Abstract
Monoclonal B-cell lymphocytosis (MBL) is detectable in > 3% of the general population. Recent data are compatible, at least in a proportion of cases, with MBL being a progenitor lesion for chronic lymphocytic leukemia (CLL) and a surrogate for inherited predisposition. Common single nucleotide polymorphisms (SNPs) at 2q13 (rs17483466), 2q37.1 (rs13397985), 2q37.3 (rs757978), 6p25.3 (rs872071), 8q24.21 (rs2456449), 11q24.1 (rs735665), 15q21.3 (rs7169431), 15q23 (rs7176508), 16q24.1 (rs305061), and 19q13.32 (rs11083846) have been shown to confer a modest but significant increase in CLL risk. To examine the impact of these 10 SNPs on MBL, we analyzed 3 case-control series totaling 419 cases and 1753 controls. An association between genotype and MBL risk was seen for 9 SNPs, 6 of which were statistically significant: rs17483466 (odds ratio [OR] =1.27; P = .02), rs13397985 (OR = 1.40; P = 1.72 × 10−3), rs757978 (OR = 1.38; P = .02), rs872071 (OR = 1.27; P = 7.75 × 10−3), rs2456449 (OR = 1.31; P = 3.14 × 10−3), and rs735665 (OR = 1.63; P = 6.86 × 10−6). Collectively, these data provide support for genetic variation influencing CLL risk through predisposition to MBL.
Introduction
The strong familial risk of chronic lymphocytic leukemia (CLL) provides evidence for inherited susceptibility to the disease.1 Diagnosis of CLL is based on a B-cell count of more than or equal to 5 × 109/L and the presence of CD19, CD5, and CD23, weak expression of CD20, CD79b, and immunoglobulin receptor on B-cells.2 Persons with peripheral CLL-phenotype monoclonal B cells less than 5 × 109/L but no clinical features of CLL are classified as having monoclonal B-cell lymphocytosis (MBL).3 Approximately 10% to 15% of persons with a lymphocytosis have MBL, and 1% to 5% per year require treatment for progressive disease.4,5 Very low levels of CLL-phenotype cells (< 0.01 × 109/L) are detected in more than 5% of adults.6
A genome-wide association study7,8 has identified single nucleotide polymorphisms (SNPs) mapping to 2q13(rs17483466), 2q37.1(rs13397985, SP140), 2q37.3(rs757978, FARP2), 6p25.3(rs872071, IRF4), 8q24.21(rs2456449), 11q24.1(rs735665), 15q21.3(rs7169431), 15q23(rs7176508), 16q24.1(rs305061), and 19q13.32(rs11083846, PRKD2), conferring a modest but significant increase in CLL risk.7 Recent studies are compatible with MBL being a surrogate marker for inherited genetic susceptibility to CLL.9 To explore whether the 10 SNPs influence CLL risk through predisposition to MBL, we analyzed 3 independent case-control series totaling 419 MBL cases and 1753 controls.
Methods
The first 2 series were ascertained through laboratory investigations for an asymptomatic lymphocytosis. The series ascertained by the Hematological Malignancy Diagnostic Service (HMDS) was composed of 264 MBL cases (139 male; mean age, 71 years; SD, 11 years; mean lymphocyte count, 5.9 × 109/L; SD, 2 × 109/L; mean B-cell count, 3.0 × 109/L; SD, 1 × 109/L), who were all United Kingdom residents. Blood samples from 891 healthy persons (340 male; mean age, 59 years; SD, 11 years) collected through the National Study of Colorectal Cancer Genetics10 and Genetic Lung Cancer Predisposition Study11 studies served as controls. These persons were all United Kingdom residents and had self-reported European ancestry. Collection of samples and clinical information was undertaken with informed consent in accordance with the Declaration of Helsinki and with ethical review board approval from the Institute of Cancer Research. The second series were ascertained through the Amedeo Avogadro University of Eastern Piedmont (AAUEP) and consisted of 78 Italian MBL cases from Novara (42 male; mean age, 68 years; SD, 10 years; mean lymphocyte count, 4.7 × 109/L; SD, 2 × 109/L; mean B-cell count, 2.7 × 109/L; SD, 1.1 × 109/L).4 Blood samples from 170 healthy persons (95 male; mean age, 68 years; SD, 10 years) from the same geographic area with normal hematology served as controls. The third series included unrelated persons identified through the Italian Network on Genetic Isolates–Val Borbera project, a study of 1664 persons from the genetically isolated population of Val Borbera, Northwest Italy,12 and Illumina 370CNV-Quad Array data provided no evidence of population stratification. Pruning of the dataset to remove closely related persons was performed using Jenti,13 at a kinship threshold of 0.0625 (ie, exclusion of second cousins). The cohort included 77 MBL cases (42 male; mean age, 67 years; SD, 12 years; mean lymphocyte count, 2.2 × 109/L; SD, 1 × 109/L; mean absolute B-cell count, 0.2 × 109/L). Controls were 692 unrelated persons with normal hematology (292 male; mean age, 54 years; SD, 18 years) from the same region.
In all series, CLL-phenotype cells met accepted criteria for MBL.3 Procedures for each study have been previously reported.4-6 DNA was extracted from samples using standard methodologies and Picogreen quantified (Invitrogen).
Genotyping of the HMDS and AAUEP cohorts was conducted using allele-specific polymerase chain reaction KASPar chemistry (KBiosciences; primer sequences available on request). Genotyping of the Val Borbera Project cohort was performed with the Illumina 370CNV-Quad array. Imputation of missing data (rs7176508 and rs7169431) was performed using MACH Version 1.0.
Results and discussion
With the exception of rs11083846 in AAUEP controls, the frequency of SNP genotypes in cases and controls satisfies the criterion for the Hardy-Weinberg equilibrium (ie, P > .05). The risk of MBL associated with each of the SNPs was calculated on the basis of odds ratios (ORs) by unconditional logistic regression adjusting for age and sex. To provide increased power to demonstrate a relationship between SNP genotype and MBL, we pooled data from the 3 case-control series. Meta-analysis was performed under a fixed-effects model, estimating Cochran Q-statistic to test for heterogeneity14 and the I2 statistic to quantify the proportion of the total variation because of heterogeneity between studies.15
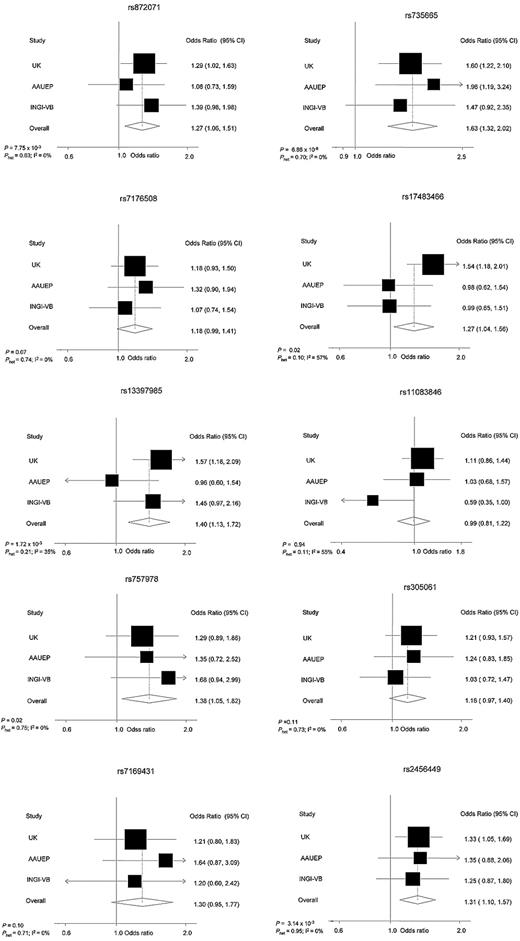
In the pooled analysis, an association between genotype and MBL risk was seen for 9 SNPs, which was statistically significant for rs17483466 (OR = 1.27; P = .02), rs13397985 (OR = 1.40; P = 1.72 × 10−3), rs757978 (OR = 1.38; P = .02), rs872071 (OR = 1.27; P = 7.75 × 10−3), rs2456449 (OR = 1.31; P = 3.14 × 10−3), and rs735665 (OR = 1.63; P = 6.86 × 10−6), with little evidence of between-study heterogeneity (Table 1; Figure 1). On the basis of the lower estimate of the effect size of SNPs for CLL risk,7,8,16 our study had high power to demonstrate a relationship between rs872071, rs735665, and rs2456449, and MBL at the 5% threshold. In contrast, we had less than 65% power to show associations for rs7176508, rs7169431, rs305061, rs11083846, rs757978, rs13397985, and rs17483466; hence, failure to demonstrate statistically significant associations for all 10 loci may simply reflect restricted study power.
Association between SNPs and MBL risk
| SNP . | Genotype . | HMDS cohort . | AAUEP cohort . | INGI-VB cohort . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases . | Controls . | OR . | 95% CI . | P . | Cases . | Controls . | OR . | 95% CI . | P . | Cases . | Controls . | OR . | 95% CI . | P . | ||
| rs872071 | AA | 48 | 196 | 1.00 | Ref | 14 | 39 | 1.00 | Ref | 15 | 192 | 1.00 | Ref | |||
| 6p25.3 | AG | 119 | 474 | 1.02 | 0.70-1.48 | .93 | 42 | 80 | 1.46 | 0.72-3.00 | .30 | 41 | 357 | 1.44 | 0.79-2.72 | .22 |
| (IRF4) | GG | 83 | 220 | 1.53 | 1.02-2.30 | .04 | 22 | 49 | 1.25 | 0.57-2.76 | .58 | 21 | 143 | 1.88 | 0.94-3.77 | .08 |
| ORtrend | 1.29 | 1.02-1.63 | .03 | 1.08 | 0.74-1.59 | .66 | 1.39 | 0.98-1.98 | .06 | |||||||
| rs735665 | GG | 125 | 579 | 1.00 | Ref | 40 | 112 | 1.00 | Ref | 49 | 492 | 1.00 | Ref | |||
| 11q24.1 | AG | 107 | 282 | 1.76 | 1.31-2.36 | 1.77 × 10−4 | 34 | 53 | 1.80 | 1.02-3.15 | .04 | 26 | 189 | 1.38 | 0.83-2.88 | .21 |
| AA | 16 | 27 | 2.75 | 1.44-5.25 | 2.29 × 10−3 | 4 | 2 | 5.60 | 0.99-31.76 | .05 | 2 | 11 | 1.83 | 0.39-8.47 | .44 | |
| ORtrend | 1.60 | 1.22-2.10 | 6.99 × 10−4 | 1.96 | 1.19-3.24 | .01 | 1.47 | 0.92-2.35 | .11 | |||||||
| rs17483466 | AA | 137 | 563 | 1.00 | Ref | 46 | 102 | 1.00 | Ref | 48 | 436 | 1.00 | Ref | |||
| 2q13 | AG | 88 | 278 | 1.30 | 0.96-1.77 | .09 | 29 | 53 | 1.21 | 0.69-2.15 | .51 | 26 | 223 | 1.06 | 0.64-1.75 | .82 |
| GG | 23 | 41 | 2.31 | 1.34-3.97 | 2.61 × 10−3 | 3 | 11 | 0.60 | 0.16-2.27 | .46 | 3 | 33 | 0.83 | 0.24-2.79 | .76 | |
| ORtrend | 1.54 | 1.19-2.01 | 1.15 × 10−3 | 0.98 | 0.63-1.54 | .94 | 0.99 | 0.65-1.51 | .10 | |||||||
| rs13397985 | TT | 126 | 577 | 1.00 | Ref | 50 | 113 | 1.00 | Ref | 42 | 461 | 1.00 | Ref | |||
| 2q37.1 | TG | 107 | 271 | 1.81 | 1.35-2.43 | 8.49 × 10−5 | 27 | 45 | 1.36 | 0.76-2.43 | .31 | 31 | 198 | 1.72 | 1.05-2.81 | .03 |
| (SP140) | GG | 11 | 29 | 1.74 | 0.85-3.57 | .13 | 1 | 10 | 0.23 | 0.03-1.81 | .16 | 4 | 33 | 1.33 | 0.45-3.94 | .61 |
| ORtrend | 1.57 | 1.20-2.09 | 1.28 × 10−3 | 0.96 | 0.59-1.54 | .85 | 1.45 | 0.97-2.16 | .07 | |||||||
| rs11083846 | GG | 132 | 539 | 1.00 | Ref | 47 | 105 | 1.00 | Ref | 59 | 460 | 1.00 | Ref | |||
| 19q13.32 | AG | 99 | 291 | 1.39 | 1.03-1.87 | .03 | 26 | 44 | 1.32 | 0.73-2.39 | .36 | 17 | 218 | 0.61 | 0.35-1.07 | .08 |
| (PRKD2) | AA | 14 | 52 | 1.10 | 0.59-2.04 | .77 | 5 | 15 | 0.74 | 0.26-2.17 | .59 | 1 | 14 | 0.56 | 0.07-4.31 | .56 |
| ORtrend | 1.11 | 0.85-1.44 | .46 | 1.03 | 0.68-1.57 | .89 | 0.59 | 0.34-1.00 | .05 | |||||||
| rs7176508 | GG | 77 | 307 | 1.00 | Ref | 24 | 64 | 1.00 | Ref | 25 | 244 | 1.00 | Ref | |||
| 15q23 | AG | 118 | 352 | 1.34 | 0.97-1.85 | .08 | 38 | 79 | 1.28 | 0.70-2.36 | .42 | 40 | 349 | 1.12 | 0.66-1.89 | .68 |
| AA | 42 | 133 | 1.26 | 0.82-1.93 | .29 | 16 | 24 | 1.78 | 0.81-3.91 | .15 | 12 | 99 | 1.18 | 0.57-2.45 | .65 | |
| ORtrend | 1.18 | 0.93-1.50 | .16 | 1.32 | 0.90-1.94 | .16 | 1.07 | 0.75-1.54 | .70 | |||||||
| rs757978 | GG | 185 | 717 | 1.00 | Ref | 64 | 144 | 1.00 | Ref | 63 | 592 | 1.00 | Ref | |||
| 2q37.3 | AG | 49 | 146 | 1.30 | 0.91-1.87 | .15 | 12 | 21 | 1.29 | 0.60-2.77 | .52 | 12 | 95 | 1.19 | 0.62-2.28 | .61 |
| (FARP2) | AA | 2 | 12 | 0.65 | 0.14-2.91 | .57 | 2 | 2 | 2.25 | 0.31-16.33 | .42 | 2 | 5 | 3.76 | 0.71-19.77 | .12 |
| ORtrend | 1.29 | 0.89-1.86 | .17 | 1.35 | 0.72-2.52 | .34 | 1.68 | 0.94-2.99 | .08 | |||||||
| rs305061 | CC | 18 | 87 | 1.00 | Ref | 13 | 26 | 1.00 | Ref | 7 | 104 | 1.00 | Ref | |||
| 16q24.1 | TC | 103 | 371 | 1.34 | 0.77-2.33 | .30 | 33 | 91 | 0.73 | 0.33-1.58 | .42 | 42 | 310 | 2.01 | 0.88-4.62 | .10 |
| TT | 113 | 407 | 1.34 | 0.78-2.32 | .29 | 32 | 51 | 1.26 | 0.56-2.79 | .58 | 28 | 278 | 1.50 | 0.63-3.53 | .36 | |
| ORtrend | 1.21 | 0.94-1.57 | .14 | 1.24 | 0.83-1.85 | .30 | 1.03 | 0.72-1.47 | .88 | |||||||
| rs7169431 | GG | 193 | 746 | 1.00 | Ref | 59 | 137 | 1.00 | Ref | 66 | 611 | 1.00 | Ref | |||
| 15q21.3 | AG | 42 | 129 | 1.26 | 0.86-1.84 | .24 | 18 | 28 | 1.49 | 0.77-2.91 | .24 | 11 | 80 | 1.27 | 0.65-2.51 | .49 |
| AA | 1 | 3 | 1.29 | 0.13-12.46 | .83 | 1 | 0 | 6.93 | 0.28-172.6 | .24 | 0 | 1 | 3.10 | 0.12-76.00 | .50 | |
| ORtrend | 1.21 | 0.81-1.83 | .35 | 1.64 | 0.87-3.09 | .12 | 1.20 | 0.59-2.42 | .61 | |||||||
| rs2456449 | AA | 80 | 367 | 1.00 | Ref | 32 | 85 | 1.00 | Ref | 39 | 377 | 1.00 | Ref | |||
| 8q24.21 | AG | 111 | 400 | 1.27 | 0.92-1.75 | .14 | 38 | 70 | 1.44 | 0.82-2.54 | .21 | 29 | 260 | 1.08 | 0.65-1.79 | .77 |
| GG | 40 | 107 | 1.71 | 1.11-2.65 | .02 | 8 | 13 | 1.63 | 0.62-4.31 | .32 | 9 | 55 | 1.58 | 0.73-3.44 | .25 | |
| ORtrend | 1.33 | 1.04-1.69 | .02 | 1.35 | 0.89-2.06 | .16 | 1.25 | 0.87-1.80 | .22 | |||||||
| SNP . | Genotype . | HMDS cohort . | AAUEP cohort . | INGI-VB cohort . | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases . | Controls . | OR . | 95% CI . | P . | Cases . | Controls . | OR . | 95% CI . | P . | Cases . | Controls . | OR . | 95% CI . | P . | ||
| rs872071 | AA | 48 | 196 | 1.00 | Ref | 14 | 39 | 1.00 | Ref | 15 | 192 | 1.00 | Ref | |||
| 6p25.3 | AG | 119 | 474 | 1.02 | 0.70-1.48 | .93 | 42 | 80 | 1.46 | 0.72-3.00 | .30 | 41 | 357 | 1.44 | 0.79-2.72 | .22 |
| (IRF4) | GG | 83 | 220 | 1.53 | 1.02-2.30 | .04 | 22 | 49 | 1.25 | 0.57-2.76 | .58 | 21 | 143 | 1.88 | 0.94-3.77 | .08 |
| ORtrend | 1.29 | 1.02-1.63 | .03 | 1.08 | 0.74-1.59 | .66 | 1.39 | 0.98-1.98 | .06 | |||||||
| rs735665 | GG | 125 | 579 | 1.00 | Ref | 40 | 112 | 1.00 | Ref | 49 | 492 | 1.00 | Ref | |||
| 11q24.1 | AG | 107 | 282 | 1.76 | 1.31-2.36 | 1.77 × 10−4 | 34 | 53 | 1.80 | 1.02-3.15 | .04 | 26 | 189 | 1.38 | 0.83-2.88 | .21 |
| AA | 16 | 27 | 2.75 | 1.44-5.25 | 2.29 × 10−3 | 4 | 2 | 5.60 | 0.99-31.76 | .05 | 2 | 11 | 1.83 | 0.39-8.47 | .44 | |
| ORtrend | 1.60 | 1.22-2.10 | 6.99 × 10−4 | 1.96 | 1.19-3.24 | .01 | 1.47 | 0.92-2.35 | .11 | |||||||
| rs17483466 | AA | 137 | 563 | 1.00 | Ref | 46 | 102 | 1.00 | Ref | 48 | 436 | 1.00 | Ref | |||
| 2q13 | AG | 88 | 278 | 1.30 | 0.96-1.77 | .09 | 29 | 53 | 1.21 | 0.69-2.15 | .51 | 26 | 223 | 1.06 | 0.64-1.75 | .82 |
| GG | 23 | 41 | 2.31 | 1.34-3.97 | 2.61 × 10−3 | 3 | 11 | 0.60 | 0.16-2.27 | .46 | 3 | 33 | 0.83 | 0.24-2.79 | .76 | |
| ORtrend | 1.54 | 1.19-2.01 | 1.15 × 10−3 | 0.98 | 0.63-1.54 | .94 | 0.99 | 0.65-1.51 | .10 | |||||||
| rs13397985 | TT | 126 | 577 | 1.00 | Ref | 50 | 113 | 1.00 | Ref | 42 | 461 | 1.00 | Ref | |||
| 2q37.1 | TG | 107 | 271 | 1.81 | 1.35-2.43 | 8.49 × 10−5 | 27 | 45 | 1.36 | 0.76-2.43 | .31 | 31 | 198 | 1.72 | 1.05-2.81 | .03 |
| (SP140) | GG | 11 | 29 | 1.74 | 0.85-3.57 | .13 | 1 | 10 | 0.23 | 0.03-1.81 | .16 | 4 | 33 | 1.33 | 0.45-3.94 | .61 |
| ORtrend | 1.57 | 1.20-2.09 | 1.28 × 10−3 | 0.96 | 0.59-1.54 | .85 | 1.45 | 0.97-2.16 | .07 | |||||||
| rs11083846 | GG | 132 | 539 | 1.00 | Ref | 47 | 105 | 1.00 | Ref | 59 | 460 | 1.00 | Ref | |||
| 19q13.32 | AG | 99 | 291 | 1.39 | 1.03-1.87 | .03 | 26 | 44 | 1.32 | 0.73-2.39 | .36 | 17 | 218 | 0.61 | 0.35-1.07 | .08 |
| (PRKD2) | AA | 14 | 52 | 1.10 | 0.59-2.04 | .77 | 5 | 15 | 0.74 | 0.26-2.17 | .59 | 1 | 14 | 0.56 | 0.07-4.31 | .56 |
| ORtrend | 1.11 | 0.85-1.44 | .46 | 1.03 | 0.68-1.57 | .89 | 0.59 | 0.34-1.00 | .05 | |||||||
| rs7176508 | GG | 77 | 307 | 1.00 | Ref | 24 | 64 | 1.00 | Ref | 25 | 244 | 1.00 | Ref | |||
| 15q23 | AG | 118 | 352 | 1.34 | 0.97-1.85 | .08 | 38 | 79 | 1.28 | 0.70-2.36 | .42 | 40 | 349 | 1.12 | 0.66-1.89 | .68 |
| AA | 42 | 133 | 1.26 | 0.82-1.93 | .29 | 16 | 24 | 1.78 | 0.81-3.91 | .15 | 12 | 99 | 1.18 | 0.57-2.45 | .65 | |
| ORtrend | 1.18 | 0.93-1.50 | .16 | 1.32 | 0.90-1.94 | .16 | 1.07 | 0.75-1.54 | .70 | |||||||
| rs757978 | GG | 185 | 717 | 1.00 | Ref | 64 | 144 | 1.00 | Ref | 63 | 592 | 1.00 | Ref | |||
| 2q37.3 | AG | 49 | 146 | 1.30 | 0.91-1.87 | .15 | 12 | 21 | 1.29 | 0.60-2.77 | .52 | 12 | 95 | 1.19 | 0.62-2.28 | .61 |
| (FARP2) | AA | 2 | 12 | 0.65 | 0.14-2.91 | .57 | 2 | 2 | 2.25 | 0.31-16.33 | .42 | 2 | 5 | 3.76 | 0.71-19.77 | .12 |
| ORtrend | 1.29 | 0.89-1.86 | .17 | 1.35 | 0.72-2.52 | .34 | 1.68 | 0.94-2.99 | .08 | |||||||
| rs305061 | CC | 18 | 87 | 1.00 | Ref | 13 | 26 | 1.00 | Ref | 7 | 104 | 1.00 | Ref | |||
| 16q24.1 | TC | 103 | 371 | 1.34 | 0.77-2.33 | .30 | 33 | 91 | 0.73 | 0.33-1.58 | .42 | 42 | 310 | 2.01 | 0.88-4.62 | .10 |
| TT | 113 | 407 | 1.34 | 0.78-2.32 | .29 | 32 | 51 | 1.26 | 0.56-2.79 | .58 | 28 | 278 | 1.50 | 0.63-3.53 | .36 | |
| ORtrend | 1.21 | 0.94-1.57 | .14 | 1.24 | 0.83-1.85 | .30 | 1.03 | 0.72-1.47 | .88 | |||||||
| rs7169431 | GG | 193 | 746 | 1.00 | Ref | 59 | 137 | 1.00 | Ref | 66 | 611 | 1.00 | Ref | |||
| 15q21.3 | AG | 42 | 129 | 1.26 | 0.86-1.84 | .24 | 18 | 28 | 1.49 | 0.77-2.91 | .24 | 11 | 80 | 1.27 | 0.65-2.51 | .49 |
| AA | 1 | 3 | 1.29 | 0.13-12.46 | .83 | 1 | 0 | 6.93 | 0.28-172.6 | .24 | 0 | 1 | 3.10 | 0.12-76.00 | .50 | |
| ORtrend | 1.21 | 0.81-1.83 | .35 | 1.64 | 0.87-3.09 | .12 | 1.20 | 0.59-2.42 | .61 | |||||||
| rs2456449 | AA | 80 | 367 | 1.00 | Ref | 32 | 85 | 1.00 | Ref | 39 | 377 | 1.00 | Ref | |||
| 8q24.21 | AG | 111 | 400 | 1.27 | 0.92-1.75 | .14 | 38 | 70 | 1.44 | 0.82-2.54 | .21 | 29 | 260 | 1.08 | 0.65-1.79 | .77 |
| GG | 40 | 107 | 1.71 | 1.11-2.65 | .02 | 8 | 13 | 1.63 | 0.62-4.31 | .32 | 9 | 55 | 1.58 | 0.73-3.44 | .25 | |
| ORtrend | 1.33 | 1.04-1.69 | .02 | 1.35 | 0.89-2.06 | .16 | 1.25 | 0.87-1.80 | .22 | |||||||
ORtrend values have been corrected for age and sex.
CI indicates confidence interval; and Ref, referent category.
Forest plots of effect size and direction for the 10 SNPs. Boxes represent allelic OR point estimates, their areas being proportional to the inverse variance weight of the estimate. Horizontal lines represent 95% confidence intervals. The diamond (and broken line) represents the summary OR computed under a fixed-effects model, with 95% confidence interval given by its width. The unbroken vertical line is at the null value (OR = 1.0).
Forest plots of effect size and direction for the 10 SNPs. Boxes represent allelic OR point estimates, their areas being proportional to the inverse variance weight of the estimate. Horizontal lines represent 95% confidence intervals. The diamond (and broken line) represents the summary OR computed under a fixed-effects model, with 95% confidence interval given by its width. The unbroken vertical line is at the null value (OR = 1.0).
We investigated the combined effect of pairs of loci using HMDS and AAUEP data by logistic regression modeling, evidence for interactive effects between SNPs assessed by a likelihood ratio test. This analysis provided no evidence of interactive effects between any of the loci (P > .05), consistent with each locus having an independent role in defining MBL risk. Although the risk of MBL associated with each of the variants is modest, the carrier frequencies of risk alleles are high in the European population; hence, the loci make a major contribution to the development of MBL. Moreover, risk of MBL increases with increasing numbers of risk alleles for the loci (ORper-allele = 1.42; 95% confidence interval, 1.28-1.57; P = 1.95 × 10−11). Persons with more than or equal to 6 risk alleles have a more than 3-fold increase in MBL risk compared with those with a median number of risk alleles.
Collectively, these data are consistent with polymorphic variation at these loci influencing the risk of CLL through development of MBL. The SNPs examined here are not necessarily strong candidates for being directly causal. Whereas rs735665, rs1743466, and rs2456449 do not annotate genes directly, rs757978, rs872071, and rs13397985 map to FARP2, IRF4, and SP140, respectively. rs757978 is the only SNP leading to a coding change, with the substitution of threonine for isoleucine at amino acid 260, which is predicted to be functionally deleterious. FARP2 (rs757978) is an important regulator of MYC gene activity through interaction with far upstream element and far upstream element-binding protein.17,18
As rs872071 and rs13397985 are not correlated with coding changes, these associations are probably mediated through noncoding variation impacting on gene expression. This is supported by the observation that IRF4 and SP140 expression is significantly associated with their respective genotypes in a dose-dependent fashion.7 IRF4 has been shown to play a role during plasma cell commitment,19 and deregulation at the germinal center/plasma cell checkpoint could therefore play a role in MBL/CLL development. Genetic variation in SP140 could influence risk through differing response to antigenic challenge as its coactivation may be important for establishment of latent viral infections and B-cell immortalization.20
Genome-wide association studies have shown that the 128- to 130-Mb interval at 8q24.21 harbors multiple independent loci with different tumor specificities, even though it is bereft of genes. The colorectal-prostate cancer association affects TCF4 binding to an enhancer for MYC, providing a mechanistic basis for this 8q24.21 association.21 Thus, it is possible that the effect of the other 8q24.21 cancer risk loci is via MYC through similar long-range cis-acting mechanisms.
For most of the SNP frequencies of risk alleles in MBL, cases were similar to those previously documented in CLL, but there are some differences: the rs11083846 risk allele was under-represented in the low-count MBL cases, suggesting that this SNP may be associated with disease progression. Further investigation with larger defined series will help to elucidate the contribution of different SNPs.
In conclusion, these data provide further evidence for MBL being a CLL precursor lesion and implicate FARP2, IRF4, and SP140 genes in the etiology of MBL risk.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by Leukemia and Lymphoma Research (United Kingdom); Cancer Research United Kingdom (C1298/A8362 supported by the Bobby Moore Fund); the Arbib Foundation; Progetto Fondo per gli Investimenti della Ricerca di Base–Programma Futuro in Ricerca 2008, Ministero della Universitá e della Ricerca (MIUR), Rome, Italy; Associazione Italiana per la Ricerca sul Cancro, Milan, Italy; Associazione Italiana per la Ricerca sul Cancro (Investigator Grant and Special Program Clinical Oncology 5 per mille n. 9965), Milano, Italy; Cassa di Risparmio delle Province Lombarde Foundation, Milano, Italy; Programma di Ricerca di Interesse Nazionale–MIUR, Rome, Italy; Compagnia di San Paolo, Torino, Italy (D.T.); Fondazione Cassa di Risparmio, Alessandria, Italy (D.T.); and Ministry of Health, Ricerca Finalizzata 2008 (D.T.).
Authorship
Contribution: D.C.-S. and T.C. conducted research and analyzed results; A.L., B.O., C.D., and F.L.B. conducted research; D.R. and P.G. contributed SNP genotyping; F.C.-C., G.G., and A.C.R. collected clinical data; D.T. analyzed results; D.C. and R.S.H. conceived the research idea; and R.S.H. wrote the paper with contributions from D.C.-S. and A.C.R.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Richard S. Houlston, Institute of Cancer Research, 15 Cotswold Rd, Sutton, Surrey SM2 5NG, United Kingdom; e-mail: richard.houlston@icr.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal