolation and HpaII tiny fragment enrichment by ligation-mediated PCR (HELP) profiling on GC B cells and DLBCL primary cells was performed and analyzed as previously described.26
Results
The EZH2 transcriptional program in GC B cells
EZH2 is critical for proper execution of the early lymphopoiesis program in mammals. However, the function of EZH2 during the terminal stage of human B-cell development in GC, from which almost all B-cell lymphomas arise,27 is unknown. We first examined EZH2 protein expression in naive and GC B cells. EZH2 protein expression is undetectable in resting NBCs, but greatly increases during transition to rapidly proliferating GC B cells (ie, centroblasts; supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). In contrast, EZH2 level decreased in the late GC B cells called centrocytes. These data confirm a previous finding13 and show that expression of EZH2 is linked to proliferation and differentiation status of lymphocytes.
To determine the genomic targets of EZH2 in centroblasts, we performed ChIP coupled to a promoter array (ChIP-chip) covering approximately 24 000 human promoters and found that EZH2 binds reproducibly to > 1800 promoters (see “Methods” for details about ChIP-chip peak discovery and statistical significance). We also performed ChIP-chip in centroblasts for trimethylated histone 3 at lysine 27 (H3K27me3). Consistent with EZH2 catalyzing H3K27me3, a majority (66%) of EZH2 target promoters contain a strong H3K27me3 peak (supplemental Dataset 1). Even for the remaining 34% of genes without a sharply delimited H3K27me3 peak, the abundance of H3K27me3 was still significantly higher than non-EZH2 target promoters (supplemental Figure 2).
To determine the biologic functions associated with EZH2 target genes, we performed a gene set enrichment analysis using Gene Ontology functional annotations28 (see “Methods”). This analysis showed that EZH2 preferentially binds to receptor kinases (eg, EGFR, TGFBR1), transcription factors, components of signal transduction pathways (WNT, TGFβ), and genes involved in core developmental processes (Table 2). This was confirmed by performing a similar analysis using KEGG pathway annotations (Table 2). A useful approach to identify critical biologic functions of transcriptional regulators like EZH2 is to identify gene networks in which multiple promoters are bound. An examination of gene networks most heavily affected by EZH2 binding (using Ingenuity Pathway Analysis) revealed a highly connected network of genes involved in cellular growth, proliferation, and cell differentiation (supplemental Figure 3). This network contained several tumor suppressor genes (eg, CDKN1A/p21, CDKN1B/p27, CDKN2A/p16, and HOXA5). Of note, the same network includes the JUN and FOS genes, whose products form the AP1 transcriptional activator. AP1 drives B lymphocyte maturation by activating Blimp1,29 which promotes the exit of B cells from the GC reaction and their differentiation into plasma cells.30 Thus, these results point at EZH2 possibly acting to facilitate cellular proliferation as well as preventing premature B-cell differentiation in GC B cells. This is consistent with the reported differentiation-blocking role of EZH2 in other tissues (eg, in epidermal progenitor stem cells via direct repression of certain genes such as CDKN2A/p16 and interference with AP1 function)31 and in undifferentiated myoblasts.32
Pathways and biological functions associated with EZH2 target genes in GC B cells
| . | No. of genes (expected) . | Overrepresentation P . | Example genes . |
|---|---|---|---|
| GO biological processes | |||
| Skeletal system development | 33 (10) | < 10−9 | TWIST1 |
| Synaptic transmission | 34 (15) | < 10−5 | CHRNB1 |
| Central nervous system development | 25 (10) | < 10−5 | SHH, PAX6 |
| Regulation of cell differentiation | 21 (8) | < 10−4 | HOXA7, NOTCH2 |
| Positive regulation of cell proliferation | 37 (16) | < 10−5 | EGFR,TGFBR1,VEGFA |
| Positive regulation of ossification | 8 (1) | < 10−4 | TGFB2, BMP7 |
| Enzyme-linked receptor protein signaling pathway | 44 (17) | < 10−8 | FLT3, TGFBR1, PDGFRA |
| GO molecular function | |||
| RNA polymerase II transcription factor activity | 37 (18) | < 10−4 | MYOD1, NEUROD1 |
| Sequence-specific DNA binding | 22 (7) | < 10−5 | TBX1, TBX3 |
| Voltage-gated cation channel activity | 17 (7) | < .001 | CACNG4 |
| Potassium channel activity | 14 (5) | < .001 | KCNK1 |
| Vascular endothelial growth factor receptor activity | 6 (1) | < 10−5 | FLT1, FLT3 |
| Transmembrane receptor protein kinase activity | 20 (5) | < 10−6 | ERRB4,EGFR,TGFBR1, FLT1 |
| KEGG pathway | |||
| Calcium signaling pathway | 40 (21) | < 10−4 | CACNA1E |
| Neuroactive ligand-receptor interaction | 65 (36) | < 10−6 | GABRA2 |
| Wnt signaling pathway | 37 (17) | < 10−5 | FZD1, WNT1 |
| TGF-β signaling pathway | 22 (10) | < .001 | TGFBR1, SMAD3 |
| . | No. of genes (expected) . | Overrepresentation P . | Example genes . |
|---|---|---|---|
| GO biological processes | |||
| Skeletal system development | 33 (10) | < 10−9 | TWIST1 |
| Synaptic transmission | 34 (15) | < 10−5 | CHRNB1 |
| Central nervous system development | 25 (10) | < 10−5 | SHH, PAX6 |
| Regulation of cell differentiation | 21 (8) | < 10−4 | HOXA7, NOTCH2 |
| Positive regulation of cell proliferation | 37 (16) | < 10−5 | EGFR,TGFBR1,VEGFA |
| Positive regulation of ossification | 8 (1) | < 10−4 | TGFB2, BMP7 |
| Enzyme-linked receptor protein signaling pathway | 44 (17) | < 10−8 | FLT3, TGFBR1, PDGFRA |
| GO molecular function | |||
| RNA polymerase II transcription factor activity | 37 (18) | < 10−4 | MYOD1, NEUROD1 |
| Sequence-specific DNA binding | 22 (7) | < 10−5 | TBX1, TBX3 |
| Voltage-gated cation channel activity | 17 (7) | < .001 | CACNG4 |
| Potassium channel activity | 14 (5) | < .001 | KCNK1 |
| Vascular endothelial growth factor receptor activity | 6 (1) | < 10−5 | FLT1, FLT3 |
| Transmembrane receptor protein kinase activity | 20 (5) | < 10−6 | ERRB4,EGFR,TGFBR1, FLT1 |
| KEGG pathway | |||
| Calcium signaling pathway | 40 (21) | < 10−4 | CACNA1E |
| Neuroactive ligand-receptor interaction | 65 (36) | < 10−6 | GABRA2 |
| Wnt signaling pathway | 37 (17) | < 10−5 | FZD1, WNT1 |
| TGF-β signaling pathway | 22 (10) | < .001 | TGFBR1, SMAD3 |
Each row of data corresponds to a pathway or function enriched among EZH2 target genes in GC B cells. The first column indicates the name of the pathway or function. The second column indicates the number of genes in that pathway/function, with expected number (by chance) in parentheses. The third column shows the overrepresentation P values, calculated using the hypergeometric distribution. The fourth column shows example EZH2 target genes from the enriched categories.
EZH2 binds to PRE-like DNA elements in centroblasts
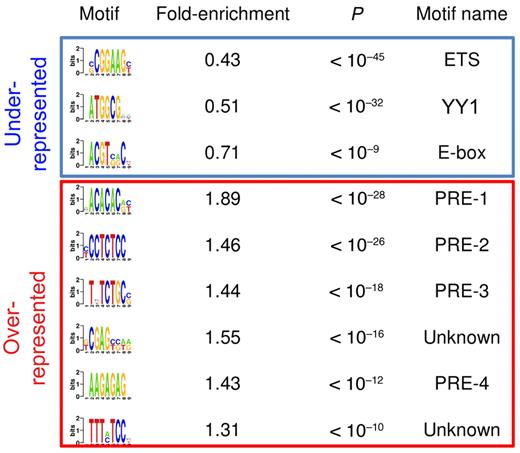
Very little is known about how EZH2 is recruited to specific locations within mammalian genomes. Therefore we explored the genomic features of EZH2 binding sites for sequences that might help to explain its localization in centroblasts. We first found that 71% of EZH2 target promoters contained CpG islands, a fraction that is significantly more than expected by chance (47% expected, P < 1e-111). Then, using an unbiased motif discovery algorithm,22 we found several DNA motifs that are either overrepresented or underrepresented in EZH2 target promoters (Figure 1). As previously reported in embryonic stem cells,18 EZH2-bound promoters in centroblasts tend to lack certain motifs typically bound by transcriptional activators, including an ETS (E-twenty six)-family motif (CGGAAG) and an E-box (Figure 1). Another underrepresented motif in Figure 1 (ATGGCG) was previously found to be overrepresented in promoters bound by the pleiohomeotic (PHO) and pleiohomeotic-like (PHOL) proteins in Drosophila.19 Of note, both PHO and PHOL bind to promoters of active, H3K4-trimethylated, and Trithorax-bound genes. Moreover, vertebrate genomes contain a unique homolog of PHO/PHOL named YY1. Despite reports showing that YY1 coimmunoprecipitates with EZH2 in Xenopus33 and at certain muscle-specific and transcriptionally inactive mouse promoters,32 our results suggest that most EZH2 binding in centroblasts is independent of YY1. Thus, other factors might be needed to recruit PRC2 to the chromatin in centroblasts. Accordingly, our analysis revealed several DNA elements that were highly enriched in EZH2-bound promoters (Figure 1). For example, a CA-repeat motif was present in 31% of EZH2-bound CpG promoters, but only in 17% of nonbound promoters (P < 10−28). A GA-repeat motif is present in 53% of EZH2-bound CpG-island containing promoters, but only in 36% of nonbound promoters (P < 10−26). None of these enriched motifs resembles any of the known motifs in JASPAR or TRANSFAC (see Methods). However, these motifs are highly reminiscent of Polycomb response elements (PRE) in Drosophila.19 We therefore conclude that the human and Drosophila EZH2 orthologs are recruited to similar DNA sequences, with the exception of PHO/YY1 sequences and with a possible additional role for CpG islands.
Putative DNA regulatory sequences enriched in and depleted from promoters bound by EZH2 binding in GC B cells. The first column shows the DNA motifs discovered by FIRE as underrepresented (top 3 motifs) and overrepresented (remaining motifs) in EZH2-bound promoters. The second column indicates the fold over/underrepresentation of each motif in EZH2-bound promoters versus nonbound promoters. The third column shows enrichment and depletion P values, calculated using the hypergeometric distribution. The last column shows motif names, when they could be identified using DNA motif databases such as TRANSFAC or JASPAR. PRE-like sequences were identified based on their similarity to Drosophila PREs.
Putative DNA regulatory sequences enriched in and depleted from promoters bound by EZH2 binding in GC B cells. The first column shows the DNA motifs discovered by FIRE as underrepresented (top 3 motifs) and overrepresented (remaining motifs) in EZH2-bound promoters. The second column indicates the fold over/underrepresentation of each motif in EZH2-bound promoters versus nonbound promoters. The third column shows enrichment and depletion P values, calculated using the hypergeometric distribution. The last column shows motif names, when they could be identified using DNA motif databases such as TRANSFAC or JASPAR. PRE-like sequences were identified based on their similarity to Drosophila PREs.
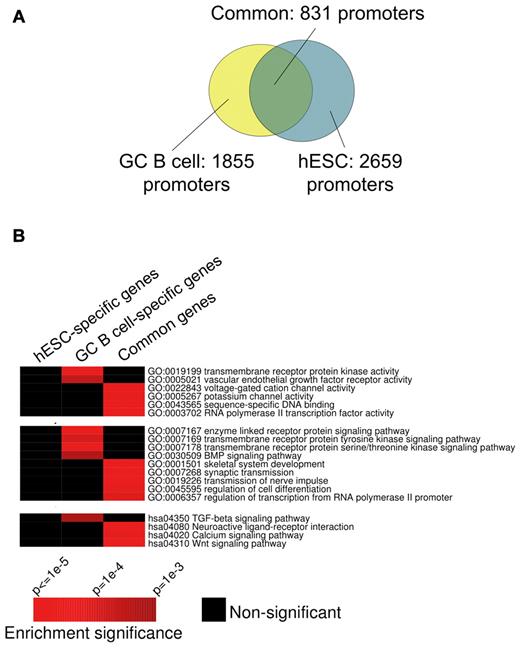
EZH2 targets partially overlapping gene sets in centroblasts and hESCs
PcG binding and function has been studied in embryonic stem cells.18 We wondered whether in the centroblast context EZH2 targets differ from its known targets in hESCs.18 After annotating and matching the centroblast binding sites with those in a previously published study (see “Methods”), we determined that approximately 45% of promoters bound by EZH2 in centroblasts are also bound by EZH2 in hESCs (Figure 2A). We next divided the promoters included in our ChIP-chip studies into 4 groups: (1) those bound in both hESCs and centroblasts; (2) those bound only in centroblasts, (3) those bound only in hESCs, and (4) those not bound in either of these cell types. We then determined whether particular pathways from GO and KEGG were specifically enriched in any of these subgroups and not in others. We found that the centroblast-specific targets were enriched in genes in specific functional classes including kinases and components of specific signal transduction pathways such as TGFβ (Figure 2B). The fact that centroblast-specific EZH2 promoter binding is associated with particular functions and is not randomly distributed suggests that the observed difference between hESC and centroblast genomic localization is likely biologically relevant to the distinct phenotypes of these cells.
Comparing the GC B-cell and hESC EZH2 regulons. (A) Overlap between sets of promoters targeted by EZH2 in GC B cells (yellow) and hESCs (turquoise). EZH2 target promoters in GC B cells were obtained from 4 ChIP-chip replicate experiments. FDR (10%) was used as threshold in each replicate, and only statistically significant peaks observed in > 50% of 4 replicates were retained; see Methods for details. EZH2 target promoters in hESCs were obtained from a published ChIP-seq study.18 (B) Certain processes, pathways, and biochemical functions from the Gene Ontology and KEGG databases are specifically enriched in GC B-cell or common EZH2 target genes. In these heatmaps, rows correspond to pathways and biochemical functions, while columns correspond to gene sets targeted by EZH2 in distinct cell types. Red entries in the heatmap correspond to significant (P < .01) enrichment of specific pathways/functions. Black entries indicate no significant enrichment. Enrichment P values were calculated using the hypergeometric distribution.
Comparing the GC B-cell and hESC EZH2 regulons. (A) Overlap between sets of promoters targeted by EZH2 in GC B cells (yellow) and hESCs (turquoise). EZH2 target promoters in GC B cells were obtained from 4 ChIP-chip replicate experiments. FDR (10%) was used as threshold in each replicate, and only statistically significant peaks observed in > 50% of 4 replicates were retained; see Methods for details. EZH2 target promoters in hESCs were obtained from a published ChIP-seq study.18 (B) Certain processes, pathways, and biochemical functions from the Gene Ontology and KEGG databases are specifically enriched in GC B-cell or common EZH2 target genes. In these heatmaps, rows correspond to pathways and biochemical functions, while columns correspond to gene sets targeted by EZH2 in distinct cell types. Red entries in the heatmap correspond to significant (P < .01) enrichment of specific pathways/functions. Black entries indicate no significant enrichment. Enrichment P values were calculated using the hypergeometric distribution.
EZH2 target genes are repressed in centroblasts
As indicated previously, EZH2 expression is high in centroblasts and undetectable in NBCs. RNA expression levels of EZH2 measured by microarray profiling of NBCs and centroblasts25 confirmed this observation (supplemental Figure 4). Using these centroblast expression profiles, we found that EZH2 target genes were generally expressed at lower levels than nontargets (P < 10−67, Wilcoxon rank-sum test; supplemental Figure 5A). Because EZH2 is not expressed in NBCs, we initially hypothesized that EZH2 target genes would be repressed in centroblasts but not in NBCs. However, this was not the case, and in fact, centroblast EZH2 targets also tend to be expressed at lower levels than non-EZH2 target genes in NBCs (P < 10−78; supplemental Figure 5B). Differentially repressed genes in centroblasts versus NBCs (determined such that FDR = 5%) were also not particularly enriched in EZH2 target genes (P = .43). Thus, most EZH2 target genes in centroblasts are already repressed in NBC, despite undetectable EZH2 expression in these cells, thus suggesting the existence of an EZH2-independent repression mechanism in NBCs.
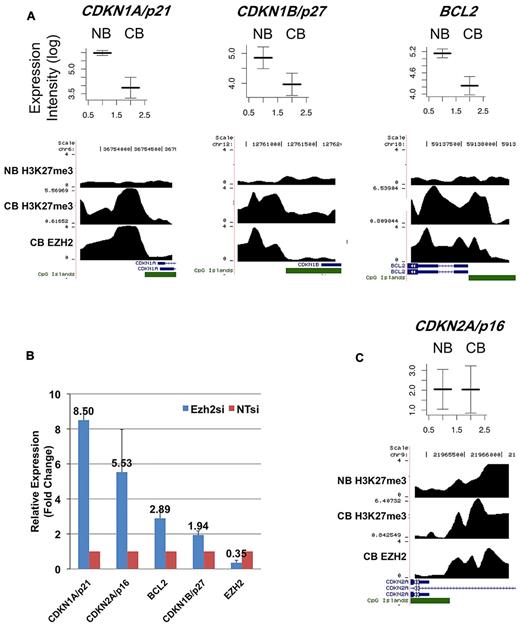
To further investigate this observation, we identified H3K27me3-marked promoters in NBCs using ChIP-chip. Despite the lack of EZH2 expression in NBCs, we found > 2000 promoters with a strong peak of H3K27me3 (see “Methods”). These promoters largely overlapped with H3K27me3-associated and EZH2-bound promoters in centroblasts (supplemental Dataset 1). We nonetheless found > 600 promoters that were not H3K27-trimethylated in NBCs but that were in contrast targeted by EZH2 and H3K27-trimethylated in centroblasts. Strikingly, many of these de novo EZH2 target genes were down-regulated in centroblasts compared with NBCs (19% of the genes for which we had array expression data, while only 8% were expected by chance; P < 10−8, hypergeometric test; supplemental Dataset 1). These down-regulated genes included the CDKN1A/p21 and CDKN1B/p27 tumor suppressors as well as the BCL2 antiapoptotic gene (Figure 3A). To determine whether EZH2 actively represses these genes, we proceeded to knockdown EZH2 expression in the SUDHL4 (GC-derived B-cell lymphoma) cell line. EZH2 mRNA and protein levels were greatly reduced in SUDHL4 cells transfected with an EZH2-specific siRNA (supplemental Figure 6A-B). We then measured the mRNA transcript abundance of CDKN1A, CDKN1B, and BCL2 in the SUDHL4 cell line using Q-PCR. Expression of these 3 genes increased by between 2- and 8-fold upon EZH2 siRNA knockdown (Figure 3B).
Representative genes from 2 classes of EZH2 targets. (A) This figure shows mRNA expression values in NBCs (NB) and centroblasts (CB) (from microarray analyses) and EZH2 binding and H3K27me3 ChIP intensity profiles across probeset-covered regions for 3 genes. In the expression plots (top portion), the y-axis corresponds to log-transformed intensity values from microarray experiments. The x-axis correspond to NB and CB. Expression intensities from 5 CB and 5 NB microarray profiles were used to draw these figures, where both average intensities and standard error (error bars) are shown. In the binding profiles, the x-axis corresponds to genomic position around the transcription start site of the considered genes. RefSeq genes overlapping with the covered regions are shown in blue. CpG islands are shown in green. The y-axis corresponds to smoothed and log-transformed ChIP intensities. The figure shows that CDKN1A/p21, CDKN1B/p27, and BCL2 are not marked by H3K27me3 in NB, and become bound by EZH2 in centroblasts and consequently acquire H3K27me3. Accordingly, these 3 genes are down-regulated at the mRNA expression level in centroblasts compared with NBCs. (B) These 3 genes (together with CDKN2A/p16) are up-regulated at the mRNA level (albeit to a different extent) upon siRNA knockdown of EZH2 in SUDHL4 cells, as measured by Q-PCR. In this figure, mRNA expression levels in siRNA-treated cells are normalized with respect to untreated cells (NTsi). Error bars were drawn using data obtained from 3 replicate experiments. (C) This figure shows average expression levels in NB and CB and binding profiles for CDKN2A/p16. The CDKN2A/p16 promoter is associated with H3K27me3 in NB and expressed at low level; in CB, the same gene is associated with EZH2/H3K27me3 and its expression level does not change.
Representative genes from 2 classes of EZH2 targets. (A) This figure shows mRNA expression values in NBCs (NB) and centroblasts (CB) (from microarray analyses) and EZH2 binding and H3K27me3 ChIP intensity profiles across probeset-covered regions for 3 genes. In the expression plots (top portion), the y-axis corresponds to log-transformed intensity values from microarray experiments. The x-axis correspond to NB and CB. Expression intensities from 5 CB and 5 NB microarray profiles were used to draw these figures, where both average intensities and standard error (error bars) are shown. In the binding profiles, the x-axis corresponds to genomic position around the transcription start site of the considered genes. RefSeq genes overlapping with the covered regions are shown in blue. CpG islands are shown in green. The y-axis corresponds to smoothed and log-transformed ChIP intensities. The figure shows that CDKN1A/p21, CDKN1B/p27, and BCL2 are not marked by H3K27me3 in NB, and become bound by EZH2 in centroblasts and consequently acquire H3K27me3. Accordingly, these 3 genes are down-regulated at the mRNA expression level in centroblasts compared with NBCs. (B) These 3 genes (together with CDKN2A/p16) are up-regulated at the mRNA level (albeit to a different extent) upon siRNA knockdown of EZH2 in SUDHL4 cells, as measured by Q-PCR. In this figure, mRNA expression levels in siRNA-treated cells are normalized with respect to untreated cells (NTsi). Error bars were drawn using data obtained from 3 replicate experiments. (C) This figure shows average expression levels in NB and CB and binding profiles for CDKN2A/p16. The CDKN2A/p16 promoter is associated with H3K27me3 in NB and expressed at low level; in CB, the same gene is associated with EZH2/H3K27me3 and its expression level does not change.
The results above indicate that while a subset of EZH2 target genes in centroblasts are de novo H3K27-trimethylated in this cell type and are actively transcriptionally repressed compared with NBCs, an additional set of EZH2 target genes are already repressed in NBCs, because their promoters already carry H3K27me3. Because the apparent absence of EZH2 in NBCs does not seem to strongly impact H3K27me3 of this subset of genes, one could wonder whether EZH2 actively represses genes in centroblasts that are already repressed in NBCs. For example, CDKN2A is an EZH2 target in centroblasts, but its promoter is also H3K27-trimethylated in NBCs (Figure 3C). We found that EZH2 knockdown significantly increased CDKN2A expression (Figure 3B), suggesting that, at least for certain genes, EZH2 is required for centroblast repression of genes already H3K27-trimethylated in NBCs.
Of note, we found that approximately 54% of the promoters that are targeted by EZH2 in centroblasts but not significantly H3K27-trimethylated in NBCs are also H3K27-trimethylated in hESCs according to the genomic localization data from Ku et al.18 These results suggest that EZH2 up-regulation in centroblasts may induce a stem-cell like repression program that is not present in NBCs. We also found that a significant fraction of centroblast EZH2 target genes (15% while 7% are expected, P < 10−14) that are not H3K27-trimethylated in NBCs are also bound by the BCL6 transcriptional repressor in centroblasts.34 BCL6 and EZH2 may thus cooperate in creating a stem cell–like repression program in centroblasts (see “Discussion”).
EZH2 controls cellular proliferation in DLBCL, and its expression is cell cycle–regulated
Gene expression profiling showed that EZH2 mRNA expression levels are variable in DLBCL (supplemental Figure 4). However, as previously demonstrated for prostate cancer,15 we found that the mRNA abundance of EZH2 positively correlates with cellular proliferation rate, measured using Ki67 staining in a published dataset of primary DLBCL cells24 (Pearson correlation = 0.3, P < .001). This is consistent with a previous observation that EZH2 overexpression increases cell proliferation in Burkitt lymphoma cells, which are also derived from GC B cells.17 Moreover, we observed that siRNA depletion of EZH2 leads to cell cycle arrest at the G1/S transition in SUDHL4 cells (Figure 4). Consistent with this observation, we also found that EZH2 levels fluctuate during cell cycle peaking at the G1 phase (supplemental Figure 7; see “Methods”).
The impact of EZH2 on cell cycle in the SUDHL4 DLBCL cell line. siRNA-mediated EZH2 knockdown in SUDHL4 results in G1/S block and accumulation of cells in S phase. In this experiment, SUDHL4 cells were incubated for 24 hours in complete RPMI medium, followed by a 24-hour incubation in serum-free medium. Starved cells were released into complete medium, and cell cycle progression was measured in 24 hours. The extent of the S phase arrest was measured by FACS using propidium iodide staining. Cell cycle was monitored using FACS 24 hours in serum-starved SUDHL4 cells and 24 hours after reentry.
The impact of EZH2 on cell cycle in the SUDHL4 DLBCL cell line. siRNA-mediated EZH2 knockdown in SUDHL4 results in G1/S block and accumulation of cells in S phase. In this experiment, SUDHL4 cells were incubated for 24 hours in complete RPMI medium, followed by a 24-hour incubation in serum-free medium. Starved cells were released into complete medium, and cell cycle progression was measured in 24 hours. The extent of the S phase arrest was measured by FACS using propidium iodide staining. Cell cycle was monitored using FACS 24 hours in serum-starved SUDHL4 cells and 24 hours after reentry.
EZH2 target promoter DNA is hypomethylated in centroblasts and hypermethylated in DLBCL
We examined the extent of promoter DNA methylation of EZH2 targets in centroblasts. DNA was isolated from tonsils of 8 healthy patients and methylation was assayed using the HELP assay and a high-density oligonucleotide microarray representing 50 000 CpGs across approximately 14 000 promoters.26,35 HELP methylation profiles for the 8 samples were then averaged. Using these data, we found that EZH2 target promoters are significantly less methylated than nontargets (Figure 5; P = 0, Wilcoxon test). These results are consistent with studies in other normal cell types that showed that H3K27-trimethylated regions are generally unmethylated at the DNA level.7-9 Because not all EZH2-bound promoters are equally hypomethylated (Figure 5), we wondered whether the level of DNA methylation measured by HELP correlates with absolute expression level of the nearby gene, in centroblasts. We found that this was not the case (Pearson correlation = 0.01, P = .77). In contrast, we found a highly significant negative correlation between expression and DNA methylation in genes not bound by EZH2 (Pearson correlation between expression levels and HELP log-ratios = 0.21, P < 10−26). We conclude that DNA methylation, if present, does not play a role in regulating gene expression at EZH2-bound promoters, unlike at EZH2-free promoters, and that therefore EZH2-mediated repression is DNA methylation-independent in normal B cells.
EZH2 target promoters are less DNA-methylated than nontargets in GC B cells. This figures shows the distribution of DNA methylation values (HELP log-ratios) for all EZH2 target promoters (red) and nontargets (black) represented on our HELP arrays. The HELP assay was performed from tonsil DNA from 8 healthy patients as described previously26 and using a high density oligonucleotide microarray representing 50 000 CpGs across approximately 14 000 promoters. Methylation profiles for the 8 patients were averaged out. Importantly, in HELP, higher and positive HELP log-ratios correspond to lower DNA methylation.
EZH2 target promoters are less DNA-methylated than nontargets in GC B cells. This figures shows the distribution of DNA methylation values (HELP log-ratios) for all EZH2 target promoters (red) and nontargets (black) represented on our HELP arrays. The HELP assay was performed from tonsil DNA from 8 healthy patients as described previously26 and using a high density oligonucleotide microarray representing 50 000 CpGs across approximately 14 000 promoters. Methylation profiles for the 8 patients were averaged out. Importantly, in HELP, higher and positive HELP log-ratios correspond to lower DNA methylation.
We next compared and contrasted the DNA methylation levels of EZH2 target genes in centroblasts versus the DNA methylation profiles (obtained by HELP) of 60 primary DLBCL samples.36 We first derived the set of differentially methylated promoters in DLBCL compared with centroblasts (using 2-tailed t tests with stringent P value threshold of 10−7). Most were hypermethylated in DLBCL compared with centroblasts (924 hypermethylated vs 16 hypomethylated). However, we also observed that > 27% of aberrantly hypermethylated promoters in DLBCL were also targets of EZH2 in centroblasts, a fraction that is much higher than expected by chance (10% expected, P < 10−48). Some of these hypermethylated genes include tumor suppressors (eg, CDKN1A and CDKN2B). Of note, genes targeted by EZH2 in both centroblasts and hESCs were often hypermethylated (22%). These results are consistent with previous reports that hypermethylated genes in tumors tend to be PcG targets in stem cells37,38 and suggest that genes broadly targeted by EZH2 are more likely to be hypermethylated in tumors than cell type–specific EZH2 targets. However, we also found that 12% of hypermethylated EZH2 target genes are centroblast-specific EZH2 targets (ie, they are targeted in centroblasts but not in hESCs). Thus, these results show for the first time that many non-hESC PcG targets are also hypermethylated in tumors.
Discussion
Our report is the first to explore EZH2-mediated transcriptional programming and functions in normal GC B cells and DLBCL. First, we find that EZH2 binds to a specific set of functionally related target genes in centroblasts that are distinct from EZH2 targets in embryonic stem cells. This result suggests that certain EZH2 functions may be specific to GC B cells. However, EZH2 target genes in GC B cells and embryonic stem cells are still partially overlapping. Importantly from the functional standpoint, > 50% of EZH2 target genes that are uniquely H3K27 trimethylated in centroblasts (versus NBCs) are also targeted by H3K27me3/EZH2 in hESCs. This suggests that EZH2 up-regulation during the NBCs to centroblast transition reactivates a stem cell–like repression program not present in NBCs and possibly featuring increased self-renewal and proliferative potential. Second, we found that EZH2 target genes that are de novo H3K27 trimethylated in centroblasts compared with NBCs are also down-regulated at the mRNA level in centroblasts. Several of these genes (ie, CDKN1A, CDKN1B and BCL2) are derepressed upon siRNA knockdown of EZH2 in a DLBCL cell line, consistent with a proproliferative function for EZH2 in GC B cells. Third, EZH2 also binds to the promoters of many centroblast genes that are already H3K27 trimethylated and silenced in NB cells; thus, EZH2 may also maintain the repressive state of these genes and perhaps reinforces that repression in proliferating cells (EZH2 knockdown in DLBCL cells derepressed one of these genes, CDKN2A). It is presently unclear why H3K27me3 is present at thousands of promoters in NBCs, despite no detectable EZH2 protein expression. Because these cells are mostly in a resting state, it is possible that low and undetectable levels of EZH2 maintain H3K27me3 levels. Other proteins may also catalyze H3K27me3 deposition (eg, EZH1).39 Finally, and consistent with the derepression of genes that restrain cellular proliferation (CDKN1A, CDKN1B, and CDKN2A), EZH2 knockdown leads to significant cell cycle arrest in G2/S in DLBCL cells. EZH2-mediated silencing in normal GC B cells therefore likely contributes to the ability of these cells to undergo massive clonal expansion. This function of EZH2 uncovered in our study may contribute to the malignant transformation of GC B cells into DLBCL and facilitate their proliferative phenotype.
Our observation that EZH2 targets are associated with low DNA methylation in centroblasts is consistent with several previous reports. Kondo et al showed a minimal overlap between genes silenced by H3K27me3 and DNA methylation in prostate and breast cancer cell lines.40 Gal-Yam et al41 showed that gene silencing in a prostate cancer cell line is either associated with PcG binding or DNA methylation but rarely with both mechanisms. Because we also showed that EZH2 tends to localize at promoters with CpG islands, our data are consistent with the lack of methylation observed at CpG islands in normal cells.42 This observation indicates that PcG-mediated repression is independent of DNA methylation in normal B cells (even in proliferating centroblasts). Interestingly, several recent studies have provided evidence that this may be different in lymphoma cells, where it was shown that many hypermethylated genes are targeted by PcG in stem cells.37 Our study extends these observations to show that hypermethylated genes in DLBCL are targeted by PcG in centroblasts. We hypothesize that lymphoma cells may have achieved a selective advantage by recruiting DNMTs to PcG-bound or/and H3K27me3-marked promoters and that increased promoter DNA methylation may consolidate and stabilize PcG-mediated repression of one or several of the tumor suppressors targeted by PcG or make them less capable of responding to antioncogenic signals. In this model, increased DNA methylation would indiscriminately affect the many promoters regulated by PcG, even if few of them can positively contribute to the tumor phenotype.
Of note, we found that a significant fraction of the promoters (15% while only 7% are expected, P < 10−14) that are targeted by EZH2 and H3K27-trimethylated in centroblasts but not NBCs are also bound by BCL6 in centroblasts.34 Moreover, EZH2 and BCL6 have a highly similar expression pattern: they are both up-regulated in centroblasts compared with NBCs and both down-regulated in centrocytes (supplemental Figure 1). This striking pattern of coexpression and target sharing points to a possible cooperation between these 2 repressors. It is for example possible that EZH2 helps maintain BCL6-mediated repression in centroblasts. Genes repressed by these 2 mechanisms, which include BCL2, CDKN1A, and CDKN1B,34,43,44 may be particularly important for the GC B-cell phenotype. Nonetheless, the overlap between BCL6 and EZH2 targets is limited and many of the known direct and important BCL6 targets (eg, TP53, CHEK1, and ATR) are not EZH2 targets. Likewise, and despite the coexpression of these 2 genes, we have not been able to show a physical interaction between BCL6 and EZH2 (data not shown).
In aggregate, these results point to several possibly overlapping functional roles for EZH2 in normal GC B-cells: (1) to favor cellular proliferation by directly repressing several tumor suppressor genes; (2) to create a repression state similar to that found in stem cells that might foster self-renewal and prevent premature differentiation; and (3) to maintain and stabilize a transcriptional repression program already in place in NBCs. Our results point toward a strong impact of EZH2 on gene expression in normal and malignant cells. While the exact binding pattern of EZH2 in malignant cells needs to be further studied, we found that in a large dataset of DLBCL expression arrays, the mRNA expression profiles of centroblast EZH2 target genes tend to be significantly anticorrelated with expression of EZH2 itself (supplemental Figure 8). Together with our functional data, this suggests that EZH2 binding is largely conserved between GC B cell and DLBCL. Of note, the recently reported Tyr641 mutation in the EZH2 SET domain was shown to reduce H3K27 methylation activity in vitro,14 although the function of mutant EZH2 has not been characterized yet. Only a fraction (9.7%) of all DLBCL primary tumors harbor the mutation, indicating that loss of EZH2 activity (if present) is specific to a subset of tumors whose physiology and additional genetic and epigenetic alterations are still to be determined. More generally, the in vivo impact of Tyr641 is presently unknown. The observation that all Tyr641 mutations are heterozygous14 suggests that EZH2 activity is still required in these tumors and is not incompatible with an overall gain-of-function effect of the mutation. Altogether, these results suggest that therapeutic targeting of EZH2 might have significant antilymphoma effects and support the rationale for development of inhibitors of the EZH2 SET domain. Our observations also suggest that genes whose EZH2-mediated repression favors self-renewal in centroblasts (and in hESCs) could become constitutively silenced by DNA methylation in tumors, thus turning reversible repressed genes in centroblast into more stably and less reversibly repressed genes. Thus, combining SET domain inhibitors with DNA demethylating agents (eg, decitabine) may represent a more efficient strategy to kill lymphoma cells compared with either drug alone.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Francesca Demichelis for her critical reading of the manuscript and 2 anonymous reviewers for their suggestions and constructive comments.
I.V. was supported by a Fellowship Award from the Lymphoma Research Foundation. A.M. is supported by R01 CA104538, the chemotherapy foundation, and is a Scholar of the Leukemia and Lymphoma Society. O.E. is supported by startup funds from the Institute for Computational Biomedicine.
National Institutes of Health
Authorship
Contribution: O.E., A.M.M., and I.V. conceived and designed the study; I.V. and R.S. performed experiments; H.G., N.A.J., and R.D.G. provided reagents, samples, and analytic tools; I.V., A.M.M., and O.E. analyzed the data; and I.V., A.M.M., and O.E. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ari M. Melnick, Weill Cornell Medical College, Hematology/Oncology Division, Department of Medicine, 1300 York Avenue, New York, NY, 10021; e-mail: amm2014@med.cornell.edu; or Olivier Elemento, Weill Cornell Medical College, Institute for Computational Medicine, Department of Physiology and Biophysics, 1305 York Avenue, New York, NY, 10021; e-mail: ole2001@med.cornell.edu.