Hodgkin lymphoma is still in many aspects enigmatic. In this issue of Blood, Steidl and colleagues present exciting novel insights into pathogenesis and clinical behavior of classical Hodgkin lymphoma by performing array comparative genomic hybridization studies to detect genomic gains and losses in the HRS tumor cells of this malignancy.1
The treatment of Hodgkin lymphoma is a success story with about 80% to 90% of patients reaching long-term cure with current treatment strategies.2 Nevertheless, about 10% to 20% of patients cannot be cured and will die of the disease. Therefore, there is much interest in identifying characteristics of resistant tumors upfront. Patients with such tumors may benefit from an early treatment intensification, whereas patients with a good prognosis may benefit from reduced treatment intensity and consequently less toxic side effects of the chemo- and/or radiation therapy.2
Prognostic markers can be identified in various ways. An attractive approach is to search for genetic lesions in the tumor cells that are associated with good or bad clinical outcome. This track was followed by Steidl et al, who performed a high-resolution array-based comparative genomic hybridization study with isolated Hodgkin and Reed-Sternberg (HRS) cells from 53 cases of classical Hodgkin lymphoma. The cases included 23 patients in whom the disease progressed or relapsed at any time. Multiple recurrent genomic gains and losses were found, including several known from other works. Interestingly, one of the gains, involving the chromosomal region 16p11.2-13.3, was significantly associated with the group of treatment failures. Thus, the authors present here the first genetic feature of HRS cells that may serve as a prognostic marker for Hodgkin lymphoma. Clearly, this finding needs validation in an independent group of homogenously treated patients. Moreover, a weakness of the study is that the treatment failure group included patients with disease progression and relapse at any time. In clinical practice, patients who do not respond to treatment at all or have an early relapse within several months after therapy have a worse prognosis than patients with a relapse after several years.2 Likewise, as the reasons for relapse may be different in those with early versus late recurrence, it will be important to find out whether the 16p gain helps to identify the therapy resistant group. Preliminary findings in the study indicate that 16p gains may indeed be most frequent in therapy refractory patients.
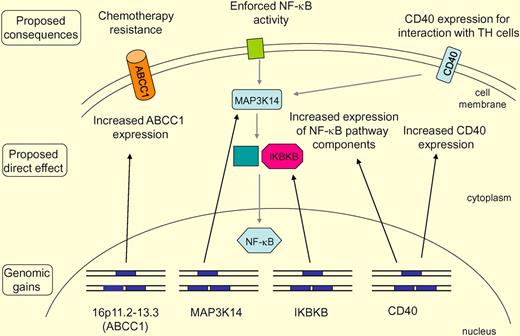
Main findings of the study and their potential consequences are summarized. Recurrent gains (indicated by an additional gene copy on one allele) of 16p11.2-13.3 include the ABCC1 gene. This may lead to increased ABCC1 expression, and the increased expression of this multidrug transporter may cause chemoresistance of the lymphoma cells. Increased gene copy numbers for components of the NF-κB pathway, including MAP3K14 (NIK) and IKBKB, may cause increased expression of these positive regulators of NF-κB activation and consequently contribute to the strong constitutive NF-κB activity in HRS cells. The NF-κB pathway is displayed in a simplified way, not discriminating the canonical and noncanonical pathway. The CD40 gains may contribute to strong CD40 expression in a setting where most B cell–typical genes are down-regulated. CD40 promotes the interaction of HRS cells with surrounding CD40 ligand-expressing T cells and likely also contributes to NF-κB activity in HRS cells.
Main findings of the study and their potential consequences are summarized. Recurrent gains (indicated by an additional gene copy on one allele) of 16p11.2-13.3 include the ABCC1 gene. This may lead to increased ABCC1 expression, and the increased expression of this multidrug transporter may cause chemoresistance of the lymphoma cells. Increased gene copy numbers for components of the NF-κB pathway, including MAP3K14 (NIK) and IKBKB, may cause increased expression of these positive regulators of NF-κB activation and consequently contribute to the strong constitutive NF-κB activity in HRS cells. The NF-κB pathway is displayed in a simplified way, not discriminating the canonical and noncanonical pathway. The CD40 gains may contribute to strong CD40 expression in a setting where most B cell–typical genes are down-regulated. CD40 promotes the interaction of HRS cells with surrounding CD40 ligand-expressing T cells and likely also contributes to NF-κB activity in HRS cells.
As we still have only a fragmentary knowledge of the transforming events involved in Hodgkin lymphoma pathogenesis, it is another aim of the study to identify novel recurrent imbalances that may point to oncogenes or tumor suppressor genes contributing to Hodgkin lymphoma development. Most genetic lesions identified in HRS cells affect the NF-κB and JAK/STAT signaling pathways.3 Regarding lesions contributing to the constitutive NF-κB activity in HRS cells, somatic mutations in the NF-κB inhibitors NFKBIA and NFKBIE are recurrently found, as are gains of the REL and BCL3 gene.3 Recently, the TNFAIP3 gene, encoding the negative NF-κB regulator A20, was found to be somatically mutated and/or deleted in the HRS cells of about 40% of Hodgkin lymphomas.4-6 In the study by Gascoyne's group, additional candidate genes were identified that may contribute to the strong NF-κB activity of HRS cells, that is, MAP3K14 (NIK), IKBKB, and CD40. Although it remains to be determined whether the gains observed in these genes indeed result in their increased expression and have a pathogenetic effect, the present findings further add to the picture that activation of the NF-κB pathway is a central event in Hodgkin lymphoma pathogenesis and that multiple genetic lesions contribute in various combinations to this activation.3
The detection of recurrent gains of the CD40 gene is also of interest in a further regard. HRS cells have largely lost the expression of B cell–specific genes, but a small fraction of such genes remain expressed.7 These include key components for an interaction of B cells with T cells, that is, CD80, MHC class II, and CD40.3 The interaction of HRS cells with surrounding T helper cells through these stimulating pathways presumably promotes their survival and proliferation. Thus, in a fraction of Hodgkin lymphomas gains of the CD40 gene may contribute to the high expression of CD40 by HRS cells in a setting where most B-cell typical genes are down-regulated, and thereby have a pathogenetic effect.
Among the many genes located in the large gained region on 16p, the authors turned their interest to one specific gene, ABCC1, which is a member of the ATP binding cassette (ABC) transporter family and encodes an efflux pump that functions as a multidrug resistance factor. One Hodgkin lymphoma line, KMH2, also had a gain of the ABCC1 locus, and inhibition of the high ABCC1 expression in KMH2 cells sensitized them to killing by doxorubicin. Hence, ABCC1 is an attractive candidate for a pathogenetically relevant gene in the gained region on 16p. This finding is of particular interest also in light of another recent study, which identified small subpopulations of so-called “side population cells” in 2 other Hodgkin lymphoma cell lines, L428 and HDLM2 (KMH2 was not analyzed).8 Side population cells are characterized by their extrusion of the Hoechst dye 33342, due to expression of multidrug resistance genes.9 The side population cells of the L428 and HDLM2 lines expressed the ABC transporter family members ABCB1 (MDR1) and ABCG2 and were more resistant to chemotherapy than the nonside population cells.8 Although the patterns and mechanisms of ABC transporter expression appears to be different in KMH2 versus L428 and HDLM2 cells, both studies nevertheless point to a potentially important role of these molecules in chemotherapy resistance and treatment failure in Hodgkin lymphoma. Importantly, as side population cells share features with cancer stem cells,9 these findings will certainly also stimulate further work to elucidate potential relationships among expression of multidrug transporters, chemotherapy resistance, and putative HRS lymphoma “stem” cells.10
Conflict-of-interest disclosure: The author declares no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal