The hyper-IgM syndromes (HIGMs) are immunodeficiencies that have taught us much about the details of humoral immunity. In this issue of Blood, Lanzi et al characterize a rare subset of HIGM patients and demonstrate that these disorders may also teach us about endoplasmic reticulum (ER) storage diseases.1
The primary HIGMs comprise genetic deficiencies in CD154 (HIGM1), CD40 (HIGM3), or AICDA (HIGM2) and share a phenotype of absent or reduced serum immunoglobulin G (IgG) and IgA, normal to elevated levels of IgM, impaired humoral responses to T cell–dependent antigens and generation of the B-cell memory compartments, and recurrent bacterial infections. Curiously enough, HIGM2 patients also present with systemic autoimmune disease.2
CD154, a type II transmembrane protein of the tumor necrosis factor (TNF) superfamily, is transiently expressed by activated CD4 T cells. CD154 is the ligand of CD40, a type I TNFR family member, constitutively expressed on B lymphocytes and myeloid antigen-presenting cells. Cognate interactions between antigen-activated T and B lymphocytes include CD154:CD40 ligation, leading to CD40 trimerization and the generation of NF-κB signals crucial for B-cell survival, proliferation, and AICDA expression. In the absence of CD154:CD40 interaction, incomplete cognate interactions abrogate B-cell responses to T cell–dependent antigens. In HIGM1 and HIGM3 patients, germinal centers do not form and memory B-cell generation is greatly impaired. In contrast, HIGM2 patients fully support cognate T:B lymphocyte interaction but cannot express the AICDA gene product, AID, and are unable to initiate Ig class-switch recombination (CSR) and somatic hypermutation (SHM). Thus, whereas all HIGM patients are unable to produce IgG and IgA antibody, plasmacytes, and memory B cells, HIGM2 patients are characterized by lymphadenopathy associated with exaggerated germinal center responses.
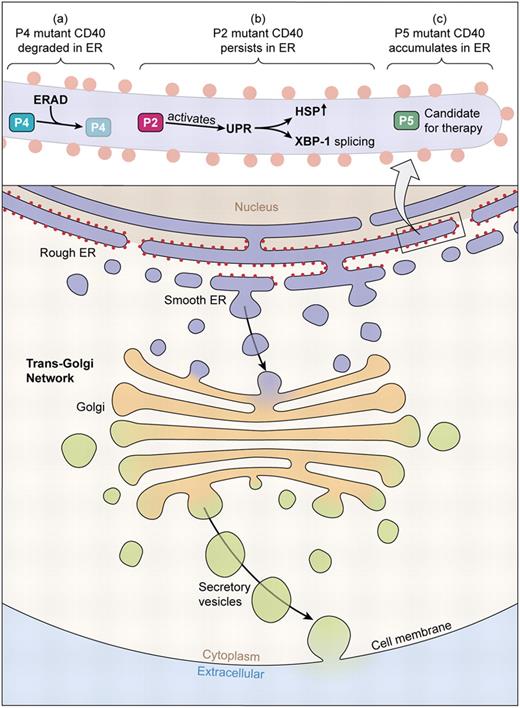
In this issue, Lanzi and colleagues have now detailed the molecular characteristics of 5 rare HIGM3 patients and identify a subset of genetic hypomorphs carrying missense mutations or small in-frame deletions.1 In the 3 hypomorphs, mutant CD40 polypeptide is synthesized but retained—to various degrees—in the ER due to improper protein folding. In one patient, mutant CD40 polypeptide is efficiently removed from the ER and degraded; in a second patient, the mutant protein persists in the ER and triggers a stress response that can lead to apoptosis. Whereas the third CD40 hypomorph patient showed ER retention and no evidence of Ig CSR, the mutation was permissive for residual CD40 expression and the mutant CD40 was capable of NF-κB signaling. This spectrum of CD40 mutations raises the possibility that some HIGM mutants might respond to therapies that mitigate the retention of misfolded protein in the ER or ER stress (see figure).
Secretory and endocytic pathways of mammalian cells. CD40 molecules synthesized in the ER are delivered to the cell surface via the trans-Golgi network and vesicles that subsequently fuse with the plasma membrane. Some CD40 mutants carrying missense mutations or small in-frame deletions cannot complete this pathway but are retained instead in the ER. The 3 novel CD40 mutants identified by Lanzi et al in this issue exhibit distinct fates in the ER: (1) the P4 mutant does not leave the ER and is promptly degraded by ERAD; (2) the P2 mutant CD40 persists in the ER and activates UPR as determined by increased HSP production and XBP-1 splicing; and (3) the P5 mutant accumulates in the ER but neither is degraded nor elicits UPR. The P5 CD40 mutant is a candidate for therapies based on pharmacologic chaperones. (Professional illustration by Kenneth X. Probst.)
Secretory and endocytic pathways of mammalian cells. CD40 molecules synthesized in the ER are delivered to the cell surface via the trans-Golgi network and vesicles that subsequently fuse with the plasma membrane. Some CD40 mutants carrying missense mutations or small in-frame deletions cannot complete this pathway but are retained instead in the ER. The 3 novel CD40 mutants identified by Lanzi et al in this issue exhibit distinct fates in the ER: (1) the P4 mutant does not leave the ER and is promptly degraded by ERAD; (2) the P2 mutant CD40 persists in the ER and activates UPR as determined by increased HSP production and XBP-1 splicing; and (3) the P5 mutant accumulates in the ER but neither is degraded nor elicits UPR. The P5 CD40 mutant is a candidate for therapies based on pharmacologic chaperones. (Professional illustration by Kenneth X. Probst.)
The ER is the site of synthesis, folding, oligomerization, and posttranslational modification for both secreted and membrane-bound proteins. In the ER, nascent peptides fold into their proper confirmations before entering other cellular compartments or the extracellular space. Protein folding in the ER requires the assistance of molecular chaperones, including the heat shock proteins (HSPs; eg, GRp78/BiP), lectin-like chaperones, and thiol oxidoreductases. Proteins that reach their native, functional state are released from chaperones and transported out of the ER. Misfolded proteins are retained in the ER and degraded by reverse translocation into the cytosol where their ubiquitination leads to proteolysis within the proteosome. This ER-associated degradation (ERAD) pathway removes as much as 30% of newly synthesized proteins that become denatured due to translational or posttranslational error in addition to removing mutant polypeptides that are intrinsically unstable. Although it is efficient, ERAD can be overwhelmed (reviewed in Kaufman et al3 ).
ER chaperones do not dissociate from misfolded proteins; accumulation of misfolded protein in the ER decreases chaperone availability and, if severe, triggers the unfolded protein response (UPR), a highly conserved signaling pathway that allows the cell to cope with ER stress.
Activation of the UPR selectively activates genes that expand the capacity of the ER to fold and degrade protein; at the same time, cellular translation is attenuated, slowing the flux of nascent polypeptides into the ER. Together, these mechanisms give the cell a chance to deal with protein-folding defects. There is, however, a limit to the cell's capacity to deal with misfolded proteins, and chronic ER stress eventually triggers apoptosis.
The UPR is triggered by translumenal proteins that monitor chaperone concentration in the ER. UPR signals are generated via 3 protein sensors: IRE1 (inositol-requiring enzyme 1), PERK (double-stranded RNA-dependent protein kinase–like ER kinase), and ATF6 (activating transcription factor 6).
IRE1 bears an endonuclease site that, when activated by autophosphorylation, splices a 26-bp intron of X-box–binding protein 1 (XBP-1) mRNA.4 This splicing event results in a frame shift that encodes an alternate C-terminus and produces a (spliced) XBP-1 transcription factor that activates chaperone and ERAD genes. PERK is maintained in an inactive state when bound to HSP. Misfolded proteins sequester HSP leading to PERK homodimers that phosphorylate eukaryotic translation-initiation factor 2 (eIF2); phosphorylation of eIF2 inhibits protein translation by preventing ribosome assembly.5 Like PERK, ATF6 is inactive when bound to HSP but free ATF6 localizes to the Golgi, where it is cleaved by specific proteases to generate a transcription factor that activates genes encoding chaperones and ERAD enzymes.6
As knowledge of the distinct signaling components of the UPR increases, it may be possible to improve existing (or design novel) pharmacologic chaperones to mitigate diseases associated with abnormal accumulations of misfolded protein. HIGM3 patients, though rare, may provide new insight into how defective protein folding and trafficking might be controlled.
Conflict-of-interest disclosure: The authors declare no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal