Abstract
The Spi1/Pu.1 transcription factor plays a crucial role in myeloid cell development in vertebrates. Despite extensive studies of Spi1, the controlled gene group remains largely unknown. To identify genes dependent on Spi1, we used a microarray strategy using a knockdown approach in zebrafish embryos combined with fluorescence-activated cell sorting of myeloid cells from transgenic embryos. This approach of using knockdowns with specific green fluorescent protein-marked cell types was highly successful in identifying macrophage-specific genes in Spi1-directed innate immunity. We found a gene group down-regulated on spi1 knockdown, which is also enriched in fluorescence-activated cell-sorted embryonic myeloid cells of a spi1:GFP transgenic line. This gene group, representing putative myeloid-specific Spi1 target genes, contained all 5 previously identified Spi1-dependent zebrafish genes as well as a large set of novel immune-related genes. Colocalization studies with neutrophil and macrophage markers revealed that genes cxcr3.2, mpeg1, ptpn6, and mfap4 were expressed specifically in early embryonic macrophages. In a functional approach, we demonstrated that gene cxcr3.2, coding for chemokine receptor 3.2, is involved in macrophage migration to the site of bacterial infection. Therefore, based on our combined transcriptome analyses, we discovered novel early macrophage-specific marker genes, including a signal transducer pivotal for macrophage migration in the innate immune response.
Introduction
The SPI1 gene, also known as PU.1, encodes a member of the E26-transformation-specific family of transcription factors and is expressed specifically in cells of the hematopoietic lineage.1 SPI1/PU.1 transcription factor activity is crucial for development of cells of the myeloid and lymphoid lineages.2-7 PU.1-null mice lack macrophages, neutrophils, and B and T cells, and they die within the first 48 hours.4,5 Distinct hematopoietic cell fates depend on graded expression levels of SPI1/PU.1, with a high concentration driving macrophage differentiation, and a lower concentration promoting B lymphocyte development.2 Several target genes of SPI1/PU.1 have already been identified in mammals, including PTPN6,8 PTPRC,9 CSF1R,10 IL-1β,11 MPO,12 and PU.113 itself. Based on the presence of cofactors and interaction with other transcription factors, PU.1-regulated gene groups are expected to be cell type and condition dependent. Microarray technology has been used to demonstrate reprogramming of PU.1-dependent expression during wound-induced inflammation in mouse neonates and to determine PU.1-regulated gene sets in cultured cells committed to macrophage differentiation and in preleukemic hematopoietic stem cells.14-16 However, which precise gene groups are under control of PU.1 at different stages of myeloid and lymphoid lineage commitment remains unknown.
The transcriptional regulation of hematopoiesis is largely conserved in human, mice, and zebrafish. The spi1 (pu.1) gene of zebrafish is first expressed at 12 hours postfertilization (hpf) in cells of the anterior lateral plate mesoderm (LPM).17 The second site of spi1 expression is observed at 14 hpf in the posterior LPM.17,18 In that region, expression of spi1 overlaps with cells expressing transcription factor gata1. The cells expressing spi1 in the anterior LPM develop into myeloid cells, whereas the gata1-expressing cells in the posterior LPM differentiate into erythroid cells. The expression of spi1 decreases gradually with only a limited number of spi1-positive cells in the erythromyeloid progenitors (EMPs)19 in the posterior blood island and sometimes over the yolk at 30 hpf.17 SPI1/PU.1 and GATA1 have been shown to antagonize each other in human myeloerythroid progenitor cells, and the reciprocal negative regulation of these transcription factors has been recapitulated in the zebrafish embryo model.20 Furthermore, expression of exogenous spi1 mRNA in zebrafish cloche mutant embryos, which lack hematopoietic and vascular tissues, was sufficient to rescue myelopoiesis but not erythropoiesis.20 The morpholino-gene knockdown of spi1 resulted in a complete loss of primitive macrophage development and a reduced development of neutrophils.21 Similar to mammalian models, these observations indicate the crucial role for spi1 in myeloid cell development in zebrafish.
Blood circulation in zebrafish embryos begins between 24 and 26 hpf. The blood cells present in the 1- to 2-day-old zebrafish embryos consist of erythrocytes and 2 distinct populations of myeloid cells that both express the pan-leukocytic marker lcp1 (l-plastin).22,23 One of these myeloid cell populations is marked by expression of mpx (coding for myeloperoxidase), which is a marker of differentiated neutrophils at later stages.21,24,25 The other population represents the early zebrafish macrophages that are able to sense, migrate toward, and phagocytose bacteria injected into body cavities.22 The csf1r (fms) gene is the only known specific marker of the macrophage lineage, but this gene is also expressed in neural crest cells.26 There is a growing need for identification of macrophage-specific marker genes, which will facilitate the understanding of the development, behavior, and role of macrophages in the immune system.
The zebrafish embryo, because of its transparency, is an excellent model to study live progression of hematopoiesis and innate immune responses. Importantly, innate immunity can be studied in separation from adaptive immunity in zebrafish embryos, as development and maturation of lymphoid cells require several weeks.27 The generation of a number of transgenic zebrafish with fluorescent proteins under the control of promoters of genes involved in distinct aspects of hematopoiesis greatly facilitated studies on cell-fate mapping and understanding the development of various hematopoietic lineages.28,29 Spi1:GFP transgenic zebrafish embryos between 12 and 28 hpf show a myeloid-specific green fluorescent protein (GFP) expression pattern that largely overlaps with expression of endogenous spi1 mRNA.30 Flow cytometric analysis of GFP-expressing cells from adult zebrafish kidney of the spi1:GFP line revealed that spi1 is also expressed in cells exhibiting myeloid or lymphoid morphology, thus indicating that this gene may be involved in definitive hematopoiesis in zebrafish adults.30,31
Recognizing the importance of the Spi1 transcription factor in regulation of macrophage and neutrophil development, we set out to identify the genes that directly or indirectly depend on Spi1 and are expressed in embryonic myeloid cells. Microarray-based expression analysis of spi1 knockdown embryos and fluorescence-activated cell sorter (FACS)–sorted myeloid cells from embryos of spi1:GFP transgenic zebrafish made it possible to identify 249 putative Spi1 target genes. These include several known genes with roles in hematopoiesis and immune system development as well as novel genes with immune-related functions. We demonstrate 4 of these genes (cxcr3.2, mfap4, mpeg1, and ptpn6) to be macrophage specific in early zebrafish embryos. Furthermore, we show that the function of gene cxcr3.2, coding for a chemokine receptor, is necessary for macrophage migration to local bacterial infection with Salmonella typhimurium.
Methods
Zebrafish strains
Zebrafish (Danio rerio) were handled in compliance with the local animal welfare regulations and maintained according to standard protocols (www.ZebrafishInformationNetwork.org). The study was approved by the Institutional Review Board of Leiden University. The spi1:GFP transgenic line used in this study (Pu1-lynEGFP, a gift from Franscesca Peri, EMBL, Heidelberg) contains 4 kb of spi1/pu.1 promoter sequence driving myeloid expression of membrane-targeted enhanced green fluorescent protein (EGFP), as validated by immunostaining with antibodies against L-plastin and Pu.1/Spi1 (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Morpholino knockdown experiments
Morpholino oligonucleotides (Gene Tools) were diluted in 1× Danieau buffer (58mM NaCl, 0.7mM KCl, 0.4mM MgSO4, 0.6mM Ca(NO3)2, 5.0mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid; pH 7.6) containing 1% phenol red (Sigma-Aldrich), and approximately 1 nL was injected into the 1- or 2-cell–stage embryo using a Femtojet injector (Eppendorf).
Embryo dissociation and FACS
Dissociation of embryos was performed according to Covassin et al.32 In short, embryos were dechorionated using pronase treatment, rinsed in calcium-free Ringer solution, followed by digestion with 0.25% trypsin. The obtained cell suspension was centrifuged, rinsed with phosphate-buffered saline (PBS), and resuspended in Leibovitz medium L15 without phenol red, 1% fetal calf serum, 0.8mM CaCl2, penicillin 50 U/mL, and streptomycin 0.05 mg/mL. The single-cell suspension was subjected to FACS at room temperature using a FACSAria (BD Biosciences) with the BD FACSDiva software, Version 5.0.3 and a Coherent Sapphire solid-state laser 488 nm with 13 mW power. The GFP+ and GFP− cell fractions were collected as in L15 medium but with 10% fetal calf serum. The standard yield from approximately 300 embryos was approximately 2 × 105 GFP+ cells.
Microarray experiment design and analysis
Embryos injected either with 1mM spi1 morpholino20 or 1mM Standard Control morpholino were harvested at 28 hpf. As a control, embryos injected with an equal volume of 1% phenol red in Danieau buffer (1 times) were used. RNA samples from pools of 20 to 30 spi1 and standard control morphants were labeled with Cy5 and hybridized against a Cy3-labeled common reference (a mixture of all samples injected with 1 times phenol red in Danieau buffer). This experiment was performed in triplicate. FACS-sorted spi1 GFP embryos were analyzed as follows: RNA of the GFP+ fraction was labeled with Cy5 and hybridized against Cy3-labeled RNA of the corresponding GFP− cell fraction from the same pool of embryos. This experiment was performed in duplicate. RNA isolation, synthesis of amino allyl-labeled aRNA, dye coupling, and hybridization conditions were as described.33 Microarray analysis was performed using custom-designed 44k Agilent chips (Gene Expression Omnibus platform accession no. GPL7735) described elsewhere.33 Microarray data were processed and analyzed as described.33 Significance cutoffs for differentially expressed probe sequences were set at 1.2-fold change at P less than .01. The data were submitted to the Gene Expression Omnibus database (National Center for Biotechnology Information; www.ncbi.nlm.nih.gov/geo) under accession no. GSE19206.
Whole-mount in situ hybridization
Whole-mount in situ hybridization using alkaline phosphatase detection with BM Purple substrate (Roche Diagnostics) was carried out as described using phenylthiourea-treated embryos.33 Colocalization experiments by whole-mount fluorescent in situ hybridization were performed as described.23 Primer sequences used for polymerase chain reaction (PCR) amplification of templates for probe synthesis are in supplemental Table 1.
Quantitative RT-PCR of embryonic RNA
Real-time PCR was performed as described33 using the primer sequences in supplemental Table 1.
Bacterial infection experiments
S typhimurium strain SF1592 expressing red fluorescent protein was grown as described.33 Morpholino-injected embryos were anesthetized at 28 hpf with 200 μg/mL tricaine (Sigma-Aldrich) and microinjected in the tail muscle with 5 nL of bacterial suspension in PBS. Injected embryos were transferred into fresh egg water, incubated at 28°C for 3 hours, and fixed with 4% paraformaldehyde in PBS. In situ hybridization was performed with digoxigenin-labeled mfap4 and fluorescein-labeled mpx probes. Leukocytes accumulated within a 50-μm radius from the injection site were counted using a Leica MZ16FA stereo microscope.
Results
Microarray-based identification of early myeloid-specific genes regulated by Spi1
In this study, we used transcriptome analysis to identify the subset of genes that are directly or indirectly dependent on the Spi1 transcription factor and are expressed in myeloid cells of the zebrafish embryo. Two distinct microarray experiments were carried out. First, mRNA isolated from 28-hpf embryos injected with spi1 morpholino was analyzed. The efficiency of spi1 knockdown was confirmed by whole-mount in situ hybridization with a probe against lcp1 (coding for L-plastin), a gene known to be regulated by Spi120 (Figure 1A). In the second approach, we profiled the transcriptome of GFP+ early myeloid cells against the GFP− remaining cell fraction. Both fractions were obtained from FACS sorting of a single-cell suspension that resulted from dissociation of embryos of a transgenic zebrafish line, spi1:GFP (Figure 1A).
Microarray experiment setup and analysis. (A) The experimental setup of the microarray study. The 28-hpf time point for microarray analysis in both experiments was chosen after the onset of blood circulation in the embryo and to ensure the abundance of myeloid cells of the innate immune system as shown by lcp1 in situ staining of embryos treated with the standard control (SC) morpholino. The lack of lcp1 staining in spi1 morphants confirms the absence of myeloid cells. GFP+ myeloid cells were isolated from embryos expressing membrane-targeted EGFP under control of the spi1 promoter. The 2-step approach eliminated false positives because of leaky expression of the spi1:GFP transgene in the brain and allowed determination of the specific effect of spi1 knockdown on myeloid cells. (B) Primary analysis of microarray data. For initial comparative analysis we chose an arbitrary cut-off of absolute fold change larger than or equal to 1.2 and a P value ≤ .01. We found 3551 sequences to be down-regulated and 3684 sequences to be up-regulated in the spi1 knockdown embryos. In the GFP+ myeloid cell fraction, 6327 sequences were enriched and 6335 sequences were reduced compared with the GFP fraction. The overlap between the sequences found down-regulated in spi1 morphants and enriched in the GFP+ myeloid cell fraction was estimated and filtered against the sequence set found down-regulated in control morpholino-injected embryos. (C) GO term annotation analysis of the 249 genes found in the overlap. Category 1 indicates zebrafish genes with at least one GO term annotation; category 2, genes without any annotation in zebrafish but having a homolog with known GO term annotation in humans; and category 3, genes without any GO term annotation either in zebrafish or in humans.
Microarray experiment setup and analysis. (A) The experimental setup of the microarray study. The 28-hpf time point for microarray analysis in both experiments was chosen after the onset of blood circulation in the embryo and to ensure the abundance of myeloid cells of the innate immune system as shown by lcp1 in situ staining of embryos treated with the standard control (SC) morpholino. The lack of lcp1 staining in spi1 morphants confirms the absence of myeloid cells. GFP+ myeloid cells were isolated from embryos expressing membrane-targeted EGFP under control of the spi1 promoter. The 2-step approach eliminated false positives because of leaky expression of the spi1:GFP transgene in the brain and allowed determination of the specific effect of spi1 knockdown on myeloid cells. (B) Primary analysis of microarray data. For initial comparative analysis we chose an arbitrary cut-off of absolute fold change larger than or equal to 1.2 and a P value ≤ .01. We found 3551 sequences to be down-regulated and 3684 sequences to be up-regulated in the spi1 knockdown embryos. In the GFP+ myeloid cell fraction, 6327 sequences were enriched and 6335 sequences were reduced compared with the GFP fraction. The overlap between the sequences found down-regulated in spi1 morphants and enriched in the GFP+ myeloid cell fraction was estimated and filtered against the sequence set found down-regulated in control morpholino-injected embryos. (C) GO term annotation analysis of the 249 genes found in the overlap. Category 1 indicates zebrafish genes with at least one GO term annotation; category 2, genes without any annotation in zebrafish but having a homolog with known GO term annotation in humans; and category 3, genes without any GO term annotation either in zebrafish or in humans.
We reasoned that the overlap between genes down-regulated in the spi1 knockdown and enriched in the GFP+ myeloid cell fraction (Figure 1B) contains genes that are both downstream of Spi1 and expressed in early myeloid cells. After filtering against control morpholino microarray data, 249 distinct genes were found in the overlapping group (Figure 1B; supplemental Table 2). Previously, 5 genes were shown to be downstream of Spi1 in zebrafish embryos, namely, lcp1, mpx, lyz, coro1a, and ncf1.20 All 5 genes were found to be significantly down-regulated in the spi1 knockdown embryos, while they were also significantly enriched in the GFP+ myeloid cell fraction (supplemental Figure 2), thus providing an internal control and confirming our analysis approach. In addition, our gene set contained several homologs of previously validated target genes of SPI1/PU.1 in mammalian systems, including ptpn6, ptprc, il1β, and mpx, as well as the homologs of 2 genes, ctss and rgs18, recently identified as novel SPI1/PU.1 target genes in an integrated microarray and chromatin immunoprecipitation (ChIP)-chip analysis of murine macrophages and hematopoietic cell lines.8,9,11,12,16 Furthermore, spi1 itself was identified in our 2-step microarray approach, consistent with its myeloid-specific expression pattern and known autoregulatory function.1
Novel genes downstream of Spi1 play a role in hematopoiesis and immune system development
In the microarray data, 249 genes were identified that are expressed in embryonic myeloid cells and downstream of the Spi1 transcription factor. Next, using Zebrafish Information Network and National Center for Biotechnology Information databases, we determined which of the 249 genes have at least one Gene Ontology (GO) term annotation (Figure 1C). Interestingly, we found that 93 of the 249 genes lack any GO term annotation either in zebrafish or when using human homologs of zebrafish genes. To characterize the annotated gene group (categories 1 and 2, Figure 1C), a GO enrichment analysis was performed using DAVID tools for functional classification and clustering (Table 1). To this extent, we converted the zebrafish gene identifiers to their human homologs, taking advantage of the fact that the GO annotations of human genes are more complete.34 DAVID analysis of GO-Biological process of the myeloid genes downstream of Spi1 revealed significant enrichment in functional groups involved in “hemopoiesis,” “immune system development,” and “myeloid cell differentiation.” In addition, several GO terms related to “apoptosis” and “anion transport” were also enriched within the investigated gene group. In the GO-Cellular Component analysis, we found significant enrichment in several categories associated with “vacuole,” “lysosome,” and “extracellular matrix,” all consistent with myeloid cell functions. In summary, the DAVID functional analysis supports our microarray approach and confirms that genes dependent on Spi1 and expressed in myeloid cells share functions required for the innate immune system.
GO enrichment analysis of the myeloid cell–specific gene set downstream of Spi1
| Term . | Count . | % . | P . |
|---|---|---|---|
| GO_biological process | |||
| GO:0006817—phosphate transport | 7 | 4.49 | < .001 |
| GO:0030097—hemopoiesis | 8 | 5.13 | < .001 |
| GO:0006820—anion transport | 8 | 5.13 | .001 |
| GO:0048513—organ development | 22 | 14.10 | .001 |
| GO:0048534—hemopoietic or lymphoid organ development | 8 | 5.13 | .001 |
| GO:0065008—regulation of biologic quality | 17 | 10.90 | .002 |
| GO:0002520—immune system development | 8 | 5.13 | .002 |
| GO:0015698—inorganic anion transport | 7 | 4.49 | .002 |
| GO:0006915—apoptosis | 15 | 9.62 | .004 |
| GO:0012501—programmed cell death | 15 | 9.62 | .005 |
| GO:0030099—myeloid cell differentiation | 5 | 3.21 | .006 |
| GO:0008219—cell death | 15 | 9.62 | .007 |
| GO:0016265—death | 15 | 9.62 | .007 |
| GO:0032502—developmental process | 39 | 25.00 | .009 |
| GO_cellular component | |||
| GO:0005737—cytoplasm | 77 | 49.36 | < .001 |
| GO:0044424—intracellular part | 100 | 64.10 | < .001 |
| GO:0005622—intracellular | 104 | 66.67 | < .001 |
| GO:0005764—lysosome | 9 | 5.77 | < .001 |
| GO:0000323—lytic vacuole | 9 | 5.77 | < .001 |
| GO:0005829—cytosol | 14 | 8.97 | < .001 |
| GO:0005773—vacuole | 9 | 5.77 | < .001 |
| GO:0044445—cytosolic part | 7 | 4.49 | < .001 |
| GO:0044420—extracellular matrix part | 6 | 3.85 | .002 |
| GO:0044444—cytoplasmic part | 44 | 28.21 | .003 |
| GO_molecular function | |||
| GO:0005515—protein binding | 73 | 46.79 | < .001 |
| GO:0005198—structural molecule activity | 15 | 9.62 | .007 |
| Term . | Count . | % . | P . |
|---|---|---|---|
| GO_biological process | |||
| GO:0006817—phosphate transport | 7 | 4.49 | < .001 |
| GO:0030097—hemopoiesis | 8 | 5.13 | < .001 |
| GO:0006820—anion transport | 8 | 5.13 | .001 |
| GO:0048513—organ development | 22 | 14.10 | .001 |
| GO:0048534—hemopoietic or lymphoid organ development | 8 | 5.13 | .001 |
| GO:0065008—regulation of biologic quality | 17 | 10.90 | .002 |
| GO:0002520—immune system development | 8 | 5.13 | .002 |
| GO:0015698—inorganic anion transport | 7 | 4.49 | .002 |
| GO:0006915—apoptosis | 15 | 9.62 | .004 |
| GO:0012501—programmed cell death | 15 | 9.62 | .005 |
| GO:0030099—myeloid cell differentiation | 5 | 3.21 | .006 |
| GO:0008219—cell death | 15 | 9.62 | .007 |
| GO:0016265—death | 15 | 9.62 | .007 |
| GO:0032502—developmental process | 39 | 25.00 | .009 |
| GO_cellular component | |||
| GO:0005737—cytoplasm | 77 | 49.36 | < .001 |
| GO:0044424—intracellular part | 100 | 64.10 | < .001 |
| GO:0005622—intracellular | 104 | 66.67 | < .001 |
| GO:0005764—lysosome | 9 | 5.77 | < .001 |
| GO:0000323—lytic vacuole | 9 | 5.77 | < .001 |
| GO:0005829—cytosol | 14 | 8.97 | < .001 |
| GO:0005773—vacuole | 9 | 5.77 | < .001 |
| GO:0044445—cytosolic part | 7 | 4.49 | < .001 |
| GO:0044420—extracellular matrix part | 6 | 3.85 | .002 |
| GO:0044444—cytoplasmic part | 44 | 28.21 | .003 |
| GO_molecular function | |||
| GO:0005515—protein binding | 73 | 46.79 | < .001 |
| GO:0005198—structural molecule activity | 15 | 9.62 | .007 |
The sets of significant ENTREZ Gene IDs were converted to human ENSEMBL gene IDs using g:orth function from G:profiler (http://biit.cs.ut.ee/gprofiler/) and subjected to GO enrichment analysis using the DAVID database (http://david.abcc.ncifcrf.gov/).33 The resulting categories were considered significant with P ≤ .001 and an E value of 1.32 or higher. The gene groups with fewer than 5 members were discarded.
Quantitative RT-PCR and in situ hybridization confirm the expression of immune-related genes downstream of Spi1
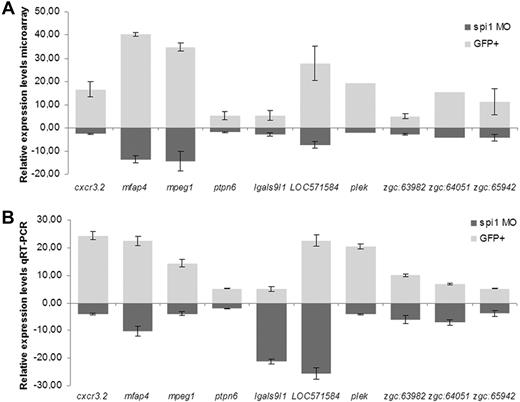
Quantitative RT-PCR was used to confirm a subset of 10 novel genes downstream of Spi1. The genes, cxcr3.2, mfap4, mpeg1, LOC571584, and plek were among the highest expressed in the GFP+ myeloid fraction, showing approximately 20- to 40-fold increased expression compared with the GFP− remaining cells of the embryo. In addition, their expression levels were strongly reduced in spi1 knockdown embryos compared with wild-type embryos (Figure 2). Gene cxcr3.2 codes for chemokine-C-X-C motif-receptor 3.2. Its potential human homolog, CXCR3, is a G protein-coupled receptor involved in chemotactic migration of leukocytes.35 Gene mfap4 (which we renamed from zgc:77076) is homologous to human MFAP4 coding for microfibril-associated glycoprotein 4, which has been implicated in inflammation through its binding capacity for surfactant proteins.36 Gene mpeg1 (formerly named zgc:66409) codes for macrophage-expressed gene 1 in human and mice. Gene LOC571584 is described as similar to macrophage receptor MARCO (macrophage receptor with collagenous structure), which belongs to the murine class A scavenger receptor family and is a part of the innate antimicrobial immune system with its ability to bind Gram-negative and Gram-positive bacteria.37 Gene plek encodes pleckstrin, which is involved in intracellular signaling and actin cytoskeleton reorganization.38
Quantitative RT-PCR validation of novel genes downstream of Spi1 identified in microarray data. (A) Relative expression levels in microarray. Values are mean ± SEM of all oligos for each gene present on the array. (B) Relative expression levels in quantitative RT-PCR. For normalization, peptidylprolyl isomerase A-like (ppial), which showed no changes over the mRNA samples used, was taken as reference. Results were analyzed using the ΔΔCt method. Gene expression values for spi1 MO (morpholino)-injected group were calculated relative to the phenol red-injected control group, whereas values for GFP+ cells were calculated relative to values of the GFP− cell fraction. Values of quantitative RT-PCR data are the mean ± SEM of 2 or 3 biologic replicates.
Quantitative RT-PCR validation of novel genes downstream of Spi1 identified in microarray data. (A) Relative expression levels in microarray. Values are mean ± SEM of all oligos for each gene present on the array. (B) Relative expression levels in quantitative RT-PCR. For normalization, peptidylprolyl isomerase A-like (ppial), which showed no changes over the mRNA samples used, was taken as reference. Results were analyzed using the ΔΔCt method. Gene expression values for spi1 MO (morpholino)-injected group were calculated relative to the phenol red-injected control group, whereas values for GFP+ cells were calculated relative to values of the GFP− cell fraction. Values of quantitative RT-PCR data are the mean ± SEM of 2 or 3 biologic replicates.
Similarly, our quantitative RT-PCR corroborated the differential expression of genes ptpn6, lgals9l1, zgc:63982, zgc:64051, and zgc:65942 (Figure 2B) as found in the microarray. Gene ptpn6 encodes protein tyrosine phosphatase, nonreceptor type 6, also known as shp1. Human PTPN6 is implicated in negative regulation of signaling from immune cell receptors.8 Gene lgals9l1 encodes a galactoside-binding, soluble lectin. A human homolog of gene zgc:63982, ALOX5AP, is required for synthesis of leukotrienes, implicated in various types of inflammatory responses.39 CD53− leukocyte surface antigen, the homolog of zebrafish Zgc:64051, is a cell surface glycoprotein whose familial deficiency has been linked to an immunodeficiency associated with infectious diseases.40 The homolog of gene zgc:65942, GRAP2, encodes a member of the GRB2/Sem5/Drk family involved in leukocyte-specific signaling.41
Further validation of 4 of the novel Spi1-dependent genes was performed by whole-mount in situ hybridization. In wild-type embryos at 28 hpf, genes mfap4, mpeg1, and ptpn6 were expressed in cells localizing anteriorly over the yolk and in the posterior blood island (Figure 3A,C,E,G,I,K). This expression pattern is very similar to that of spi1 or the pan-leukocytic marker lcp1 and typical for myeloid cells. Expression of cxcr3.2 could only be detected by fluorescent in situ hybridization but also showed a similar pattern (Figure 3M,O). The knockdown of spi1 resulted in a total loss of cells expressing genes mfap4, mpeg1, ptpn6, and cxcr3.2 (Figure 3B,D,F,H,J,L,N,P), demonstrating that Spi1 is required for myeloid-specific expression of these genes. All 4 genes as well as 3 of the myeloid-specific genes confirmed by quantitative RT-PCR (lgals9l1, LOC571584, and plek) contain Spi1 consensus binding sites in their upstream promoter regions, suggesting that they might be direct targets of Spi1.
Expression of mfap4, mpeg1, ptpn6, and cxcr3.2 is dependent on Spi1 transcription factor. (A-L) Whole-mount in situ hybridization. Wild-type expression of genes mfap4 (A,C), mpeg1 (E,G), and ptpn6 (I,K) as shown in whole embryos and enlarged tail regions shows a pattern typical for myeloid cells. Expression of genes mfap4 (B,D), mpeg1 (F,H), and ptpn6 (J,L) is absent in spi1 knockdown embryos. Images were taken with a Leica M165C stereomicroscope equipped with DFC420 camera. Composite images of different focal planes were generated using Adobe Photoshop. (M-P) Single fluorescent in situ hybridization using TSA Cy3 Plus detection. Wild-type expression of gene cxcr3.2, as shown in the tail region (M) and in the magnified region of the blood island (O), shows a pattern typical for myeloid cells. Expression of gene cxcr3.2 (N,P) is absent in spi1 knockdown embryos. Images were taken with Leica TCS SPE confocal microscope, using the 532-nm laser line for scanning of signal in the red channel. The HC PL Fluotar 10.0×/0.30 dry objective was used. The images were processed in ImageJ Version 1.43 software (National Institute of Mental Health). Bars represent 100 μm (M-N) and 60 μm (O-P).
Expression of mfap4, mpeg1, ptpn6, and cxcr3.2 is dependent on Spi1 transcription factor. (A-L) Whole-mount in situ hybridization. Wild-type expression of genes mfap4 (A,C), mpeg1 (E,G), and ptpn6 (I,K) as shown in whole embryos and enlarged tail regions shows a pattern typical for myeloid cells. Expression of genes mfap4 (B,D), mpeg1 (F,H), and ptpn6 (J,L) is absent in spi1 knockdown embryos. Images were taken with a Leica M165C stereomicroscope equipped with DFC420 camera. Composite images of different focal planes were generated using Adobe Photoshop. (M-P) Single fluorescent in situ hybridization using TSA Cy3 Plus detection. Wild-type expression of gene cxcr3.2, as shown in the tail region (M) and in the magnified region of the blood island (O), shows a pattern typical for myeloid cells. Expression of gene cxcr3.2 (N,P) is absent in spi1 knockdown embryos. Images were taken with Leica TCS SPE confocal microscope, using the 532-nm laser line for scanning of signal in the red channel. The HC PL Fluotar 10.0×/0.30 dry objective was used. The images were processed in ImageJ Version 1.43 software (National Institute of Mental Health). Bars represent 100 μm (M-N) and 60 μm (O-P).
Cell-type specificity of genes cxcr3.2, mpeg1, mfap4, and ptpn6 in zebrafish embryos
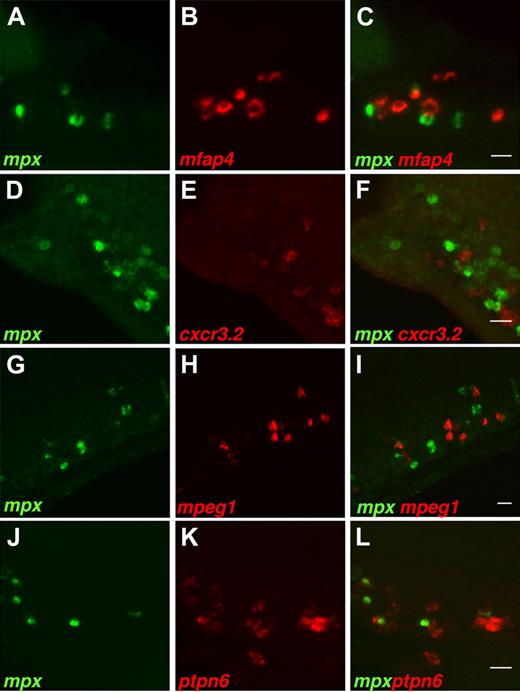
Double fluorescent in situ hybridization was used to determine the identity of the myeloid cells expressing 4 novel Spi1-regulated genes found in this study, namely, cxcr3.2, mfap4, mpeg1, and ptpn6. Colocalization studies were performed using csf1r as a marker for macrophages and mpx as a marker for a distinct myeloid population at 28 hpf and for differentiated neutrophils at 2 dpf. The expression of these 2 genes was previously shown to have no overlap at 28 hpf and at later stages.21,23
In 28 hpf wild-type embryos, the expression of mfap4 colocalized completely with that of csf1r (Figure 4A-C), although no overlap was observed with expression of mpx (Figure 5A-C). Similarly, expression of genes cxcr3.2, mpeg1, and ptpn6 did not overlap with expression of mpx (Figure 5D-L), and their expression overlapped almost completely (> 90%) with expression of macrophage marker csf1r (Figure 4D-L). The minor difference with the csf1r pattern may be the result of the differences of expression levels between the genes. The important observation is that expression of mpx and the 4 genes investigated is mutually exclusive in myeloid cells in 28-hpf embryos. In 2-dpf embryos, the same results of lack of colocalization with mpx and almost complete overlap with csf1r expression were obtained for genes mfap4 and mpeg1. Gene ptpn6 showed a slight overlap with gene mpx, while we were not able to detect the fluorescent in situ signal for gene cxcr3.2 at 2 dpf (supplemental Figure 3).
Expression of genes mfap4, cxcr3.2, mpeg1, and ptpn6 colocalizes with expression of macrophage-specific marker csf1r. Double fluorescent in situ with the csf1r gene. Probes for mRNA are used as indicated in the bottom left corner of each image. Expression of genes, mfap4 (A-C), cxcr3.2 (D-F), mpeg1 (G-I), and ptpn6 (J-L) overlaps with expression of macrophage-specific marker csf1r. Images show summed Z-stacks of consecutive confocal images through the posterior blood island of 28-hpf embryos (lateral view). A Zeiss axioplan microscope with Bio-Rad MRC1024ES scanhead was used for confocal imaging. Signals in the green and red channels were scanned sequentially using, respectively, the 488-nm laser line with 522 DF32 filter for detection of emitted light, and the 568-nm laser line with 605 DF32 filter for detection of emitted light: 10×/0.30 NA and 20×/0.50 NA Plan-Neofluar objectives were used. Images were processed in ImageJ Version 1.43 software and show (from left to right) the signal in the green channel, the signal in the red channel, and the merged signals. (A-F) Bars represent 20 μm. (G-L) Bars represent 60 μm.
Expression of genes mfap4, cxcr3.2, mpeg1, and ptpn6 colocalizes with expression of macrophage-specific marker csf1r. Double fluorescent in situ with the csf1r gene. Probes for mRNA are used as indicated in the bottom left corner of each image. Expression of genes, mfap4 (A-C), cxcr3.2 (D-F), mpeg1 (G-I), and ptpn6 (J-L) overlaps with expression of macrophage-specific marker csf1r. Images show summed Z-stacks of consecutive confocal images through the posterior blood island of 28-hpf embryos (lateral view). A Zeiss axioplan microscope with Bio-Rad MRC1024ES scanhead was used for confocal imaging. Signals in the green and red channels were scanned sequentially using, respectively, the 488-nm laser line with 522 DF32 filter for detection of emitted light, and the 568-nm laser line with 605 DF32 filter for detection of emitted light: 10×/0.30 NA and 20×/0.50 NA Plan-Neofluar objectives were used. Images were processed in ImageJ Version 1.43 software and show (from left to right) the signal in the green channel, the signal in the red channel, and the merged signals. (A-F) Bars represent 20 μm. (G-L) Bars represent 60 μm.
Expression of genes mfap4, cxcr3.2, mpeg1, and ptpn6 does not colocalize with expression of neutrophil-specific marker mpx. Double fluorescent in situ with the mpx gene. Probes for mRNA are used as indicated in the bottom left corner of each image. Expression of genes, mfap4 (A-C), cxcr3.2 (D-F), mpeg1 (G-I), and ptpn6 (J-L) does not colocalize with expression of the mpx gene. Images show summed Z-stacks of consecutive confocal images through the posterior blood island of 28-hpf embryos (lateral view). In addition, no colocalization was observed in the head and yolk regions or in embryos at 48 hpf when mpx expression marks differentiated neutrophils (data not shown). Imaging conditions are as in Figure 4. Bars represent 20 μm.
Expression of genes mfap4, cxcr3.2, mpeg1, and ptpn6 does not colocalize with expression of neutrophil-specific marker mpx. Double fluorescent in situ with the mpx gene. Probes for mRNA are used as indicated in the bottom left corner of each image. Expression of genes, mfap4 (A-C), cxcr3.2 (D-F), mpeg1 (G-I), and ptpn6 (J-L) does not colocalize with expression of the mpx gene. Images show summed Z-stacks of consecutive confocal images through the posterior blood island of 28-hpf embryos (lateral view). In addition, no colocalization was observed in the head and yolk regions or in embryos at 48 hpf when mpx expression marks differentiated neutrophils (data not shown). Imaging conditions are as in Figure 4. Bars represent 20 μm.
Considering that Spi1 controls development of both the myeloid and the lymphoid lineage, we investigated whether our genes of interest are also expressed in the thymus of developing larvae. As cxcr3.2 expression was below the detection limit of in situ hybridization at 5 dpf, the timing of lymphoid expression could not be determined for this gene. Gene ptpn6 showed clear expression in the thymus at 5 dpf (supplemental Figure 4), which was confirmed by colocalization with the rag1 marker (supplemental Figure 5). Genes mfap4 and mpeg1 were expressed at lower levels at 5 dpf than at earlier stages but still displayed a myeloid-specific expression pattern and were not expressed in the thymus (supplemental Figure 4).
Based on these data, we conclude that genes mfap4 and mpeg1 are the most specific novel markers for macrophage-specific expression in zebrafish embryos.
Transcriptional regulation of hematopoiesis on spi1 knockdown and in early myeloid cells
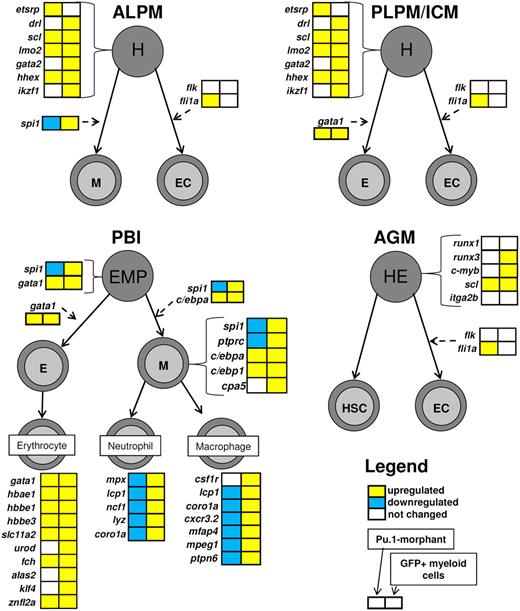
Transcriptional regulation of hematopoiesis is a well-conserved process between zebrafish and mammals, and during embryogenesis a wave of primitive hematopoiesis precedes definitive hematopoiesis in vertebrates.42 We constructed a pathway map of primitive and definitive hematopoiesis in early zebrafish embryos, on which we superimposed the microarray data obtained from 28 hpf spi1 knockdown embryos and GFP+ myeloid cells from embryos of spi1:GFP transgenic zebrafish (Figure 6; supplemental Figure 6).
Hematopoiesis pathway regulation on spi1 knockdown and in myeloid cells of a developing embryo at 28 hpf. Expression profiles of the spi1 knockdown embryos and GFP+ myeloid fractions at 28 hpf are simultaneously mapped on a schematic representation of hematopoiesis in zebrafish embryos based on the current knowledge of this process19,42,43 and including the novel macrophage-specific markers identified in this study. The primitive wave of hematopoiesis in zebrafish initiates at 2 distinct sites. At the anterior lateral plate mesoderm (ALPM), hemangioblasts (H) differentiate into myeloid cells (M) or endothelial cells (EC). At the second site, the posterior lateral plate mesoderm (PLPM), which later converts into the intermediate cell mass (ICM), hemangioblasts give rise to erythroid cells (E) and also ECs. In a transient wave of hematopoiesis occurring in posterior blood island (PBI), the EMPs, which are the first multipotent hematopoietic progenitor cells, differentiate into either myeloid or erythroid cells. The definitive wave of hematopoiesis starts in the aorta-gonad-mesonephros (AGM), where hemogenic endothelium (HE) gives rise to hematopoietic stem cells (HSC) and ECs. Gene boxes are color-coded with the microarray data from spi1 knockdown on the left and GFP+ myeloid data on the right side. Expression of gene csf1r was determined by quantitative RT-PCR as the microarray contained only one probe for that gene, and it did not return a significant result. Up-regulation is indicated in yellow, down-regulation in blue, and no significant change in white. Genes marked by asterisks are the genes that were both down-regulated in spi1 knockdown embryos and enriched in the spi1-GFP myeloid cell fraction, and are the focus of this study. It should be noted that it is currently unknown to what extent the microarray expression data from 28-hpf embryos can be extrapolated to later waves of hematopoiesis. Macrophage specificity of cxcr3.2 in embryos is based on colocalization data at 28 hpf, and macrophage specificity of mpeg1, mfap4, and ptpn6 is based on colocalization studies at 28 and 48 hpf.
Hematopoiesis pathway regulation on spi1 knockdown and in myeloid cells of a developing embryo at 28 hpf. Expression profiles of the spi1 knockdown embryos and GFP+ myeloid fractions at 28 hpf are simultaneously mapped on a schematic representation of hematopoiesis in zebrafish embryos based on the current knowledge of this process19,42,43 and including the novel macrophage-specific markers identified in this study. The primitive wave of hematopoiesis in zebrafish initiates at 2 distinct sites. At the anterior lateral plate mesoderm (ALPM), hemangioblasts (H) differentiate into myeloid cells (M) or endothelial cells (EC). At the second site, the posterior lateral plate mesoderm (PLPM), which later converts into the intermediate cell mass (ICM), hemangioblasts give rise to erythroid cells (E) and also ECs. In a transient wave of hematopoiesis occurring in posterior blood island (PBI), the EMPs, which are the first multipotent hematopoietic progenitor cells, differentiate into either myeloid or erythroid cells. The definitive wave of hematopoiesis starts in the aorta-gonad-mesonephros (AGM), where hemogenic endothelium (HE) gives rise to hematopoietic stem cells (HSC) and ECs. Gene boxes are color-coded with the microarray data from spi1 knockdown on the left and GFP+ myeloid data on the right side. Expression of gene csf1r was determined by quantitative RT-PCR as the microarray contained only one probe for that gene, and it did not return a significant result. Up-regulation is indicated in yellow, down-regulation in blue, and no significant change in white. Genes marked by asterisks are the genes that were both down-regulated in spi1 knockdown embryos and enriched in the spi1-GFP myeloid cell fraction, and are the focus of this study. It should be noted that it is currently unknown to what extent the microarray expression data from 28-hpf embryos can be extrapolated to later waves of hematopoiesis. Macrophage specificity of cxcr3.2 in embryos is based on colocalization data at 28 hpf, and macrophage specificity of mpeg1, mfap4, and ptpn6 is based on colocalization studies at 28 and 48 hpf.
The hematopoiesis map reveals increased expression of most early hematopoietic marker genes (drl, scl, lmo2, gata2, and hhex) in the GFP+ myeloid cells. We found 3 genes (c-myb, runx3, and ikzf1/ikaros) involved in differentiation and proliferation of hematopoietic progenitor cells to be enriched in the GFP+ myeloid cells. The observed increase of expression of transcription factor genes c/ebpa and c/ebp1 in GFP+ cells is concomitant with their role in myeloid development,21 similar to genes spi1 and ptprc (CD45). The increased expression levels of gene gata1 and other genes characteristic of erythrocytes (α-globin, slc11a2, urod, fch, alas2, klf4, and znfl2a) are consistent with the expression of spi1 in both the committed myeloid cells and the EMPs.20
In the spi1 knockdown embryos, we found 5 early hematopoietic marker genes (etsrp, scl, lmo2, hhex, and fli1a) to be weakly up-regulated, possibly as a transcriptional compensatory mechanism against spi1 deficiency. In addition, the myeloid-expressed c/ebp1 and c/ebpa genes21 were both up-regulated in spi1 knockdown. Analysis of the transcriptional regulation of the hematopoiesis pathway in spi1 knockdown embryos also revealed activation of expression of several genes specific to the erythrocyte population (gata1, hbae1, hbbe1, hbbe3, slc11a2, fch, and znfl2a), consistent with earlier observations that the loss of spi1 redirects myeloid progenitor cells into erythroid precursors.20
Finally, genes specific for the myeloid lineage at this embryonic stage, including pan-leukocytic markers (lcp1, coro1a), early macrophage markers (csf1r and as demonstrated in this study, mpeg1, cxcr3.2, mfap4, and ptpn6), and neutrophil markers (mpx, ncf1, and lyz), consistently showed enrichment in expression in GFP+ myeloid cells, although they were all but csf1r repressed in spi1 knockdown.
Cxcr3.2 function in the response of macrophages to local bacterial infection
The zebrafish embryonic myeloid cells, macrophages, and neutrophils mount a quick and effective defense response against wounding or invading pathogens. Although macrophage migration toward sites of bacterial infection and their phagocytotic activity have been documented in zebrafish embryos using various infection models, the receptors involved in macrophage migration to sites of infection are still unknown.
Five members of the chemokine receptor family, namely, cxcr3.1, cxcr3.2, cxcr4a, cxcr4b, and cxcr7b, were present on our microarray platform. Only one, cxcr3.2, was found to be specific for early myeloid cells and downstream of Spi1. The remaining 4 genes did not show significant change in expression in either the spi1 knockdown or the GFP+ myeloid cell fraction. Three other members of the family were tested by quantitative RT-PCR on mRNA samples used for the microarray. Genes cxcr1, cxcr2, and cxcr5 showed enriched expression in GFP+ myeloid cells compared with the GFP cell fraction; however, their expression levels were independent of Spi1 (supplemental Figure 7).
Our discovery of a member of the chemokine receptor family as an early macrophage-specific downstream target of Spi1 prompted us to investigate the role of cxcr3.2 in macrophage response to a bacterial cue.
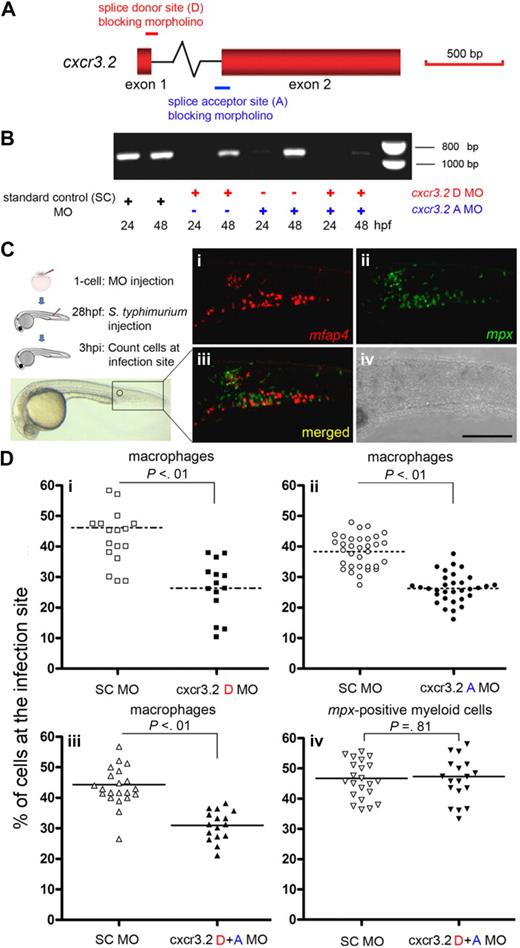
Splice donor and acceptor morpholinos, targeting the single intron of the cxcr3.2 gene, were used to obtain cxcr3.2 knockdown embryos (Figure 7A). RT-PCR on embryos injected either with cxcr3.2 donor or acceptor or combination of both morpholinos showed a significant depletion of normal cxcr3.2 transcript compared with the control embryos (Figure 7B).
Knockdown analysis of cxcr3.2 using splice donor and acceptor morpholinos revealed a significantly reduced accumulation of macrophages at sites of bacterial infection. (A) Schematic of the cxcr3.2 gene and MO targeting sites. Red boxes represent the exons in the cxcr3.2 gene. The intron (∼ 4 kb) is not drawn to scale. Red and blue bars represent the splice donor (D) and acceptor (A) MOs, which bind to the pre-mRNA on the intron/exon boundaries, inducing an insertion of the intron into the final transcripts. (B) RT-PCR detection of cxcr3.2 transcripts on MO knockdown. Altered splicing induced by D or A MOs resulted in an effective knockdown of the original cxcr3.2 mRNA at 24 hpf, whereas the combination of D + A MOs extended the knockdown effect until 48 hpf. The altered splice product with insertion of the approximately 4 kb intron is not amplified by RT-PCR. (C) Schematic of the experimental setup and representative images showing the attraction of macrophages and neutrophils to a local infection site. S typhimurium bacteria were injected into the embryo tail muscle at 28 hpf followed by a 3-hour incubation. Leukocytes were visualized at 3 hpi after double fluorescent in situ hybridization: (i) mfap4+ macrophages; (ii) mpx+ myeloid cells; (iii) merged; and (iv) bright-field image; bar represents 200 μm. (D) Migration of innate immune cells after MO knockdown. Accumulation of macrophages marked by mfap4 (i-iii) and accumulation of the nonoverlapping mpx+ myeloid cell population (iv) were quantified by determining the percentages of cells accumulated at the infection site with respect to the total number of cells in the tail. Data were collected from 3 independent experimental setups, in which the cxcr3.2 D MO (i), A MO (ii), or D + A MOs (iii-iv) were tested versus the Standard Control (SC) MO. Each data point represents an individual embryo, and each graph represents the combined results of 2 replicate experiments with the indicated MO combination. Lines indicate the mean, and P values indicate the level of statistical significance by Student t test. The knockdown of cxcr3.2 did not have a significant effect on migration of either macrophages or mpx+ myeloid cells in response to sterile injury (data not shown).
Knockdown analysis of cxcr3.2 using splice donor and acceptor morpholinos revealed a significantly reduced accumulation of macrophages at sites of bacterial infection. (A) Schematic of the cxcr3.2 gene and MO targeting sites. Red boxes represent the exons in the cxcr3.2 gene. The intron (∼ 4 kb) is not drawn to scale. Red and blue bars represent the splice donor (D) and acceptor (A) MOs, which bind to the pre-mRNA on the intron/exon boundaries, inducing an insertion of the intron into the final transcripts. (B) RT-PCR detection of cxcr3.2 transcripts on MO knockdown. Altered splicing induced by D or A MOs resulted in an effective knockdown of the original cxcr3.2 mRNA at 24 hpf, whereas the combination of D + A MOs extended the knockdown effect until 48 hpf. The altered splice product with insertion of the approximately 4 kb intron is not amplified by RT-PCR. (C) Schematic of the experimental setup and representative images showing the attraction of macrophages and neutrophils to a local infection site. S typhimurium bacteria were injected into the embryo tail muscle at 28 hpf followed by a 3-hour incubation. Leukocytes were visualized at 3 hpi after double fluorescent in situ hybridization: (i) mfap4+ macrophages; (ii) mpx+ myeloid cells; (iii) merged; and (iv) bright-field image; bar represents 200 μm. (D) Migration of innate immune cells after MO knockdown. Accumulation of macrophages marked by mfap4 (i-iii) and accumulation of the nonoverlapping mpx+ myeloid cell population (iv) were quantified by determining the percentages of cells accumulated at the infection site with respect to the total number of cells in the tail. Data were collected from 3 independent experimental setups, in which the cxcr3.2 D MO (i), A MO (ii), or D + A MOs (iii-iv) were tested versus the Standard Control (SC) MO. Each data point represents an individual embryo, and each graph represents the combined results of 2 replicate experiments with the indicated MO combination. Lines indicate the mean, and P values indicate the level of statistical significance by Student t test. The knockdown of cxcr3.2 did not have a significant effect on migration of either macrophages or mpx+ myeloid cells in response to sterile injury (data not shown).
To examine the role of cxcr3.2 in macrophage migration toward a local bacterial infection, S typhimurium was injected in the tail muscle of 28-hpf cxcr3.2 knockdown or control embryos (Figure 7C; supplemental Figure 8). In wild-type embryos at 3 hours post injection (hpi) a clear migration of macrophages was observed as detected by fluorescent in situ with mfap4, identified as a robust macrophage marker gene in this study. No significant differences were observed in the general number of mfap4+ cells in the tail region between cxcr3.2 knockdown and control embryos (data not shown). The number of macrophages that accumulated at the infection site at 3 hpi was significantly lower in the cxcr3.2 knockdown embryos compared with the control (Figure 7Di-iii). Interestingly, the migration of the distinct population of myeloid cells that expresses the neutrophil specific marker mpx was not affected by cxcr3.2 knockdown (Figure 7Div). These results demonstrate that cxcr3.2 is necessary for normal migration of macrophages, but not mpx-expressing cells, to the sites of bacterial infection and indicate a function for cxcr3.2 in the innate immune response of zebrafish embryos.
Discussion
The Spi1/Pu.1 transcription factor plays a crucial role in myeloid and lymphoid cell development and is conserved between human and zebrafish. The temporal separation of myeloid and lymphoid development during zebrafish embryogenesis offers a unique opportunity to identify the specific gene group expressed in early myeloid cells and dependent on Spi1.43 Our microarray-based analysis of this gene group gives insight into the transcriptional program of developing myeloid cells and resulted in the identification of specific markers of the macrophage lineage, which have thus far been lacking in zebrafish embryos. Furthermore, it resulted in the discovery of an early macrophage-specific chemokine receptor gene, which is the first shown to be involved in migration of macrophages during the innate immune response of zebrafish embryos to bacterial infection.
To identify the putative gene group dependent on Spi1 and expressed in early embryonic myeloid cells, we used a 2-step microarray approach, where we determined the overlapping group between genes enriched in GFP+ myeloid cells isolated from 28-hpf embryos of spi1:GFP transgenic zebrafish and genes down-regulated in embryos that lack myeloid cells resulting from knockdown of Spi1. The 2-step approach was applied to lower the rate of false positive discovery. The overlap group consisted of 249 genes. Six of these were known previously to be dependent for their expression on active Spi1 in the zebrafish embryo,20 thus validating our approach. These genes included spi1 itself, which is known to have an autoregulatory function.13 In addition to down-regulation of known myeloid cell markers and immune-related genes, the transcriptional regulation of hematopoiesis in spi1 knockdown embryos resulted in the induction of genes involved in early steps of hematopoiesis and genes regulating erythroid development (eg, gata1), suggesting that a transcriptional compensatory mechanism is initiated in the absence of spi1. These data are consistent with the finding that gata1 drives erythrocyte differentiation by antagonizing spi1 activity in both zebrafish and mammalian models.6,7,20 GO analysis of the Spi1-dependent gene group identified by our 2-step microarray approach showed enrichment of genes functionally linked to the innate immune response. In addition, approximately one-third of the genes have not yet been functionally characterized either in zebrafish or in humans; therefore, they represent novel genes potentially involved in myeloid cell development or function, which are of great interest for further research. Presently, we do not know which genes identified in our study are direct targets of Spi1; however, several contain the consensus Spi1 binding sites44 in the upstream promoter region or represent validated target genes in mammalian systems. This includes genes, such as ctss and rgs18, recently shown to be part of a PU.1 regulatory network in cultured murine macrophages,16 suggesting that these genes are also important during early myeloid development in the embryo model. Further studies using ChIP-microarray or ChIP-Seq may assist in discovering which of the genes identified here are indeed the direct targets of Spi1. In addition, we found that csf1r, which is also part of the PU.1 regulatory network in murine macrophages, was enriched in early myeloid cells isolated from zebrafish embryos. This gene was not selected in our strict 2-step microarray approach, as Spi1-dependent down-regulation was not detected, probably because of Spi1-independent expression of csf1r in neural crest cells of developing embryos.26 Further insight into the precise gene groups under control of Spi1 or other hematopoietic transcription factors at different stages of myeloid and lymphoid development might be obtained by extending our approach of knockdown analysis in combination with fluorescent marker lines of other leukocyte cell types.
In situ hybridization results for genes cxcr3.2, mfap4, mpeg1, and ptpn6 confirmed that all 4 genes are expressed in embryonic myeloid cells in an Spi1-dependent manner as their expression was lost in spi1 morphants. In addition, quantitative RT-PCR results validated the microarray data for 7 new myeloid-specific and Spi1-dependent genes with suggested functions in the immune response. By double fluorescent in situ colocalization experiments, we determined that genes cxcr3.2, mfap4, mpeg1, and ptpn6 all were macrophage specific in early zebrafish embryos before the onset of adaptive immunity, as their expression did not overlap with the expression of neutrophil-specific marker mpx, although they showed clear colocalization with macrophage marker csf1r. Although no differentiated neutrophils are present at 28 hpf in the developing embryo, we clearly show that there are 2 distinct myeloid cell populations present at this stage, where one expresses markers, such as csf1r, cxcr3.2, mfap4, mpeg1, and ptpn6, whereas the second cell population is characterized by expression of mpx. Gene ptpn6 showed some overlap with mpx activity at 2 dpf, suggesting that this gene is also expressed in a subset of neutrophils from this stage. The same may be true for cxcr3.2, of which we could not detect in situ expression at 2 dpf. However, our preliminary results of fluorescent reporter gene expression driven by the cxcr3.2 promoter suggest that this gene is also expressed in a subset of mpx-expressing cells at later stages of embryo development (M. van der Vaart, unpublished results, January 2010). At 5 dpf, gene ptpn6 was also expressed in the thymus consistent with its murine homolog, which is found in cells of other hematopoietic lineages and is known as a direct target of Spi1 functioning as a negative regulator of the immune response.8,45 Expression of genes mfap4 and mpeg1 was not observed in the larval thymus at 5 dpf but still remained myeloid specific. Compared with the only known macrophage-specific marker csf1r, these markers show more robust expression, and they are not expressed in neural crest cells. Although expression of mfap4 in mammalian macrophages is not known, the perforin-like protein encoded by murine mpeg1 is expressed in mature macrophages and prion-infected brain cells.46 Gene mfap4 encodes a protein with a fibrinogen receptor-like domain. A role in inflammation for human MFAP4 was suggested based on its interaction with D-type lectin surfactant proteins,36 but it might also have a direct function in pathogen recognition similar to the fibrinogen-related proteins of invertebrates.47 Whereas GFP transgenic lines marking the early myeloid, neutrophil, and lymphoid lineages are already available to facilitate real-time analyses in the zebrafish model, macrophage-specific marker lines are in great demand. The embryonic macrophage markers downstream of Spi1 discovered here can greatly facilitate future research into the role of macrophages in the innate immune response by GFP transgenic lines that are currently in development.
To initiate the investigation of the novel Spi1-dependent genes in macrophage function, we chose to study gene cxcr3.2 in the context of the well-established zebrafish embryo model of bacterial infection with S typhimurium.34,48 We found that cxcr3.2 is the only zebrafish CXC chemokine receptor gene that is both Spi1-dependent and expressed specifically in early myeloid cells. The local infection of zebrafish embryos with S typhimurium in the cxcr3.2 knockdown resulted in the reduced migration of macrophages, although the total number of macrophages was not altered in the absence of cxcr3.2. These results led us to conclude that cxcr3.2 plays a role in macrophage migration toward bacterial cues. As cxcr3.2 knockdown did not completely abolish macrophage migration, it is probable that other currently unknown receptors are also involved in the response to S typhimurium infection. cxcr3.2 is most similar to human CXCR3 and CXCR5, which are expressed predominantly on T and B lymphocytes. Consistent with the role of cxcr3.2 in cell migration during bacterial infection in zebrafish, it has been shown that CXCR3-positive cells are recruited to sites of Mycobacterium tuberculosis infection in primates.49 To our knowledge, the possible function of CXCR3/CXCR5 receptors in the embryonic immune system of vertebrates has not been addressed, and our study is the first that directly implicates a CXCR3/5 homolog in the innate immune response to bacterial infection. However, it has been shown that CXCR3 expression can be induced on CD34+ hematopoietic progenitors from human cord blood by stimulation with granulocyte-macrophage colony-stimulating factor and that chemotactic activity of these progenitor cells could subsequently be triggered by CXC chemokines.50 These in vitro studies suggest that CXCR3 may also have an innate immune function in mammalians, as we have shown here in vivo for its homolog in the zebrafish embryo model. In conclusion, this study contributes to our understanding of the Spi1-dependent development of myeloid cells and has identified novel early macrophage-specific marker genes for zebrafish, including a chemokine receptor gene with a pivotal role in the migration of macrophages during the innate immune response.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Francesca Peri for the gift of the Pu1-lynEGFP transgenic line, Ulrike Nehrdich, Davy de Witt, and Karen Bosma for fish care, Zakia Kanwal and Michiel van der Vaart for their help with in situ experiments, Gerda Lamers for assistance with microscopy, Menno van der Hoorn for help with FACS sorting, and David Traver for many helpful suggestions.
This work was supported by the European Commission 6th framework project ZF-TOOLS (LSHG-2006-037220) and the Smart Mix Program of The Netherlands Ministry of Economic Affairs and the Ministry of Education, Culture and Science.
Authorship
Contribution: A.Z. and C.C. designed and conducted experiments and wrote the manuscript; O.W.S. and E.L.B. contributed to the experiments; and H.P.S. and A.H.M. designed and supervised the study and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Annemarie H. Meijer, Institute of Biology, Leiden University, Einsteinweg 55, 2333 CC Leiden, The Netherlands; e-mail a.h.meijer@biology.leidenuniv.nl.
References
Author notes
A.Z. and C.C. contributed equally to this study.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal