Abstract
The expression of interferon-β (IFN-β) in virus-infected HeLa cells established a paradigm of multifactorial gene regulation, in which cooperative assembly of transcription factors (TFs) at the composite DNA element (enhanceosome), is central for amplification of weak activating signals provided by individual TFs. However, whether the same TFs and the same DNA element are essential for IFN-β induction in response to bacterial stimuli are less well understood. Here we report that rapid and transient transcription of IFN-β in response to TLR4 stimulation with bacterial lipopolysaccharide (LPS) follows nuclear factor-κB (NF-κB) RelA activation and recruitment to the IFN-β genomic locus at multiple spatially separated regulatory regions. We demonstrate that the IFN-β enhanceosome region is not sufficient for maximal gene induction in response to LPS and identify an essential cluster of homotypic κB sites in the 3′ downstream of the gene. The cluster is characterized by elevated levels of histone 3 lysine 4 mono-methylation, a chromatin signature of enhancers, and efficiently binds RelA-containing NF-κB complexes in vitro and in vivo. These findings demonstrate that IFN-β gene activation via multifactorial enhanceosome assembly is potentiated in LPS-stimulated cells by NF-κB interactions with all functional κB sites in the locus.
Introduction
Type I interferons (IFNs) comprise a large gene family (with IFN-β and IFN-αs playing the major role) that in response to viral infection can stimulate the synthesis of a large set of antiviral products, including enzymes that bind double-stranded RNA, GTPases that affect the intracellular transport of viral particles, transcription and translational regulators, proapoptotic genes, etc.1 Type I IFNs are also produced in response to other pathogens and play an important role in the protection against bacterial infection.2 A considerable part of the gene expression signature in cells stimulated with bacterial lipopolysaccharide (LPS) is formed by IFN-stimulated genes.3,4 Another group of antiviral factors, which exhibit several features in common with type I IFNs, are type III IFNs.5,6 These include IFN-λ1 (IL29), exclusively found in primate genomes, and virtually identical IFN-λ2 (IL28a)/IFN-λ3 (IL28b), which are the results of a recent gene-duplication event.7
The early studies of the human IFN-β gene regulation in a model system of virus-infected human epithelial carcinoma HeLa cell line established a paradigm of multifactorial gene regulation.8 The highly evolutionarily conserved enhancer of IFN-β comprises 4 regulatory cis-elements: namely the positive regulatory domains (PRDs) I-IV. Upon virus infection, the PRDs have been shown to facilitate the cooperative assembly of a multiprotein regulatory complex involving nuclear factor κB (NF-κB) RelA/p50, IFN regulatory factors (IRF) 3/7 and activating transcription factor (ATF) 2/c-Jun to form a stable enhanceosome complex.9,10 This subsequently leads to the recruitment of histone acetyl transferases (HATs) and other chromatin remodelling activities that are necessary for the initiation of IFN-β gene expression.11
In contrast, the proximal promoter region of the IFN-λ1 gene, which resembles that of IFN-β and is essential for its induction by IRF3/7,12 is not sufficient for maximal gene activation in response to bacterial LPS. IFN-λ1 is regulated via a cluster of homotypic NF-κB binding sites in the distal promoter that efficiently bind NF-κB and function independently of the proximal IRF3/7 binding site.13 The differences in the organization of regulatory elements of IFN-β and IFN-λ1 genes are intriguing considering their remarkable resemblance in the pattern of induction and biological activity.14 For example, in human monocyte-derived dendritic cells (MDDCs) and plasmacytoid dendritic cells (pDCs), IFN-β and IFN-λs are the only IFNs strongly induced by stimulation of Toll-like receptor 4 (TLR4) with bacterial LPS.15 Moreover, the kinetics and magnitude of their induction are highly similar,15 indicating that these IFNs might be under a common transcriptional regulatory mode.
Here we sought to address the question whether in response to LPS stimulation, regulation of IFN-β follows the transcription mode of IFN-λ1 (ie, uses alternative κB sites in addition to PRDII in the proximal promoter region). Using short-interfering RNA (siRNA) and a dominant negative mutant of IκBα (super repressor of canonical NF-κB signaling), we demonstrate that NF-κB RelA is a major factor regulating expression of the human IFN-β gene in response to bacterial LPS. We inspect the molecular architecture of the human IFN-β locus and map putative κB sites outside the enhanceosome region of IFN-β. Our data reveal that a cluster of κB sites in the 3′ downstream region of the human IFN-β gene is essential for gene induction in response to LPS. RelA is efficiently recruited to this cluster as well as to the proximal promoter region. Moreover, both regions are characterized by elevated levels of histone 3 lysine 4 monomethylation (H3K4me1), a chromatin modification associated with enhancers.16 Multiple RelA binding events in the IFN-β locus are closely followed by the recruitment of PolII to the transcription start site (TSS) of the gene and by expression of IFN-β mRNA. Our findings suggest that activation of IFN-β in response to LPS extends beyond the conventional enhanceosome assembly.
Methods
Plasmids
NF-κB and IRF expression constructs were generated by cloning cDNAs encoding these factors in the pENTR vector (Invitrogen) modified to contain the CMV promoter and IRES-linked GFP (pBent). IFN-β promoter fragments were obtained by PCR of genomic DNA and cloned into the pGL3 Basic vector (Promega). The sequences and restriction maps of all constructs are available on request.
Cell culture
All reagents used for cell culture were tested for endotoxin and only in use if the endotoxin levels were < 20 pg/mL (Lonza). All cell cultures were maintained at 37°C in 5% CO2 and 95% humidity in the appropriate media supplemented with 10% fetal calf serum (Gibco) and 1% penicillin/streptomycin (PAA). HEK293-TLR4/CD14-Md2 cells (Invivogen) were cultured in DMEM (PAA) supplemented with 10 mg/mL and 50 mg/mL Blasticidin and HygroGold (Invivogen), respectively, for selection pressure. Enriched populations of human monocytes were obtained from the blood of healthy donors by elutriation essentially as described previously.17 Monocyte-derived dendritic cells (MDDCs) were obtained by culturing the human monocytes in RPMI (PAA) supplemented with of 50 ng/mL granulocyte macrophage colony-stimulating factor and 10 ng/mL interleukin-4 (Peprotech) for 5 days. MDDCs were stimulated with 10 ng/mL LPS (Alexis Biochemicals) or 50 μg/mL poly I:C (Invivogen).
Transfection and luciferase reporter assays
siRNA-mediated knockdown was performed using On-Target Plus SMART pool reagents (Dharmacon) designed to target human NF-κB subunits (LQ-003533, LQ-004767, LQ-004768) and IRF3 (LQ-006875) and either Lipofectamine RNAiMAX (HEK293-TLR4/CD14-Md2; Invitrogen) or DharmaFECT I (MDDCs; Dharmacon) transfection reagents. Transfections of luciferase (100 ng) and protein expressing (1-10 ng) constructs were performed in 96-well plates in triplicate using Lipofectamine 2000 Transfection Reagent (Invitrogen). Luciferase activity was measured with Dual-Glo Luciferase Assay System (Promega) after stimulation with LPS (1 μg/mL) for 8 hours. Firefly luciferase readings were normalized to the intensity of produced from the control pRL-TK construct (10 ng; Promega) cotransfected into the same cells.
Adenovirus production and infection
Replication-deficient adenoviral constructs were prepared using the AdEasy system (Qbiogene). Adenoviral vector without insert (control), vector encoding a mutant of IκΒα (S32A/S36A) resistant to degradation (IκBαSR; kind gift from Dr Michael Karin, University of California, San Diego), and a luciferase reporter driven by tandem copies of κB site (NFκB-luc; Clontech) were utilized in this study. The adenoviruses were generated as previously described.17 Infections of MDDCs were performed in 96-well plates in triplicate at a multiplicity of infection of 50:1. Cells were seeded in serum-free, antibiotics-free RPMI containing the desired number of viral particles in a final volume of 50 μL. The plates were incubated overnight at 37°C followed by aspiration of the supernatants and replacement with 100 μL of standard media per well. Cells were allowed to recover for a further 24 hours before experimental assay.
RNA extraction and qRT-PCR
MDDCs and HEK293-TLR4/CD14-Md2 cells were stimulated with the indicated ligands for 1 hour or 4 hours, respectively, unless otherwise stated. Total RNA was extracted using a QiaAmp RNA Blood mini kit (QIAGEN). cDNA was synthesised using SuperScript III Reverse Transcriptase (Invitrogen) and 18-mer oligo dTs (Eurofins MWG Operon). The gene expression was analyzed by 2-standard curve or ΔΔCt methods where appropriate by quantitative real-time polymerase chain reaction (qRT-PCR) with TaqMan primer sets to human IFN-β (Hs00277188-s1), IFN-λ1 (Hs00601677-g1), and PO (4310879E) in a Corbett Rotor-gene 6000 machine (Corbett Research Ltd).
Western blotting
Equal amounts of proteins were resolved by Novex Tris-glycine gel electrophoresis and transferred onto Hybond-N membranes (Amersham Biosciences) using Novex X-Cell II Mini Cell (Invitrogen). The membranes were incubated overnight with the primary antibodies: sc-372 (RelA), sc-9082 (IRF3; Santa Cruz Biotechnology), 06-886 (p50), 06-413 (p52; Upstate Biotech), and 265 (c-Rel), 1319 (RelB; kindly provided by Dr Nancy Rice, National Cancer Institute), actin (Sigma-Aldrich) at 4°C, followed by 60 minutes' incubation with horseradish peroxidase-conjugated secondary antibody, and detection by chemiluminescence with ECL (GE Healthcare). The membranes were exposed to the X-ray film (Kodak) and densitometry measurements were performed using ScanMaker 9800XL scanner (Microtek) and Scion Image software (National Institutes of Health).
Nucleoplasmic and cytoplamic extraction
Cells were subjected to experimental conditions and nuclear extracts were prepared as previously described.18
ChIP
Chromatin immunoprecipitation (ChIP) assays were carried out essentially as previously described19 using antibodies against RelA, PolII (sc-899; Santa Cruz Biotechnology), or H3K4me1 (ab8895; Abcam). The im-munoprecipitated DNA fragments were then interrogated by qPCR using SYBR Premix Ex Taq master mix (Takara Bio) and the following primers for IFNβ locus: Cluster 1 (5prime]-AGCTCCTTGCATTGGCTTTTGTGCT-3′ and 5prime]-GCTCTTAGCAAGACCTTGGCCATCTCA-3′); Cluster 2 (5prime]-AGTGCCACAGACCACAACTGCTTCTAA-3′ and 5prime]-AAACCGCCCTGAACCACTTCCTGG-3′); promoter/TSS (5prime]-TGAAAGGGAGAAGTGAAAGTGGG-3′ and 5prime]-AAGGCTTCGAAAGGTTGCAGTTA-3′); Cluster 3 (5prime]-GGGGTTCCCATTCCTCCTACTGTGTGC-3′ and 5prime]-TTAAGCTTGCAGGGATGGTGGGGG-3′). Intergenic primer sequences were as follows (5prime]-GATGGAGGCTGAATGGCCTTC-3′ and 5prime]-GGGCACACAGCAGATCAGAGG-3′). Data were analyzed using Rotorgene 6000 software (Corbett Research Ltd). All primer sets were tested for specificity and efficiency of amplification by analyzing melt curves and the number of cycles required to observe a product using a range genomic DNA concentrations.
Electrophoretic mobility shift assay
Oligonucleotide probes (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) were radiolabeled with [α-32P] dCTP (PerkinElmer). Nuclear extracts from MDDCs stimulated with 100 ng/mL LPS for 1 hour or recombinant p50/RelA proteins were used in binding as previously described.20 For supershift analysis, the reaction mixture was preincubated with antibodies against RelA, p50, or c-Rel for 10 minutes before addition of the labeled probe. The gels were quantified using the PhosphorImager (FujiFilm).
Bioinformatics and statistical analyses
Genomic sequences were obtained using the publicly available UCSC hg18 human genome assembly (http://genome.ucsc.edu/). The nucleotide sequence encompassing −5 kb to +2 kb at the human IFN-β locus was inspected with JASPAR transcription factor binding sites searching software (http://jaspar.cgb.ki.se/)21 for the presence of putative NF-κB (JASPAR matrixes MA0061, MA0101) sites with a relative score profile threshold of 80%. All statistical analyses were performed using 1-way analysis of variance (ANOVA) with Dunnett multiple comparison posttest or Student t test where appropriate (*P < .05, **P < .01, ***P < .001; GraphPad Prism 5.0 software).
Results
LPS induces rapid and transient transcription of IFN genes in human MDDCs
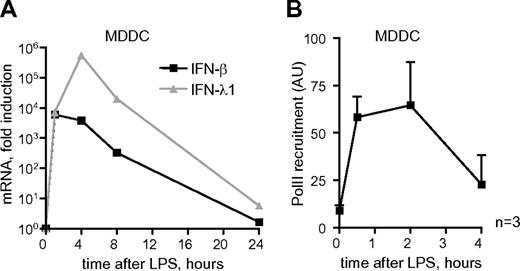
Previous studies demonstrated that mRNA of IFN-β and IFN-λ1 were coinduced in MDDCs by stimulation of TLR4 with LPS.15 To confirm these results, MDDCs were obtained from primary human monocytes and cultured for 5 days in the presence of granulocyte macrophage colony-stimulating factor and interleukin-4.22 Rapid and transient induction of both IFNs was observed, with a peak of mRNA expression between 1 and 4 hours after LPS induction (Figure 1A). Consistent with previously published data,15 no IFN-α expression was detected in these cells (data not shown). The expression of the IFN-β and IFN-λ1 genes was regulated at the transcriptional level, shown by the induced recruitment of PolII to the TSS of the genes (Figure 1B and data not shown). For IFN-β, maximal PolII levels were observed between 30 minutes and 2 hours after LPS stimulation, and decreased after 4 hours, preceding the kinetics of mRNA expression (Figure 1B).
Rapid and transient expression of IFN genes induced by LPS in MDDCs. (A) Expression of IFN-β and IFN-λ1 mRNA in LPS-stimulated MDDCs. The y-axis represents fold induction over mRNA expression in resting cells. Representative data of 2 experiments each performed in triplicate are shown. (B) PolII recruitments to the promoter/TSS regions of IFN-β. Data shown are the mean + SEM from 3 experiments conducted in the cells from independent donors, each nor-malized against the signal generated by the same primer set using no antibody control sample.
Rapid and transient expression of IFN genes induced by LPS in MDDCs. (A) Expression of IFN-β and IFN-λ1 mRNA in LPS-stimulated MDDCs. The y-axis represents fold induction over mRNA expression in resting cells. Representative data of 2 experiments each performed in triplicate are shown. (B) PolII recruitments to the promoter/TSS regions of IFN-β. Data shown are the mean + SEM from 3 experiments conducted in the cells from independent donors, each nor-malized against the signal generated by the same primer set using no antibody control sample.
Thus, TLR4 signaling resulted in rapid and transient transcription of IFN-β and IFN-λ1 genes in MDDCs. Significantly, the kinetics of IFN mRNA expression in response to TLR4 activation were distinct from those induced by virus infection or TLR3 stimulation with double-stranded RNA mimic poly I:C, which peaks at around 6-8 hours poststimulation.11,23
NF-κB RelA is a key factor regulating IFN-β gene expression in response to LPS
In MDDCs, nuclear RelA is observed as early as 10 minutes, reaching the maximum between 30 and 60 minutes after LPS stimulation (Thomson et al13 and supplemental Figure 1A), suggesting that in these cells this factor may be involved in early IFN induction in response to LPS stimulation. Of interest, consistent with Lundberg et al,24 we observed a very limited nuclear accumulation and activity of RelA in MDDCs treated with poly I:C (supplemental Figure 1A-B). To test the involvement of NF-κB RelA in regulation of the human IFN-β gene, siRNA-mediated knockdown of RelA was conducted in MDDCs extracted from 5 independent donors and in the model system of HEK293-TLR4/CD14-Md2 cells stimulated with LPS. The efficiency of RelA siRNA knockdown was estimated by serial dilutions of the mock sample run on the same gel and was normally around 50% in MDDCs (supplemental Figure 2A) and above 85% in HEK293-TLR4/CD14-Md2 cells (supplemental Figure 2B).
Consequently, in HEK293-TLR4/CD14-Md2 cells IFN-β mRNA expression was significantly reduced by RelA knockdown (down to approximately 20% of its original levels; Figure 2A). Notably, inhibition of other NF-κB Rel subunits had a less profound effect on TLR4-induced IFN-β gene expression (supplemental Figure 2C). Furthermore, ectopically expressed RelA could efficiently drive IFN-β mRNA expression, while expression of RelB and c-Rel subunits had no substantial effect (supplemental Figure 2D). To further validate the specificity of the observed IFN-β mRNA inhibition, we performed a complementation analysis whereby an exogenous RelA, encoded by a modified nucleotide sequence resistant to siRNA-mediated degradation (Figure 2B), was introduced into HEK293-TLR4/CD14-Md2 cells. This led to the complete reversal of IFN-β mRNAs inhibition by siRNA to RelA (Figure 2C).
RelA is a key transcription factor regulating IFN-β expression in response to LPS. (A-C) Depletion of RelA levels greatly reduces IFN-β gene expression in HEK-293-TLR4/CD14-Md2 cells. (A) IFN-β mRNA expression in cells transfected with siRNAs targeting NFκB RelA (siRelA) presented as percent of expression in the control cells transfected with nontargeting siRNA (siC). Data shown are the mean + SEM from 3 independent experiments. ***P < .001 (Student t test). (B) Western blots showing the siRelA induced degradation of RelAwt and unaffected expression of RelAmut and are representatives of 3 experiments. (C) LPS-induced IFN-β mRNA expression in cells complemented with RelAwt or RelAmut shown as fold induction over the level in nonstimulated control cells transfected with nontargeting siRNA (siC). Data shown are the mean + SD of a representative of 2 independent experiments each performed in triplicate. (D-F) Inhibition of RelA significantly reduces IFN-β mRNA expression in MDDCs. (D) IFN-β mRNA expression in cells transfected with siRNAs targeting NF-κB RelA (siRelA) presented as percent of expression in the control cells transfected with nontargeting siRNA (siC). Data shown are the mean + SEM from 5 independent donors. **P < .01 (Student t test). (E) IFNβ mRNA expression in MDDCs infected with increasing concentrations of adenovirus carrying IκBαSR: normalized against cells infected with adenovirus carrying an empty expression vector pBent and shown as the mean + SD of a representative of 3 independent experiments. (F) Efficiency of ΝF-κB activity suppression by IκBαSR in MDDCs estimated using luciferase reporter assay with NF-κB-luc virus: shown as the mean + SD of a representative of 3 independent experiments each performed in triplicate.
RelA is a key transcription factor regulating IFN-β expression in response to LPS. (A-C) Depletion of RelA levels greatly reduces IFN-β gene expression in HEK-293-TLR4/CD14-Md2 cells. (A) IFN-β mRNA expression in cells transfected with siRNAs targeting NFκB RelA (siRelA) presented as percent of expression in the control cells transfected with nontargeting siRNA (siC). Data shown are the mean + SEM from 3 independent experiments. ***P < .001 (Student t test). (B) Western blots showing the siRelA induced degradation of RelAwt and unaffected expression of RelAmut and are representatives of 3 experiments. (C) LPS-induced IFN-β mRNA expression in cells complemented with RelAwt or RelAmut shown as fold induction over the level in nonstimulated control cells transfected with nontargeting siRNA (siC). Data shown are the mean + SD of a representative of 2 independent experiments each performed in triplicate. (D-F) Inhibition of RelA significantly reduces IFN-β mRNA expression in MDDCs. (D) IFN-β mRNA expression in cells transfected with siRNAs targeting NF-κB RelA (siRelA) presented as percent of expression in the control cells transfected with nontargeting siRNA (siC). Data shown are the mean + SEM from 5 independent donors. **P < .01 (Student t test). (E) IFNβ mRNA expression in MDDCs infected with increasing concentrations of adenovirus carrying IκBαSR: normalized against cells infected with adenovirus carrying an empty expression vector pBent and shown as the mean + SD of a representative of 3 independent experiments. (F) Efficiency of ΝF-κB activity suppression by IκBαSR in MDDCs estimated using luciferase reporter assay with NF-κB-luc virus: shown as the mean + SD of a representative of 3 independent experiments each performed in triplicate.
IFN-β mRNA expression in MDDCs with depleted levels of RelA was also diminished although to somewhat lesser extent than in HEK293-TLR4/CD14-Md2 cells (Figure 2D). The apparent limited dependence of the IFN-β gene expression on the presence of RelA in MDDCs can be misleading and reflect on the efficiency of siRNA inhibition in primary cells (supplemental Figure 2A-B). Thus, we used a complementary approach to inhibit NF-κB. We transduced MDDCs with the adenovirus expressing a dominant-negative variant of IκBα, which is resistant to signal-induced phosphorylation and degradation and acts as a super repressor of canonical NF-κB signaling (IκBαSR). The efficiency of DNA delivery at higher multiplicities of infection approached 100% with no significant effect on the resting cells assessed by IFN response (supplemental Figure 2E). We observed a dose-dependent inhibition of IFN-β mRNA expression (Figure 2E). When NF-κB activity was inhibited to virtually background levels in the IκBαSR infected cells at multiplicity of infection = 50 plaque-forming units/cell (Figure 2F), we detected a significant decrease in IFN-β expression. This decrease was comparable to the level of IFN-β mRNA inhibition achieved by siRNA-mediated RelA knockdown in the HEK293-TLR4/CD14-Md2 cells (down to approximately 20% of its original level; Figure 2A,E).
Thus, stimulation of MDDCs with LPS but not poly I:C efficiently induce NF-κB RelA nuclear translocation and activity, which is required for expression of the IFN-β gene.
NF-κB RelA is recruited to the multiple κB sites in the human IFN-β locus
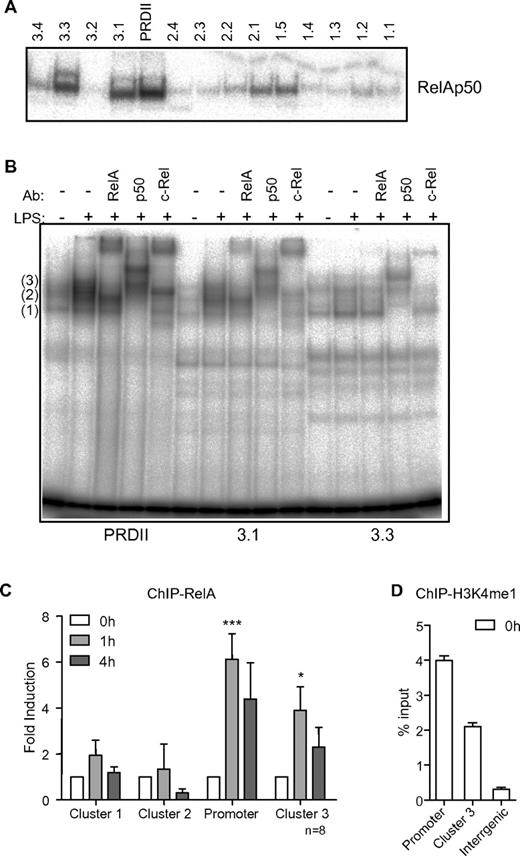
The well-described IFN-β enhancer is located in the proximal promoter region and comprises adjacent binding sites for IRF3/7 and NF-κB RelA/p50 dimers, which are essential for cooperative assembly of TFs via the unique DNA interface formed by these sites.25 Previously we have shown that κB sites in the distal part of the IFN-λ1 promoter are required for maximal levels of gene expression in response to LPS.13 Here we asked the question if there are any other κB sites outside the enhanceosome region that may serve as additional docking sites for RelA in the IFN-β locus. The human IFN-β gene locus (up to 5 kb upstream of the gene and 2 kb downstream) was inspected for the presence of such sites using JASPAR database and search engine21 and 3 clusters of putative κB sites located: cluster 1 at −2902/−2128 nucleotides upstream (5 sites); cluster 2 at −1205/−311 nucleotides upstream (4 sites); and cluster 3 at +1110/+1562 nucleotides downstream (4 sites) of the IFN-β TSS were identified (supplemental Figure 3A).
We next examined the binding affinity of these individual distal κB sites in the IFN-β locus for recombinant RelA/p50, the canonical NF-κB heterodimer. Electrophoretic mobility shift assay (EMSA) analysis revealed that 4 of 13 κB sites (1.5, 2.1, 3.1, and 3.3) exhibited prominent protein/DNA interactions (Figure 3A). Furthermore, titration assays showed that κB sites 3.1 and 3.3 had a higher binding affinity than sites 1.5 and 2.1 to RelA/p50, while the promoter PRDII site had the strongest affinity (supplemental Figure 3B). We then inspected whether these sites could interact with the nuclear contents of resting or LPS-stimulated MDDCs. Increased protein binding was observed with extracts from LPS-stimulated cells, and supershift assay with antibodies against RelA, c-Rel, and p50 revealed the binding of RelA-, c-Rel, and p50-containing complexes to sites PRDII and 3.1 and p50-containing complexes to site 3.3 (Figure 3B). In addition, competition assay using unlabeled PRDII probe confirmed that the same complexes were binding to κB sites 3.1 as to the proximal promoter of IFN-β (supplemental Figure 3C). These are likely to be: (1) p50 homodimer, (2) RelA/p50 heterodimer, and (3) RelA/c-Rel heterodimer and RelA/RelA homodimer, consistent with previously observed complex formation at the PRDII site.26 Both RelA/p50 and RelA/c-Rel heterodimers have been shown to efficiently activate PRDII.26
Multiple κB sites in the human IFN-β locus recruit NF-κB RelA. (A) Binding of recombinant purified RelA/p50 to radioactively labeled probes corresponding to the respective κB sites. (B) EMSA supershift analysis of protein-DNA binding in nuclear extracts from resting or LPS-stimulated MDDCs using antibodies against RelA, p50, and c-Rel. The identified complexes are likely to be (1) p50 homodimer, (2) RelA/p50 heterodimer, and (3) RelA/c-Rel heterodimer and RelA/RelA homodimer. (C) RelA recruitment to IFN-β ehanceosome (promoter) and clusters 1, 2, and 3 in MDDCs from multiple blood donors. Data shown are the mean ± SEM of 8 independent experiments normalized against levels of RelA recruitment at 0 hours. *P < .05, ***P < .001 (Student t test). (D) H3K4me1 modification presence at IFN-β ehanceosome (promoter), cluster 3 and a transcriptionally inactive intergenic region in MDDCs. Data are normalized to total histone 3 levels and presented as mean values ± SD from a representative of 4 independent donors.
Multiple κB sites in the human IFN-β locus recruit NF-κB RelA. (A) Binding of recombinant purified RelA/p50 to radioactively labeled probes corresponding to the respective κB sites. (B) EMSA supershift analysis of protein-DNA binding in nuclear extracts from resting or LPS-stimulated MDDCs using antibodies against RelA, p50, and c-Rel. The identified complexes are likely to be (1) p50 homodimer, (2) RelA/p50 heterodimer, and (3) RelA/c-Rel heterodimer and RelA/RelA homodimer. (C) RelA recruitment to IFN-β ehanceosome (promoter) and clusters 1, 2, and 3 in MDDCs from multiple blood donors. Data shown are the mean ± SEM of 8 independent experiments normalized against levels of RelA recruitment at 0 hours. *P < .05, ***P < .001 (Student t test). (D) H3K4me1 modification presence at IFN-β ehanceosome (promoter), cluster 3 and a transcriptionally inactive intergenic region in MDDCs. Data are normalized to total histone 3 levels and presented as mean values ± SD from a representative of 4 independent donors.
ChIP analysis was used to examine the recruitment of the common denominator for 2 active NF-κB complexes, RelA, to the IFN-β proximal promoter region and to the newly identified clusters of putative κB sites in vivo. In multiple MDDC donors, we observed enrichment in RelA binding to the proximal promoter region and to cluster 3, which peaked at 1 hour and returned back to the basal level at 4 hours after LPS stimulation (Figure 3C), corresponding to the kinetics of PolII recruitment to the TSS and of the gene transcription (Figure 1). No statistically significant enrichment in RelA recruitment was detected at clusters 1 and 2 (Figure 3C). Histone 3 lysine 4 monomethylation (H3K4me1) is now recognized as a marker for enhancers that could contribute to the induction of target genes.16 Significantly, our ChIP analysis detected the presence of this histone modification at cluster 3 and the proximal promoter regions, but not at an intergenic region (Figure 3D).
Thus, we identified a region downstream of the IFN-β gene, which is marked as an enhancer in human MDDCs, aids in recruitment of RelA-containing complexes to the locus in response to TLR4 stimulation and encompasses at least 2 sites capable of forming complexes with NF-κB proteins.
κB sites in the 3′ downstream region are required for maximal levels of IFN-β gene activation in response to LPS
To examine whether cluster 3 contributes to transcriptional regulation of the IFN-β gene in response to LPS stimulation, a series of luciferase gene-reporter constructs driven by the sequences upstream and downstream of the IFN-β gene, that encompassed the newly identified clusters of κB sites were generated (Figure 4A). All constructs demonstrated LPS-inducible luciferase gene-reporter activity, but the response was significantly stronger for the constructs that incorporated cluster 3 in HEK293-TLR4/CD14-Md2 cells (Figure 4B) and in RAW264 macrophages (supplemental Figure 4A). Furthermore, in the presence of cluster 3, overexpression of RelA (but not c-Rel) significantly induced gene-reporter activity (Figure 4C and supplemental Figure 4B), confirming the role of RelA in this context. Thus, we concluded that the proximal promoter region, which contains the previously described virus-inducible promoter of the IFN-β gene,8 was not sufficient for maximal gene induction by LPS, whereas cluster 3 was essential.
κB sites located 3′ downstream of human IFN-β loci are required for maximal gene induction. (A) The molecular organization of the luciferase constructs is shown. Construct 1: −102/+75 (promoter/enhanceosome [P]); construct 2: −2036/+75 (cluster 2 [C2] and promoter [P]); construct 3: −102/+75 and +639/+1840 (promoter [P] and cluster 3 [C3]); construct 4: −3030/+75 (cluster 1 [C1], cluster 2 [C2], and promoter [P]); construct 5: −3030/+75 and +639/+1840 (cluster 1 [C1], cluster 2 [C2], promoter [P], cluster 3 [C3]). (B) LPS-stimulated gene reporter activity of the respective constructs in HEK293-TLR4/CD14-Md2: data shown are the mean + SEM from 5 independent experiments each performed in triplicate; ***P < .001 (1-way ANOVA). (C) RelA-induced gene-reporter activity of the luciferase constructs: data shown are the mean + SEM from 4 independent experiments each performed in triplicate; ***P < .001 (1-way ANOVA). (D) The molecular organization of the luciferase construct with site-specific mutations in κB sites in cluster 3 (C3). (E) RelA-induced gene-reporter activity of the wild-type construct 5 (5.wt) and the construct with site-specific mutations in κB sites (5.mut): results are shown as mean + SEM from 3 independent experiments each performed in triplicate and expressed as percent stimulated control (wild-type = 100%), **P < .01 (Student t test).
κB sites located 3′ downstream of human IFN-β loci are required for maximal gene induction. (A) The molecular organization of the luciferase constructs is shown. Construct 1: −102/+75 (promoter/enhanceosome [P]); construct 2: −2036/+75 (cluster 2 [C2] and promoter [P]); construct 3: −102/+75 and +639/+1840 (promoter [P] and cluster 3 [C3]); construct 4: −3030/+75 (cluster 1 [C1], cluster 2 [C2], and promoter [P]); construct 5: −3030/+75 and +639/+1840 (cluster 1 [C1], cluster 2 [C2], promoter [P], cluster 3 [C3]). (B) LPS-stimulated gene reporter activity of the respective constructs in HEK293-TLR4/CD14-Md2: data shown are the mean + SEM from 5 independent experiments each performed in triplicate; ***P < .001 (1-way ANOVA). (C) RelA-induced gene-reporter activity of the luciferase constructs: data shown are the mean + SEM from 4 independent experiments each performed in triplicate; ***P < .001 (1-way ANOVA). (D) The molecular organization of the luciferase construct with site-specific mutations in κB sites in cluster 3 (C3). (E) RelA-induced gene-reporter activity of the wild-type construct 5 (5.wt) and the construct with site-specific mutations in κB sites (5.mut): results are shown as mean + SEM from 3 independent experiments each performed in triplicate and expressed as percent stimulated control (wild-type = 100%), **P < .01 (Student t test).
To examine the contribution of mapped κB sites to LPS induction of the IFN-β gene; site-specific mutations in the κB sites were generated in the background of construct 5 (Figure 4D). Removal of NF-κB docking sites in the distal region significantly reduced the level of the RelA-induced gene-reporter activity (Figure 4E), further supporting the notion that the cluster of κB sites downstream of the IFN-β gene constitutes a part of the gene transcription machinery.
In summary, κB sites in the 3′ downstream region are required for the maximal level of IFN-β expression in response to TLR4 stimulation. The region represents a novel enhancer of the IFN-β gene locus, which transmits an activating signal along with the previously mapped enhanceosome region.
NF-κB controlled expression of IFN-β and IFN-λ1 in response to LPS is proportional to the number of functional κB sites
NF-κB has been recently shown to act as an analog transcriptional regulator, whereby the degree of gene activation is proportional to the number of functional κB sites.27 We have previously mapped 5 functional κB sites in the promoter region of the IFN-λ1 gene.13 Here we identified at least 2 novel functional κB sites in 3′ downstream region of the IFN-β gene in addition to the known PRDII site in the enhanceosome region. Furthermore, during the course of this study we noticed a somewhat stronger induction of the IFN-λ1 gene compared with IFN-β gene in MDDCs in response to LPS stimulation (Figures 1A), suggesting that it may be due to a higher number of κB sites in the IFN-λ1 locus. To address this possibility, we analyzed the levels of IFN-β and IFN-λ1 gene activation over a range of RelA concentrations in LPS-stimulated HEK293-TLR4/CD14-Md2 cells by varying the amount of siRNA reagent used for inhibiting endogenous RelA (Figure 5A). Under a broad range of RelA concentrations greater activation of IFN-λ1 than IFN-β was observed (Figure 5B), with the shapes of the curves similar to the ones described by Giorgetti et al for an additive model of activating signal transmission to PolII.27 Similar results were obtained when RelA was ectopically expressed in resting HEK293-TLR4/CD14-Md2 cells (supplemental Figure 5A). We therefore concluded that activation of IFN-β, as well as IFN-λ1, by NF-κB is likely to be proportional to the number of available strong affinity κB sites and comply with the analog mode of transcriptional response.
LPS-induced expression of IFN-β and IFN-λ1 is proportional to the number of κB sites. (A) Western blot analysis of the efficiency of siRNA-mediated RelA knockdown in HEK293-TLR4/CD14-Md2 cells using a range of siRelA concentrations (0.1hyphen]30nM). (B) Dose-dependent inhibition of IFNβ and IFN-λ1 mRNA expression in HEK293-TLR4/CD14-Md2 cells and stimulated with LPS (1 μg/mL). The x-axis shows percent RelA expression relative to control (100%) based on the densitometry analysis of Western blots. The y-axis is fold induction over mRNA expression in resting cells. Data shown are the mean + SD from a representative of 2 independent experiments.
LPS-induced expression of IFN-β and IFN-λ1 is proportional to the number of κB sites. (A) Western blot analysis of the efficiency of siRNA-mediated RelA knockdown in HEK293-TLR4/CD14-Md2 cells using a range of siRelA concentrations (0.1hyphen]30nM). (B) Dose-dependent inhibition of IFNβ and IFN-λ1 mRNA expression in HEK293-TLR4/CD14-Md2 cells and stimulated with LPS (1 μg/mL). The x-axis shows percent RelA expression relative to control (100%) based on the densitometry analysis of Western blots. The y-axis is fold induction over mRNA expression in resting cells. Data shown are the mean + SD from a representative of 2 independent experiments.
Of interest, the profile of IFN-β and IFN-λ1 gene activation across the wide range of IRF3 concentrations was similar for both genes (supplemental Figures 5B), suggesting that both genes rely on comparable number of IRF-binding sites. IRF3 is another factor critical for the TLR4-induced IFN-β expression by myeloid cells.3 In fact, knockdown of IRF3 in MDDCs resulted in a similar degree of inhibition of IFN-β and IFN-λ1 expression (supplemental Figure 5C). We therefore inspected the human IFN-β gene locus for the presence of putative IRF-binding sites and detected 2 putative IRF-binding sites in cluster 1, 1 in cluster 2, and 2 in cluster 3 regions (supplemental Table 2). Although the detailed functional characterization of the IRF-binding sites is beyond the scope of this study, we noted that the reporter construct encompassing clusters 1, 2, 3 was significantly greater induced by IRF3 than the proximal promoter construct (supplemental Figure 5D). However, the mutations in the newly identified κB sites of cluster 3 had no effect on the IRF3-induced gene activation (supplemental Figure 5E), suggesting that IRF3 and RelA are unlikely to form a multicomponent complex at this region.
Discussion
Type I and type III IFNs play a critical role in the innate immune response against viral and microbial infection. In this study, we investigated the molecular mechanisms leading to the production of IFN-β in TLR4-stimulated human MDDCs. We demonstrate that NF-κB RelA plays a major role in regulating IFN-β transcription and discover a novel cluster of κB sites downstream of the gene that is essential for RelA recruitment to the IFN-β locus and for maximal levels of gene induction in response to LPS stimulation. Thus, expression of IFN-β in TLR4-stimulated MDDCs is dependent on multiple RelA binding events at spatially separated κB binding sites. This is different to regulation of its expression in virus-infected HeLa cells, which has been shown to rely on enhanceosome assembly at the proximal promoter region, as well as in TLR3-stimulated MDDCs. Our findings provide new insights into the complexity of IFN-β induction in human immune cells and demonstrate stimulus-specific elements of IFN-β regulation.
In human MDDCs, LPS induces a strong and transient mRNA expression of IFN-β and IFN-λ1, which subsides within 8 hours (Figure 1). In contrast, IFN-β mRNA expression in response to virus infection or induction with poly I:C peaks at around 6-8 hours after stimulation.11,23 One of the main distinguishing factors between these polar situations is the selective induction of high levels of NF-κB RelA in the cell nuclei; it is seen within 30 minutes after LPS-stimulation in the nuclei of MDDCs (supplemental Figure 1 and Thomson et al13 ), and its depletion by siRNA results in a statistically significant reduction of LPS-induced IFN-β mRNA expression (Figure 2). Poly I:C, in contrast, fails to efficiently induce RelA nuclear translocation in human MDDCs (supplemental Figure 1 and Lundberg et al24 ). Similarly RelA appears to be present at suboptimal concentrations in the nuclei of virus-infected HeLa cells, limiting the input signal to IFN-β.28 Overexpression of RelA significantly increases the number of IFN-β–producing cells.28 Differences in cell physiology and the type of stimuli used may favor alternative models of transcriptional response for IFN-β gene: additive binding of RelA to all available κB binding sites in the IFN-β locus at high concentrations of NF-κB (ie, LPS-stimulated MDDCs) versus cooperative binding of RelA and other enhanceosome components at low initial concentrations of key TFs (ie, virus-infected HeLa cells, poly I:C–stimulated MDDCs).27 In the latter case, the relatively low input signal can be amplified by the fixed arrangement of binding sites in the enhanceosome, controlling the expression of IFN-β in a highly nonlinear fashion.29
The human IFN-β locus encompasses many putative κB sites in addition to the previously described PRDII within the enhanceosome region (supplemental Figure 3). Spatially separated κB sites of the IFN-β locus are loosely organized in clusters (supplemental Figure 3), among which the cluster located downstream of the gene consisting of at least 2 functional κB sites (Figure 3) is essential for maximal level of gene induction in response to TLR4-stimulation (Figure 4). Similarly, a cluster of κB binding sites in the distal promoter region of IFN-λ1 gene recruits the bulk of RelA activity and is required for maximal levels of IFN-λ1 induction by LPS.13 The proximal promoter region of IFN-λ1, which is also one of the most conserved regions of the locus, has a strong resemblance to the IFN-β enhanceosome region and comprises binding sites for NF-κB as well as IRF proteins.12,13,30 Type I and type III IFN genes are believed to have arisen from a common ancestor.31 Retroposition and extended segmental DNA duplication undergone by the type I IFN system make it a particularly interesting example of evolution of regulatory elements and acquisition of functional promoters. While retrocopies can inherit basic promoters proximal to the TSS, they can also obtain enhancer elements at the new location.32 It is exciting to speculate that all type I and type III IFNs can respond to viruses via the inherited ancestoral regulatory elements (ie, IRF binding sites or specific arrangement of NF-κB and IRF binding sites) immediately upstream of the TSS, but IFN-β and IFN-λ1 are the only IFNs expressed in response non-nucleic acid ligands, such as LPS, because their genomic loci acquired multiple functional distal κB (and possibly IRF) sites.
A recent studied by Wang et al33 reported that IFN-β mRNA expression was reduced by 40% in virus infected, RelA-deficient mouse DCs, and interpreted it as the lack of an effect. We observed a similar level of reduction in human MDDCs treated with siRNA against RelA (Figure 2) and concluded that RelA plays a key role in IFN-β production in response to TLR4 stimulation. One major difference is the use of knockdown or knockout approaches to protein inhibition, with the RNA interference–mediated knockdown never achieving the complete removal of the protein. Despite the residual levels of RelA in the human cells, the reduction in IFN-β mRNA expression was: (1) reproducible in independent blood donors (Figure 2); (2) siRNA dose-dependent (Figure 5); and (3) validated by independent means (using IκBα super repressor mutant; Figure 2). The benefits of siRNA knockdown approach, however, include the possibility for modulating protein levels in fully differentiated cells over a range of concentrations allowing better assessment of the quantitative differences in transcriptional responses (Figure 5). Thus, this study aligns with other previously published work proposing a key role for NF-κB RelA in IFN-β transcription (eg, in TLR4-stimulated mouse macrophages3 or virus-induced human epithelial carcinoma derived HeLa cells).26
The studies reported here further our understanding of the complexity of IFN-β gene regulation. The assembly of multifactorial enhanceosome complex essential for gene induction in cells with limiting amounts of key TFs, or in response to stimuli that do not efficiently activate them, is further potentiated at high concentrations of active NF-κB by the involvement of extra κB sites outside of the conserved enhanceosome region. Thus, our data point to IFN-β as an example of a gene that comprises 2 modes of transcriptional response: the first being a digital mode and the second one, analog.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr David Saliba (Kennedy Institute of Rheumatology [KIR]) for his role in optimizing ChIP conditions and Miss Katrina Blazek (KIR) for technical assistance with qPCR. We are also grateful to Drs Lynn Williams, Matt Peirce, and Grigory Ryzhakov (KIR) for helpful discussions and critical reading of the manuscript.
The research leading to these results was supported by Wellcome Trust Core Award 075491/Z/04 to R.R.C. and Medical Research Council (MRC) New Investigator Award 75548 to I.A.U. and has received funding from the European Community's Seventh Framework Program FP7/2007-2013 under grant agreement number 222008.
Wellcome Trust
Authorship
Contribution: F.G.G., S.J.P.T., T.K., and A.L. performed research; F.G.G., S.J.P.T., R.R.C., and I.A.U. designed research and analyzed data; and F.G.G. and I.A.U. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Irina A. Udalova, Kennedy Institute of Rheumatology Division, Faculty of Medicine, Imperial College of Science, Technology and Medicine, 65 Aspenlea Rd, London, W6 8LH, United Kingdom; e-mail: i.udalova@imperial.ac.uk.


![Figure 4. κB sites located 3′ downstream of human IFN-β loci are required for maximal gene induction. (A) The molecular organization of the luciferase constructs is shown. Construct 1: −102/+75 (promoter/enhanceosome [P]); construct 2: −2036/+75 (cluster 2 [C2] and promoter [P]); construct 3: −102/+75 and +639/+1840 (promoter [P] and cluster 3 [C3]); construct 4: −3030/+75 (cluster 1 [C1], cluster 2 [C2], and promoter [P]); construct 5: −3030/+75 and +639/+1840 (cluster 1 [C1], cluster 2 [C2], promoter [P], cluster 3 [C3]). (B) LPS-stimulated gene reporter activity of the respective constructs in HEK293-TLR4/CD14-Md2: data shown are the mean + SEM from 5 independent experiments each performed in triplicate; ***P < .001 (1-way ANOVA). (C) RelA-induced gene-reporter activity of the luciferase constructs: data shown are the mean + SEM from 4 independent experiments each performed in triplicate; ***P < .001 (1-way ANOVA). (D) The molecular organization of the luciferase construct with site-specific mutations in κB sites in cluster 3 (C3). (E) RelA-induced gene-reporter activity of the wild-type construct 5 (5.wt) and the construct with site-specific mutations in κB sites (5.mut): results are shown as mean + SEM from 3 independent experiments each performed in triplicate and expressed as percent stimulated control (wild-type = 100%), **P < .01 (Student t test).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/25/10.1182_blood-2010-05-282285/4/m_zh89991062570004.jpeg?Expires=1769287783&Signature=oR0amnH7IgnOut3cBMeaqnjXF08gaZdM1mPd3DzAmf3dCszkuULr9f3iHQII8-3lLdCDtfpMXbwnH3RWRXH8152N75uuun-cdiexzZyCF7AjGz1dQprNyVdXQMVw3VakhPvhsqJOAf9X4Ps2-CYAoP1SZ2kzwSkPy5eyBPMA~fh7psQGky9u2k4q5mKZnumRqRPVbYSWyfXHyculwvPLxxi-isyczGFvTL-ujN5NuLFoTVslW~hXUTkch9WBPvJ7to32ciXxKXdJ1uOWIgxGjsb5MGYUBWqvjy2tJilTbh91tM6m2tWVUgfTIHWw8Xy7QbHcEPHy-TNW2LfMeXYi9w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. LPS-induced expression of IFN-β and IFN-λ1 is proportional to the number of κB sites. (A) Western blot analysis of the efficiency of siRNA-mediated RelA knockdown in HEK293-TLR4/CD14-Md2 cells using a range of siRelA concentrations (0.1hyphen]30nM). (B) Dose-dependent inhibition of IFNβ and IFN-λ1 mRNA expression in HEK293-TLR4/CD14-Md2 cells and stimulated with LPS (1 μg/mL). The x-axis shows percent RelA expression relative to control (100%) based on the densitometry analysis of Western blots. The y-axis is fold induction over mRNA expression in resting cells. Data shown are the mean + SD from a representative of 2 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/25/10.1182_blood-2010-05-282285/4/m_zh89991062570005.jpeg?Expires=1769287783&Signature=itkBAWWMT6rmftYzEtrOT-Dkqu-BKmvLSmUiyjH2cKXoqQ9UoVJoMKI9txZBAJGNcTB9r2vUdoNUn1LuNwS~ho-UncokcLUMhkzLqq6NTTwFNvb2iHJGH8BZMWkH9vmAUj232Tpcw0xfWEqDl1vRhk7jgovuo1ZopvfHeAcd6BpuEJ8RUTDDjzVfxf83vt0IfwFBmmUdt9vkZ8LbjUBxwV5jeQdIp43XcU8DYAjip5uoGxaWi~ZodvR3l4AXF99Uhfvn7NJSjsrxt4YdixxmzWdJSJayODAXQkmOlWtuBAUbOlyUp~RESJO~BXJwR-WnuSX1y~mwuj1oh-i~4rxzag__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)