Abstract
The concept of reprogramming of somatic cells has opened a new era in regenerative medicine. Transduction of defined factors has successfully achieved pluripotency. However, during the generation process of induced pluripotent stem (iPS) cells, genetic manipulation of certain factors may cause tumorigenicity, which limits further application. We report that that a single transfer of embryonic stem (ES) cell–derived proteins into primarily cultured adult mouse fibroblasts, rather than repeated transfer or prolonged exposure to materials, can achieve full reprogramming up to the pluripotent state without the forced expression of ectopic transgenes. During the process, gene expression and epigenetic status were converted from somatic to ES-equivalent status. We verified that protein-based reprogramming was neither by the contamination of protein donor ES cell nor by DNA/RNA from donor ES cell. Protein-iPS cells were biologically and functionally very similar to ES cells and differentiated into 3 germ layers in vitro. Furthermore, protein-iPS cells possessed in vivo differentiation (well-differentiated teratoma formation) and development (chimeric mice generation and a tetraploid blastocyst complementation) potentials. Our results provide an alternative and safe strategy for the reprogramming of somatic cells that can be used to facilitate pluripotent stem cell–based cell therapy.
Introduction
Stem cells are a promising source of biologic material for regenerative medicine. The concept of producing autologous or customized pluripotent stem cells from somatic cells has attracted the attention of investigators and clinicians who seek a feasible methodology for cell therapy that can be applied to the treatment of patients with degenerative diseases and organ failure, as well as experimental applications for drug discovery, screening, and toxicology.
Epoch-making discoveries in the reprogramming of mouse as well as human somatic cells into pluripotent stem cells (induced pluripotent stem, or iPS, cells) by viral transduction of certain transcription factors has opened a new era of regenerative medicine.1-6 However, there is concern that the retroviral or lentiviral introduction of defined factors could cause unexpected long-term instability and tumorigenicity due to permanent genetic integration.7,8 These critical issues remain to be addressed before clinical application. Therefore, there is currently ongoing an extensive search for new methods such as a reduced number of defined factors,9 adenoviral or plasmid-based transient gene delivery,10-12 or oocyte-free, nonviral inducers like small molecules13-15 and proteins16 that could be safely used in this context.
Meiotic oocyte and mitotic zygote cytoplasm can induce the reprogramming of a somatic cell within a short period of time and a few cell divisions after nuclear transfer, whereas ectopic expression of a limited number of certain transcription factors needed a long period and multiple cell divisions to achieve full reprogramming.17-20 In this study, we hypothesized that the proteins of actively proliferating embryonic stem (ES) cells, which has yet to be precisely determined, could have the capacity to induce reprogramming of the adult somatic cell, thereby the transfer of proteins from ES cell into adult cell, instead of nuclear transfer of adult cell into ES cell, could achieve somatic cell reprogramming.
Previous studies using various cellular proteins or extracts have shown a modest effect on reprogramming into specific lineages21,22 or dedifferentiation into the pluripotent state.23 The reprogrammed multipotent cells produced by transferring extracts of embryonic carcinoma or ES cell did not reach the pluripotent state in terms of the transcriptional state, in vivo 3 germ layer differentiation capacity, and developmental potential.23,24 Here, we investigated whether a single transfer of ES cell–derived extract proteins into primarily cultured adult somatic cells, rather than repeated transfer or prolonged exposure to materials, could achieve full reprogramming up to the pluripotent state. We demonstrate that protein-mediated reprogrammed adult fibroblasts (protein-iPS) are biologically and functionally very similar to ES cells in vitro and possess in vivo differentiation and developmental potentials including well-differentiated teratoma formation, contribution to chimeras, and tetraploid blastocyst complementation. These results may provide a simple, safe, and effective alternative strategy for dedifferentiation or reprogramming of adult somatic cells.
Methods
Animals
C57BL/6 and FVB strain wild-type mice and Oct4-promoter–driven green fluorescent protein (GFP) mice (The Jackson Laboratory) were used for primary cardiac and skin fibroblast preparations. Nonobese diabetic/severe combined immunodeficient (NOD/SCID) mice (The Jackson Laboratory) were used for teratoma formation. All animal experiments were performed after receiving approval from the Institutional Animal Care and Use Committee (IACUC) of Clinical Research Institute in Seoul National University Hospital, Korea.
Cell culture and reprogramming by proteins
The C57BL/6-background mouse ES cells (C57-mESCs, accession no. SCRC-1002; ATCC) and E14 mouse ES cells (generously provided by Jeong Mook Lim, Seoul National University, Seoul, Korea) were cultured on Mitomycin C (Sigma-Aldrich)–treated STO feeder layer in 0.1% gelatin (Sigma-Aldrich)–coated tissue culture dish. To obtain cardiac fibroblast and to minimize potential resident stem cells, the enzymatically digested heart harvested from 8-week-old mice was incubated with anti–c-kit microbeads (Miltenyi Biotec). C-kit–negative fraction was collected by magnetic separation and cultured with Dulbecco modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS) and antibiotics (C57-cFB). At least 4 passages of subcultured cells were used for experiments. Skin fibroblasts were primarily cultured from the dermis of 8-week-old mice (FVB-sFB, Oct4-promoter-GFP-sFB). In the experiments, to prepare genomic DNA, RNA, and cellular proteins without feeder cell contamination, ES cells and protein-iPS cells were passaged twice in a 0.1% gelatin-coated tissue culture dish without the feeder layer, then preplated in a tissue culture dish for 40 minutes, and then floating cells were harvested.
To induce the reprogramming of adult fibroblasts, ES cell–derived extract proteins were prepared and transferred using streptolysin O (Sigma-Aldrich)–mediated reversible permeabilization (details in supplemental Methods, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). We summarized the substantial differences versus the previous comparable studies (see supplemental Table 5).23,24 Typically, 20 to 35 mg/mL proteins were used to induce reprogramming. To fractionize proteins by molecular weight, Centricon (Millipore) was used. When primary colonies were observed, they were reseeded on a STO feeder layer and subcultured. Passage 5 to 7 cells (culture days 45-55) were used for further experiments.
Genomic DNA polymerase chain ceaction
Genomic DNA was extracted using DNeasy Blood and Tissue kit (QIAGEN). Microsatellite markers developed at the Whitehead Institute/Massachusetts Institute of Technology Center for Genome Research (Cambridge, MA) were applied to be able to amplify genomic DNA from C57, FVB, 129, and ICR mouse strains.25
Analysis of mRNA expression
Global gene expression analyses were performed using Affymetrix GeneChip Mouse Gene 1.0 ST oligonucleotide arrays (Affymetrix) or Mouse Whole-genome BeadChips (Illumina). The sample preparation was performed according to the instructions provided by the manufacturer. To draw scatterplots of ES, cFB, and cFB-protein-iPS, 3 groups were expressed into images through GCOS software. To detect different probe (differentially expressed gene or DEG) among ES, cFB, and cFB-protein-iPS, a one-way analysis of variance (ANOVA) test was operated and measured with Benjamini-Hochberg false discovery rate (FDR) for enhancing significant test in multiple testing. Significance was determined by FDR less than 5% and P value less than .01. Microarray results are accessible at the Gene Expression Omnibus (GEO) database (National Center for Biotechnology Information; accession no. series GSE13770).
Alkaline phosphatase and immunocytochemical staining
Alkaline phosphatase (ALP) staining was performed using Alkaline Phosphatase Detection kit (BCIP/NBT Substrate System; Dako). For immunocytochemical staining, cell colonies were fixed with 4% paraformaldehyde and blocking with 1% bovine serum albumin (BSA) and 0.1% Triton X. Staining was carried out using anti-SSEA1 antibody (Santa Cruz Biotechnology), anti-Oct4 antibody (Santa Cruz Biotechnology), and anti-SSEA4 antibody (Chemicon, Millipore) overnight at 4°C. Images were obtained using a confocal microscope (LSM 510 Meta; Carl Zeiss).
DNA methylation and chromatin analysis
To assess the methylation status of Oct4 and Nanog promoters, bisulfite sequencing was performed as described previously.26,27 For chromatic immunoprecipitation (ChIP) assays of Oct4 and Nanog promoters, 1 × 106 cells were cross-linked and quenched using 1% formaldehyde and glycine.1 A ChIp assay was performed by chromatin immunoprecipitation assay kit (Upstate, Millipore). Sonicated cell lysates were used for immunoprecipitations in the presence of anti–trimethyl histone 3 lysine 4 (H3K4; Abcam), anti–trimethyl histone 3 lysine 27 (H3K27; Upstate, Millipore), anti–acetyl-Histone H3 (Ach-H3; Upstate, Millipore), or anti–immunoglobulin G (IgG) antibody.1,26,27 Samples were separated by protein A agarose/salmon sperm DNA (50% slurry; Upstate, Millipore). For conventional semiquantitative reverse transcription–polymerase chain reaction (RT-PCR) and real-time PCR, an equal amount of template DNA was amplified. SYBR Green PCR master mix was used for real-time PCR. For genome-wide ChiP sequencing, chromatin immunoprecipitation with H3K4me3 and H3K27me3 antibodies and sequencing using Solexa (Illumina) were performed. The correlation between ES cell and protein-iPS cell and between adult fibroblast and protein-iPS cell was examined by Pearson correlation coefficients.
In vitro and in vivo differentiation
To assess the in vitro differentiation potential, cell aggregates generated in suspension culture (embryoid body; EB) were plated onto 0.1% gelatin-coated tissue culture dishes and cultured. Morphologically, some spontaneously contracting EBs were filmed (Olympus 1 × 71 and Olympus DP71 Digital Camera, 15 fps at 680 × 512; Olympus). The gene and protein expression for markers found in the 3 germ layers were examined by RT-PCR and immunocytochemistry. To examine in vivo differentiation potential, 1 × 107 of the protein-iPS cells were injected subcutaneously into the backs of NOD/SCID mice, and the histology was reviewed.
Blastocyst injection, chimera generation, and tetraploid complementation
To determine the in vivo developmental potentials, chimeric mice were generated using the FVB strain of protein-iPS cells with standard ES cell transfer procedures for the production of chimeras (Macrogen). Typically, 8 to 10 cells were injected into the C57BL/6 blastocyst cavity and transferred to the uterus of a pseudopregnant ICR female 2.5 days postcoitum (dpc).28 The combination of the sFB-protein-iPS cells with C57BL/6 host blastocysts resulted in black/white-colored chimeric mice. To verify the pluripotency of the reprogrammed cells, a tetraploid blastocyst complementation experiment was performed.29,30 Diploid GFP-transduced FVB strain of protein-iPS cells were injected into ICR tetraploid blastocysts. At E10.5, embryos were harvested and genotyped.
Detailed full experimental methods, materials, and associated references are described in the supplemental Methods.
Results
Generation of pluripotent stem cells from adult fibroblasts
We cultured primary cardiac fibroblasts (cFBs) from an adult C57/BL mouse. To exclude potential resident stem cells (eg, adult cardiac stem cells), we collected c-kit− cells and cultured them to at least passage 4 before inducing reprogramming. We checked that the cFBs did not exhibit stem or progenitor cells' characteristics (Figure 1A-B).
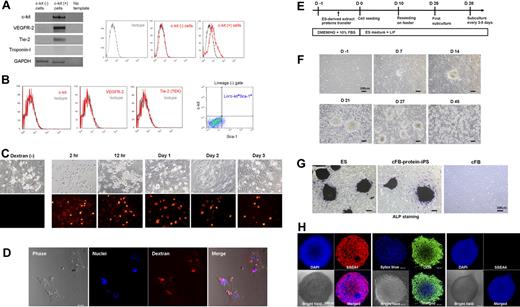
Generation of protein-iPS cells from c-kit− cardiac fibroblasts. (A) Gene and surface protein expression of c-kit− and c-kit+ cells (immediately after magnetic sorting). (B) Reevaluation of surface protein expressions on fourth-passage c-kit− cardiac fibroblasts. (C) Fibroblasts were reversibly permeabilized, incubated with 70 kDa Texas Red–conjugated dextran and resealed. Serial fluorescent images. (D) Confocal images at day 7. (E) Time line of reprogramming by proteins. Colonies developed on days 4 to 7. DMEM/HG, DMEM with high glucose (D-glucose; 4500 mg/L). (F) Phase-contrast bright-field morphology of cells and colonies. (G) ALP staining of authentic ES cell, cFB-protein-iPS cell, and adult cardiac fibroblast. (H) Immunocytochemistry showing that cFB-protein-iPS cells express SSEA1 and Oct4 but not SSEA4 (a known human ES cell marker).
Generation of protein-iPS cells from c-kit− cardiac fibroblasts. (A) Gene and surface protein expression of c-kit− and c-kit+ cells (immediately after magnetic sorting). (B) Reevaluation of surface protein expressions on fourth-passage c-kit− cardiac fibroblasts. (C) Fibroblasts were reversibly permeabilized, incubated with 70 kDa Texas Red–conjugated dextran and resealed. Serial fluorescent images. (D) Confocal images at day 7. (E) Time line of reprogramming by proteins. Colonies developed on days 4 to 7. DMEM/HG, DMEM with high glucose (D-glucose; 4500 mg/L). (F) Phase-contrast bright-field morphology of cells and colonies. (G) ALP staining of authentic ES cell, cFB-protein-iPS cell, and adult cardiac fibroblast. (H) Immunocytochemistry showing that cFB-protein-iPS cells express SSEA1 and Oct4 but not SSEA4 (a known human ES cell marker).
We first tested the efficacy of streptolysin O–mediated reversible permeabilization using fluorescent dextran (Figure 1C-D). We confirmed intracellular fluorescence stay of 10, 40, and 70 kDa dextran up to 7 days after a single transfer and resealing procedure, indicating that transferred materials would last intracellularly for a certain period of time.
The protocol for reprogramming by proteins is shown in Figure 1E. We introduced C57 background ES cell–derived extract proteins into cFB by reversible permeabilization. When we started with 1 × 106 cFBs, we observed approximately 5 to 10 colonies from day 4 to 7 after induction. On day 10, we disaggregated the colonies into a single cell and reseeded the cells onto supporting feeder cell layers. We observed numerous secondary colonies, which were similar to the ES cell colonies in morphology, on the feeder layers, 10 to 15 days after reseeding (days 20-25 from induction; Figure 1F). The ES-like cells (colonies) were expanded with subculture every 3 to 5 days using the standard ES cell culture protocol. We named these ES-like cells cFB-protein-iPS cells.
In vitro characteristics of the protein-iPS cells shows typical features of ES cells
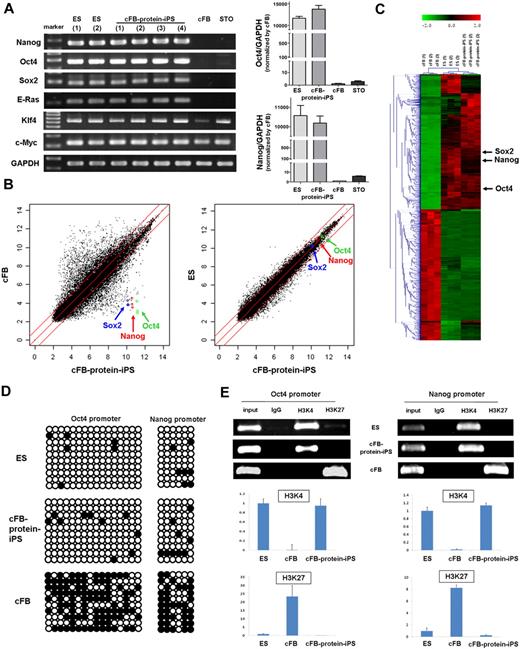
After 45 to 55 days of culture (passages 5-7), we carried out several assays to compare the cFB-protein-iPS with ES cell that is the donor of extract proteins. Both of cFB-protein-iPS and ES colonies exhibited comparably strong ALP activity, whereas cFB showed very weak ALP activity (Figure 1G). Immunocytochemistry revealed that the cFB-protein-iPS cells expressed pluripotency markers including SSEA1 and Oct4 (Figure 1H). The RT-PCR showed that the cFB-protein-iPS expressed typical pluripotent ES cell genes, such as Nanog, Oct4, Sox2, and E-Ras at consistent levels compared with the ES cell, whereas the cFB and feeder cells did not express these genes (Figure 2A right panels). Klf4 and c-Myc expression in the cFB-protein-iPS was also comparable with the ES cell. To confirm the expression levels of the pluripotency genes, we performed real-time PCR to compare the relative expression levels of Oct4 and Nanog between cFB-protein-iPS cells and fibroblasts (Figure 2A, left panels). There is no significant difference in the expression levels of Oct4 and Nanog between ES and c-FB-protein-iPS cells. The expression levels in ES and protein-iPS cells are at least 10 000-fold higher than those in fibroblasts.
In vitro characteristics of the protein-iPS cells. (A) RT-PCR of the ES cells, cFB-protein-iPS cells, cFBs, and feeder cells (STO). RNA of cFB-protein-iPS cells was harvested from 4 clones. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a loading control (right panels). The gene expressions of Oct4 and Nanog were verified by real-time PCR (left panels). There is no significant difference in the expression levels of Oct4 and Nanog between ES and protein-iPS cells. The expression levels in ES and protein-iPS cells are at least 10 000-fold higher than those in fibroblasts. (B) Scatterplots as determined by microarrays (n = 3 each). The global gene expression patterns were compared between the cFB and cFB-protein-iPS cell, and the ES cell and cFB-protein-iPS cell. The red lines indicate 2-fold changes in log scale. (C) Hierarchical clustering based on 4503 differentially expressed genes. (D) DNA methylation status of the Oct4 and Nanog promoters using bisulphite sequencing. ○ indicates unmethylated CpG nucleotides, and ● indicates methylated CpGs. (E) Histone modification status of the promoters. ChIP assays of trimethylated H3K4 and H3K27. Conventional PCR (top panel) and real-time PCR (bottom panel, n = 3, each). Expression level was adjusted by GAPDH (ES cell = 1.0 as an arbitrary unit).
In vitro characteristics of the protein-iPS cells. (A) RT-PCR of the ES cells, cFB-protein-iPS cells, cFBs, and feeder cells (STO). RNA of cFB-protein-iPS cells was harvested from 4 clones. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a loading control (right panels). The gene expressions of Oct4 and Nanog were verified by real-time PCR (left panels). There is no significant difference in the expression levels of Oct4 and Nanog between ES and protein-iPS cells. The expression levels in ES and protein-iPS cells are at least 10 000-fold higher than those in fibroblasts. (B) Scatterplots as determined by microarrays (n = 3 each). The global gene expression patterns were compared between the cFB and cFB-protein-iPS cell, and the ES cell and cFB-protein-iPS cell. The red lines indicate 2-fold changes in log scale. (C) Hierarchical clustering based on 4503 differentially expressed genes. (D) DNA methylation status of the Oct4 and Nanog promoters using bisulphite sequencing. ○ indicates unmethylated CpG nucleotides, and ● indicates methylated CpGs. (E) Histone modification status of the promoters. ChIP assays of trimethylated H3K4 and H3K27. Conventional PCR (top panel) and real-time PCR (bottom panel, n = 3, each). Expression level was adjusted by GAPDH (ES cell = 1.0 as an arbitrary unit).
Next, we performed a microarray analysis to assess the global gene expression profiles. Scatterplots demonstrated that the cFB-protein-iPS was very different from the cFB, and cFB-protein-iPS had an ES-like global gene expression pattern (Figure 2B). Hierarchical clustering also showed a similarity between the cFB-protein-iPS and ES cell, but not between the cFB-protein-iPS and the cFB (Figure 2C). In detail, the profile of DEGs selected by a fold change of more than 2 revealed that 3824 genes were differently expressed when cFB was compared with cFB-protein-iPS, whereas only 186 genes (50 genes up in ES, 136 genes up in cFB-protein-iPS) were differently expressed when cFB-protein-iPS was compared with ES cell (supplemental Figure 1; supplemental Tables 1-2). Interestingly, compared with cFB, CD44 was down-regulated, and Dnmt3a, Dnmt3b, Dnmt3l, histone methyltransferase 1 (HMT1), leukemia inhibitory factor receptor (LIF-Rc), undifferentiated embryonic cell transcription factor 1 (Utf1), and T-cell lymphoma breakpoint 1 (Tcl1) were markedly up-regulated in the cFB-protein-iPS. When we analyzed the differences between ES cells and cFB-protein-iPS cells in terms of biologic processes and molecular functions, up-regulated genes in ES cells were categorized to metabolic and cellular processes and cell cycle-related genes, and up-regulated genes in cFB-protein-iPS cells were categorized to anatomical structure development and metabolic process-related genes. Gene expression profiles focused on pluripotency-related genes revealed that there was no difference between protein donor ES cells and cFB-protein-iPS cells. Collectively, based on clustering, we could not find any meaningful difference related to reprogramming process. Interestingly, with respect to individual genes in the microarray, cFB-protein-iPS cells showed high expressions of Dnmt3a, Dnmt 3b (no change in Dnmt1 and Dnmt3l), and histone methyltransferase 1 (HMT1) compared with ES cells.
In addition to the global gene expression level, epigenetic modifications are critical for reprogramming and dedifferentiation up to the pluripotent state.27,31 Therefore, we evaluated DNA at the methylation level. Bisulfite genomic sequencing analysis showed that the promoter regions of Oct4 and Nanog were largely unmethylated in both cFB-protein-iPS and ES cell, but not in cFB (Figure 2D). In addition, we examined 2 other promoter regions, lamin A and B1, and found that both promoters were largely unmethylated. Interestingly, in contrast to lamin B1, lamin A expression was transcriptionally shutdown in ES cell and cFB-protein-iPS, but up-regulated in cFB (supplemental Figure 2). We also assessed the histone modification status.2 The ChIP assays of the Oct4 and Nanog promoters showed that cFB-protein-iPS had histone H3 lysine 4 (H3K4) methylation, whereas the cFB had histone H3 lysine 27 (H3K27) methylation (Figure 2E). Furthermore, cFB-protein-iPS showed increased acetylation of histone H3 of Oct4 and Nanog promoters, whereas cFB decreased (supplemental Figure 3). Genome-wide ChiP sequencing with H3K4me3 and H3K27me3 antibodies demonstrated that the correlation between histone modification in ES cells and cFB-protein-iPS was sound, and the pattern of cFB was substantially different from that of cFB-protein-iPS (supplemental Figure 4). Taken together, patterns of DNA methylation and histone modifications in cFB-protein-iPS were different from those of the cFB and very similar to those of the ES cell, implying that cFB-protein-iPS underwent epigenetic reprogramming.
In vitro and in vivo differentiation potentials of protein-iPS cells
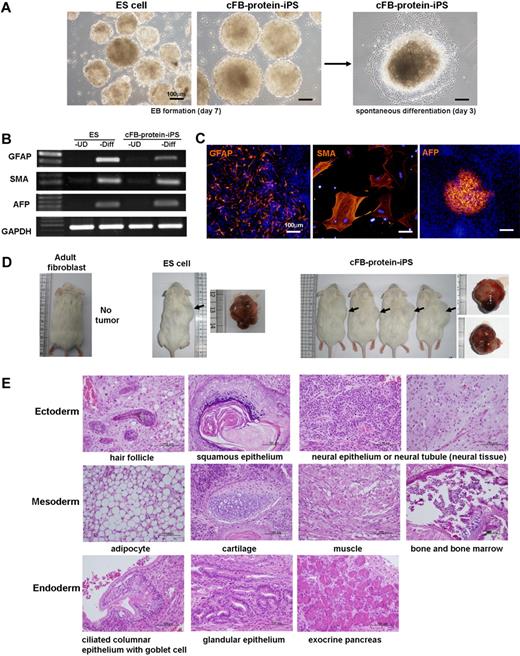
To determine the differentiation potential of the cFB-protein-iPS, we used the EB-based spontaneous differentiation protocol. After 7 days of suspension, the cFB-protein-iPS formed cell aggregates that looked like typical EBs. The ES cell–derived aggregates served as a positive control (Figure 3A). To further assess the differentiation potential, the aggregated cells (EBs) were plated onto a 0.1% gelatin-coated dish. Beginning at 4 to 7 days of differentiation, we observed some of the EBs contracting spontaneously and expressing cardiac troponin T and α-sarcomeric actinin, indicating the properties of cardiomyocytes (supplemental Video 1; supplemental Figure 5). On days 7 and 14, the cells were harvested, and the expression of 3 germ layer marker genes and proteins was evaluated. These cells expressed ectoderm (glial fibrillary acidic protein), mesoderm (α-smooth muscle actin), and endoderm (α-fetoprotein) lineage markers (Figure 3B-C).
In vitro and in vivo differentiation potentials of the protein-iPS cells. (A) EB formation by suspension culture (left), followed by the attachment of EB onto gelatin-coated dishes for the induction of spontaneous differentiation induction (right). (B-C) Seven to 14 days after commencing spontaneous differentiation, cells were harvested, and RT-PCR (B) and immunocytochemistry (C) were performed. UD, undifferentiated; Diff, spontaneous differentiation; GFAP, glial fibrillary acidic protein; SMA, α-smooth muscle actin; AFP, α-fetoprotein. DAPI (4′,6-Diamidino-2-phenylindole; blue) for nuclei. (D) cFB-protein-iPS cells were injected into NOD/SID mice. Adult fibroblasts and ES cells served as controls. Four weeks later, tumors were harvested (arrows). Gross morphologies showed well-demarcated tumors. (E) Hematoxylin and eosin (H&E) staining of a tumor from cFB-protein-iPS cells showing a well-differentiated teratoma.
In vitro and in vivo differentiation potentials of the protein-iPS cells. (A) EB formation by suspension culture (left), followed by the attachment of EB onto gelatin-coated dishes for the induction of spontaneous differentiation induction (right). (B-C) Seven to 14 days after commencing spontaneous differentiation, cells were harvested, and RT-PCR (B) and immunocytochemistry (C) were performed. UD, undifferentiated; Diff, spontaneous differentiation; GFAP, glial fibrillary acidic protein; SMA, α-smooth muscle actin; AFP, α-fetoprotein. DAPI (4′,6-Diamidino-2-phenylindole; blue) for nuclei. (D) cFB-protein-iPS cells were injected into NOD/SID mice. Adult fibroblasts and ES cells served as controls. Four weeks later, tumors were harvested (arrows). Gross morphologies showed well-demarcated tumors. (E) Hematoxylin and eosin (H&E) staining of a tumor from cFB-protein-iPS cells showing a well-differentiated teratoma.
Next, to investigate the in vivo pluripotency and differentiation potential, cFB-protein-iPS cells were injected into NOD/SCID mice. The cFB-protein-iPS gave rise to subcutaneous mass on days 14 to 21, and well-demarcated tumors were harvested on day 28 (Figure 3D). The tumors contained the various derivatives of the 3 germ layers, indicating the development of a well-differentiated teratoma (Figure 3E). These results suggest that the cFB-protein-iPS cell is comparable with the ES cell with regard to in vitro and in vivo pluripotency.
Protein-based reprogramming is neither by the contamination of donor ES cell nor by DNAs/RNAs from donor ES cell
The reprogramming of somatic cells into pluripotent stem cells has been generally considered a slow stochastic process, which require continuous ectopic expression of the defined transcription factors in a certain period and also require multiple cell divisions.19,32 In contrast, we, in the present study, observed that the transfer of ES cell–derived proteins reprogrammed somatic cells in the relatively short time window. Therefore, we next made every effort to exclude the possibility of experimental artifacts.
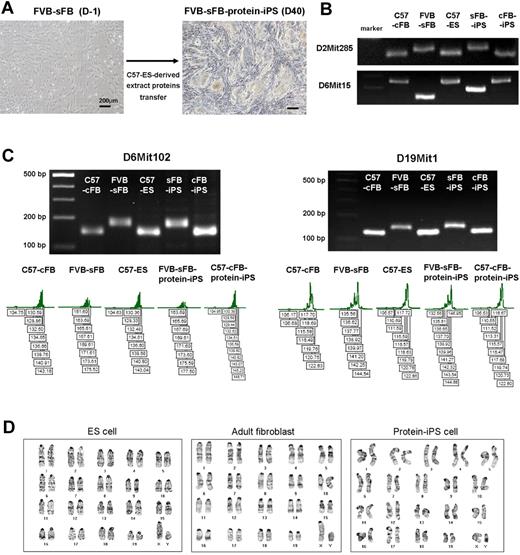
First, to rule out the possibility of ES cell contamination during the preparation of ES-derived extract proteins and the potential impurity of adult fibroblast, such as mixed-up culture with resident cardiac stem cells, we decided to generate iPS cells using adult skin fibroblasts of a different genetic strain from protein-donor ES cells. The donor of ES-derived extract proteins was a C57 background ES cell, and the recipient of ES proteins (a target cell of reprogramming) was a FVB background skin fibroblast (Figure 4A). FVB is an inbred strain. FVB strain has been widely used for the generation of transgenic animals but has considered resistant or nonpermissive strain for ES cell derivation. Recently, a few FVB-ES cell lines have been reported. However, only one wild-type ES cell line was described germ line-competent,33 whereas STAT3 overexpression in the inner cell mass achieved the germ line competency of FVB strain-derived ES cells.34 These data may suggest that inbred FVB strain is optimal to test the reliability or efficacy of new reprogramming method.
Protein-based reprogramming is not by the contamination of protein-donor ES cell. To rule out the possibility of ES cell contamination during the preparation of ES-derived extract proteins, further iPS cell generation was performed using adult somatic cells of a different genetic strain from protein-donor ES cells. The donor of ES-derived extract proteins was a C57 background, and the recipient of ES proteins (a target cell of reprogramming) was a FVB background. (A) Bright-field morphology of ES-like colonies developed from FVB background skin fibroblasts (sFB) using C57 ES cell–derived extract proteins. (B) Multiple microsatellite markers were applied to confirm the origin of the protein-iPS cells. PCR was performed using genomic DNA from each cell type. (C) To further verify the cellular origin, the sizes of amplified PCR products were validated using fluorescent 5′-FAM or 5′-HEX primers. The Genescan 500ROXTM size standard served as an internal control. (D) The chromosomes of ES cell, adult fibroblast, and protein-iPS cell were diploid and normal karyotype.
Protein-based reprogramming is not by the contamination of protein-donor ES cell. To rule out the possibility of ES cell contamination during the preparation of ES-derived extract proteins, further iPS cell generation was performed using adult somatic cells of a different genetic strain from protein-donor ES cells. The donor of ES-derived extract proteins was a C57 background, and the recipient of ES proteins (a target cell of reprogramming) was a FVB background. (A) Bright-field morphology of ES-like colonies developed from FVB background skin fibroblasts (sFB) using C57 ES cell–derived extract proteins. (B) Multiple microsatellite markers were applied to confirm the origin of the protein-iPS cells. PCR was performed using genomic DNA from each cell type. (C) To further verify the cellular origin, the sizes of amplified PCR products were validated using fluorescent 5′-FAM or 5′-HEX primers. The Genescan 500ROXTM size standard served as an internal control. (D) The chromosomes of ES cell, adult fibroblast, and protein-iPS cell were diploid and normal karyotype.
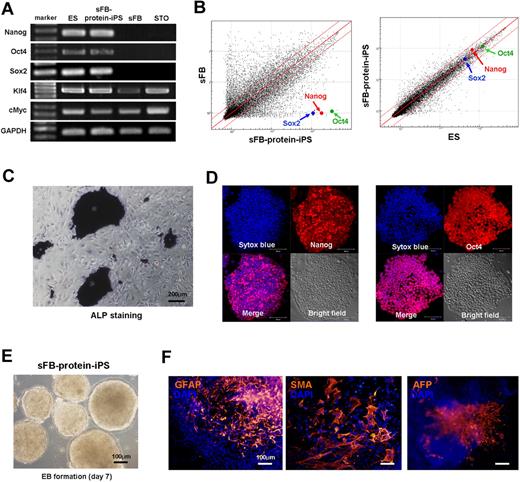
After we acquired ES-like colonies from skin fibroblasts by proteins transfer, we confirmed the origin of the sFB-protein-iPS cells by multiple microsatellite markers of genomic DNA and exact size measurement (Figure 4B-C). sFB-protein-iPS cell is a FVB background. Furthermore, to verify chromosomal stability after reprogramming as well as to rule out the addition of ES cell chromosomes to protein-iPS cells, we karyotyped the cells and confirmed diploidy (Figure 4D). We further characterized FVB background sFB-protein-iPS cells. The expression of pluripotency-related genes and proteins, global gene expression patterns, DNA methylation status, and in vitro differentiation potential of FVB-sFB-protein-iPS cells were similar to the authentic ES cells (Figure 5A-F and supplemental Figure 6), indicating that sFB-protein-iPS cells is fully reprogrammed and comparable with the ES cell in terms of pluripotency.
Characteristics of protein-iPS cell derived from a different genetic strain from protein-donor ES cell. We further characterized FVB background sFB-protein-iPS cells. (A) RT-PCR for pluripotency-related genes. GAPDH was used as a loading control. (B) Scatterplots as determined by microarrays (n = 4, each). The global gene expression patterns were compared between the sFB and sFB-protein-iPS cell, and the ES cell and sFB-protein-iPS cell. The red lines indicate 2-fold changes in log scale. (C) ALP staining of sFB-protein-iPS cells. (D) Immunocytochemistry showing that sFB-protein-iPS cells express Nanog and Oct4. (E-F) In vitro differentiation potential of sFB-protein-iPS cells was evaluated by EB formation (E) and EB-based spontaneous differentiation (F). Immunocytochemistry revealed the expression of 3 germ layer marker proteins.
Characteristics of protein-iPS cell derived from a different genetic strain from protein-donor ES cell. We further characterized FVB background sFB-protein-iPS cells. (A) RT-PCR for pluripotency-related genes. GAPDH was used as a loading control. (B) Scatterplots as determined by microarrays (n = 4, each). The global gene expression patterns were compared between the sFB and sFB-protein-iPS cell, and the ES cell and sFB-protein-iPS cell. The red lines indicate 2-fold changes in log scale. (C) ALP staining of sFB-protein-iPS cells. (D) Immunocytochemistry showing that sFB-protein-iPS cells express Nanog and Oct4. (E-F) In vitro differentiation potential of sFB-protein-iPS cells was evaluated by EB formation (E) and EB-based spontaneous differentiation (F). Immunocytochemistry revealed the expression of 3 germ layer marker proteins.
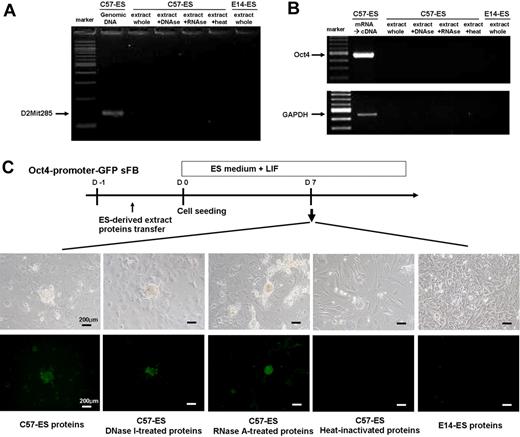
Next, to demonstrate that ES extract-mediated reprogramming is dependent upon proteins, we first checked for DNA or RNA contamination in the ES-derived extract (Figure 6A-B). Second, to further exclude the possibility of contamination by genetic material or RNA, we treated the extract with DNase or RNase before transfer. In a series of experiments, we used Oct4 promoter-driven GFP expressing fibroblast as a reporter. To verify proteins are an effector, we also inactivated the extract by heat (Figure 6C). Data showed that DNA or RNA from ES cell has no effect on the reprogramming and that ES cell–derived proteins are wholly responsible.
Effector of reprogramming. Protein-based reprogramming is not mediated by DNAs/RNAs from donor ES cell. (A-B) The absence of contaminating DNA or RNA in the extract proteins was confirmed by PCR and RT-PCR. (C) Colony formation of Oct4-promoter–driven GFP reporter cells (skin fibroblast) at 7 days after ES-derived extract proteins transfer.
Effector of reprogramming. Protein-based reprogramming is not mediated by DNAs/RNAs from donor ES cell. (A-B) The absence of contaminating DNA or RNA in the extract proteins was confirmed by PCR and RT-PCR. (C) Colony formation of Oct4-promoter–driven GFP reporter cells (skin fibroblast) at 7 days after ES-derived extract proteins transfer.
Early (4-7 days after protein transfer) colony formation and Oct4 expression are unique features of our approach, compared with reprogramming sequences used in previous studies based on 4 factor virus/plasmid/protein methods.1,2,10-12,16 Accordingly, to further assess the role of 4 factors like Oct4 in this protein-based reprogramming process, we measured the concentration of Oct4 in the ES-derived protein extracts. The total concentration of C57-ES–derived extract proteins by the BCA and Bradford assay was 32.065 mg/mL, and Oct4 concentration by enzyme-linked immunosorbent assay (ELISA) was 46.225 ng/mL, indicating that Oct4 accounted for only 0.00 014% of total extract proteins. We usually transferred 200 μL extract proteins for 106 cells, and thus, the absolute amount of Oct4 available for reprogramming was is 9.25 ng. A recent study reported that repeated transfer of 8 μg/mL (8000 ng/mL) Oct4 protein with valproic acid could induce pluripotent stem cells.16 Compared with that study, we used a substantially lower concentration of Oct4 protein and transferred proteins into adult fibroblast once, which suggests that Oct4 may not have been an essential functional effector molecule in our study, and instead, a certain group of effector proteins, which could be the downstream targets of the defined transcriptional factors such as Oct4, Sox2, Nanog, c-Myc, Lin28, C/EBP-α, and Esrrb, etc,1,3,35,36 may be responsible for ES extract protein-mediated reprogramming. Taken together, ES cell–derived proteins may have a combination of many factors like cytoplasms of meiotic oocyte and mitotic zygote,18 which can induce pluripotency of somatic cells. Future studies to characterize specific effector proteins would provide insights of the mechanistic understanding of the protein-based reprogramming process.
In vivo differentiation and developmental potential of protein-iPS cells from a different genetic strain from protein-donor ES cell
To confirm pluripotency of protein-iPS cells generated by genetically unmatched ES extract proteins, we first injected FVB-sFB-protein-iPS cells into NOD/SCID mice (Figure 7A). Four weeks later, protein-iPS cells gave rise to well-differentiated teratomas that contained the various derivatives of the 3 germ layers, indicating that protein-iPS possess in vivo differentiation potential (Figure 7B).
Teratoma formation and in vivo developmental potential of FVB background protein-iPS cell. (A) FVB-sFB-protein-iPS cells were injected into NOD/SID mice. Four weeks later, well-demarcated tumors were observed. (B) H&E staining showed well-differentiated teratomas. (C) Chimera mice produced by protein-iPS cells. FVB-sFB-protein-iPS cells contributed to the viable chimera after being injected into the C57 host blastocysts. Mice with mixed coat colors are chimeras. As a control, C57-ES cells (extract-protein donors) were injected into ICR blastocysts. (D) A fetus derived from GFP-transduced FVB background protein-iPS by tetraploid blastocyst complementation. The arrow indicates a beating heart. (E) Genomic DNA PCR showed the origin of fetus is FVB background protein-iPS cells. (F) The accurate sizes of amplified products were confirmed using fluorescent primers.
Teratoma formation and in vivo developmental potential of FVB background protein-iPS cell. (A) FVB-sFB-protein-iPS cells were injected into NOD/SID mice. Four weeks later, well-demarcated tumors were observed. (B) H&E staining showed well-differentiated teratomas. (C) Chimera mice produced by protein-iPS cells. FVB-sFB-protein-iPS cells contributed to the viable chimera after being injected into the C57 host blastocysts. Mice with mixed coat colors are chimeras. As a control, C57-ES cells (extract-protein donors) were injected into ICR blastocysts. (D) A fetus derived from GFP-transduced FVB background protein-iPS by tetraploid blastocyst complementation. The arrow indicates a beating heart. (E) Genomic DNA PCR showed the origin of fetus is FVB background protein-iPS cells. (F) The accurate sizes of amplified products were confirmed using fluorescent primers.
Next, to assess developmental potential, we injected FVB-protein-iPS cells into a C57 blastocyst cavity and transferred them into the uterus of pseudopregnant ICR mice. Twenty-four of 110 offspring showed white striped-agouti coat color, indicating chimeras (Figure 7C; experiments listed in supplemental Table 4). Moreover, up until 40 weeks of age, no tumor was grossly observed. These findings indicate that protein transfer strategy can offer a safe and effective way for the induction of reprogramming. When male chimeras were mated with wild-type FVB female mice, we did not obtain viable homozygote offspring. Therefore, as an alternative approach to verify the developmental potential and pluripotency of FVB-sFB-protein-iPS cells, we performed a tetraploid blastocyst complementation experiment, which is considered the most stringent functional assay of pluripotency.29,30 We introduced diploid GFP-transduced FVB-sFB-protein-iPS cells into ICR tetraploid blastocysts. At E10.5, we harvested and genotyped the embryos (Figure 7D-F; supplemental Video 2 demonstrates a beating heart of a harvested fetus). We confirmed fetal animals have been derived from FVB-background adult fibroblasts, indicating that the protein-iPS cells possess in vivo developmental potential. The generation of live-born animals from protein-based reprogrammed cells will be tested in the near future.
Discussion
In the present study, we demonstrate that the delivery of ES cell–derived proteins enables the reprogramming of adult fibroblasts, converting them into pluripotent stem cells without the forced expression of certain genetic factors. Our results are different in several aspects, compared with previous studies conducted using a similar protocol (supplemental Table 5 describes detailed comparisons).23,24 First, we achieved full reprogramming of adult fibroblast up to the pluripotent state and redifferentiation into 3 germ layers in vitro and in vivo. We also demonstrated in vivo developmental potential of protein-iPS cells. Second, instead of immortalized cell lines or fetal cells used in the previous studies, we used primarily cultured adult fibroblasts, which obviously possess a limited lifespan; this might have resulted in the natural selection of pluripotent stem cells during the reprogramming. Third, after the initial colony formation, we subcultured and maintained the colonies on the feeder cell layers. Fourth, we used C57 strain ES cells as a protein donor and initially transferred extract proteins to C57 background adult fibroblasts, which are considered permissive strain for ES cell derivation, resulting in the successful reprogramming. When we applied these to FVB background fibroblasts, which are considered a resistant or nonpermissive strain, we obtained the same successful results. Collectively, we succeeded in protein-based reprogramming in both permissive and nonpermissive strains. However, when we tried other genetic background cells, such as the 129 strain of ES cells (E14), as a protein donor, we failed to achieve pluripotency with the C57 adult fibroblasts.
Currently, little is known about the molecular mechanisms underlying reprogramming process by cellular proteins. Proteomic analysis of the 129 and C57 strain of ES cells using iTRAQ and Mass/Mass experiment demonstrated a marked difference of protein expressions between the 2 ES cell lines (data not shown). These findings suggest that differences in cellular proteins, between different ES cell lines, could lead to substantial differences in reprogramming process (Figure 6 and supplemental Table 3 list experiments in this study). Further studies regarding mechanistic analyses, such as which components of ES cell–derived protein(s) are essential for epigenetic modification and reprogramming and which somatic cell types would be the best candidate for protein-based reprogramming, will provide insight and technical refinements. For instance, a matrix of experiments (using C57-ES proteins into C57 somatic cells, 129-ES proteins into 129 somatic cells, C57-ES proteins into 129 somatic cells, and 129-ES proteins into C57 somatic cells) and substitution experiments using specific proteins based on proteomic analysis would be helpful. In addition, application of this technique to adult human somatic cells should be tested. Such studies will bring us closer to the application of protein-iPS cells to therapeutic purposes.
Cell fusion studies indicated that the ES cell nucleus is needed to reprogram the somatic cell nucleus.37,38 Accordingly, we performed a fractionation experiment. When we divided ES proteins into cytosolic and nuclear fraction, neither cytoplasmic nor nuclear fraction was able to induce reprogramming of adult fibroblasts, whereas whole cellular proteins reproducibly generated protein-iPS cells. Our results imply that not only nuclear proteins but also cytoplasmic proteins of ES cells are needed to induce the pluripotency of somatic cells. Regarding the molecular sizes of effector proteins, extract proteins larger than 30 kDa induced reprogramming, but proteins larger than 100 kDa did not (supplemental Figure 7), suggesting that a certain group of ES cell–derived proteins between 30 to 100 kDa could be responsible for protein-based iPS generation.
Recent studies have shown that partially reprogrammed “intermediate” cells when defined factors expressed ectopically.32,39 Therefore, we also tried to indentify intermediate cells during protein-based reprogramming experiments. We induced reprogramming using skin fibroblast from actin promoter–driven enhanced GFP (eGFP) expressing mouse and were able to pick up and distinguish fully reprogrammed cells from partially reprogrammed cells by colony morphology (well-delineated colonies versus spiculating colonies in supplemental Figure 8). Fully reprogrammed cells expressed Oct4 and redifferentiated into 3 germ layers in vitro and in vivo. Partially reprogrammed or intermediate protein-iPS cells weakly expressed Oct4 and Nanog mRNA, compared with ES cells. Interestingly, the level of Sox2 and Klf4 expression was similar between ES and partial protein-iPS cells. When cells were injected into SCID mice subcutaneously, tumors were formed with invasion into intra-abdominal space and metastasis to liver. Histology demonstrated poorly differentiated tumors. This data imply that transfer of proteins into some cells might be insufficient for reprogramming up to pluripotency or protein-based reprogramming approach might be also a stochastic rather than deterministic process.19
In summary, we apply protein-based reprogramming approach and demonstrate that a single transfer of proteins can induce reprogramming of adult fibroblasts, rather than using fetus- or newborn-origin cells, up to the pluripotent state. We confirmed that a certain group of ES-derived extract proteins, rather than DNA or RNA, acts as the effector of reprogramming. Our approach is relatively simple and reproducible and does not require repeated transfer or prolonged exposure to materials or a combinatorial approach involving proteins and chemicals. These results provide a safe and effective alternative strategy for reprogramming of adult somatic cells and suggest that the described technique could be further developed to provide tailored or patient-specific cell therapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Byoung-Kwon Kim and Yoon-Kyung Jeon for histologic analysis of tumors and Sun-Hee Lee for technical assistance.
This study was supported by a grant from the Innovative Research Institute for Cell Therapy (A062260) and Stem Cell Research Center (SC4210), Republic of Korea.
Authorship
Contribution: H.-J.C. designed the research, analyzed data, and wrote the paper. C.-S.L., J.S.P., Y.-W.K. S.-H. Lee, J.H., and E.J.L. performed experiments regarding generation and characterization of protein-iPS cells. T.-Y.R., I.-S.C., Y.S.K., and S.-H. Leem performed experiments regarding epigenetic analysis, gene expression analysis, and protein analysis. H.-J.K., Y.-B.P., and H.-S.K. planned the project, coordinated the research team, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hyo-Soo Kim or Young-Bae Park, Cardiovascular Center, Seoul National University Hospital, 28 Yongon-dong, Chongno-gu, Seoul 110-744 Korea; e-mail: hyosoo@snu.ac.kr.
References
Author notes
H.-J.C. and C.-S.L. contributed equally to this work.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal