Enforced expression of the homeobox transcription factor HOXB4 has been shown to enhance hematopoietic stem cell self-renewal and expansion ex vivo and in vivo. To investigate the downstream targets of HOXB4 in hematopoietic progenitor cells, HOXB4 was constitutively overexpressed in the primitive hematopoietic progenitor cell line EML. Two genome-wide analytical techniques were used: RNA expression profiling using microarrays and chromatin immunoprecipitation (ChIP)–chip. RNA expression profiling revealed that 465 gene transcripts were differentially expressed in KLS (c-Kit+, Lin−, Sca-1+)-EML cells that overexpressed HOXB4 (KLS-EML-HOXB4) compared with control KLS-EML cells that were transduced with vector alone. In particular, erythroid-specific gene transcripts were observed to be highly down-regulated in KLS-EML-HOXB4 cells. ChIP-chip analysis revealed that the promoter region for 1910 genes, such as CD34, Sox4, and B220, were occupied by HOXB4 in KLS-EML-HOXB4 cells. Side-by-side comparison of the ChIP-chip and RNA expression profiling datasets provided correlative information and identified Gp49a and Laptm4b as candidate “stemness-related” genes. Both genes were highly ranked in both dataset lists and have been previously shown to be preferentially expressed in hematopoietic stem cells and down-regulated in mature hematopoietic cells, thus making them attractive candidates for future functional studies in hematopoietic cells.
Introduction
Hematopoietic stem cells (HSCs) play a very important role in the establishment as well as lifelong maintenance of hematopoiesis. A critical property of HSCs is the ability not only to differentiate into cells that make up all of the formed elements of the blood, but also to self-renew and constitute a viable multipotent stem cell compartment. HSCs are by far the most well-studied adult stem cell population; but despite decades of research on this topic, the underlying process of HSC self-renewal is still not well understood. The relatively low abundance of HSCs (≤ 0.001% of nucleated bone marrow cells or ∼ 5000 per mouse)1 and the inability to effectively expand HSCs in vitro have made it difficult to study HSC self-renewal.
The ability to expand HSCs in vitro has numerous potential clinical applications and could revolutionize the way many types of blood diseases are treated.2,3 Identification of the extrinsic and intrinsic factors that control self-renewal and lineage commitment will be instrumental to the advancement of such clinical applications. Studies have shown that self-renewal is possible in vitro, but most culture conditions still lead to a net loss of HSCs, indicating that differentiation is favored over self-renewal.4 Expansion of HSCs has been achieved by culturing HSCs in the presence of high concentrations of early acting cytokines and/or by adding to various culture systems growth factors, such as bone morphogenetic protein, which plays an important role in embryonic ventral mesoderm development and in promoting hematopoiesis from mesodermal tissue, as well as sonic hedgehog, a key factor in determining embryonic segment polarity and patterning.5,–7 However, these expansions of HSCs were modest (up to 3-fold), and the relative expansion seemed to be more the result of HSC survival than to stimulation of self-renewal.
A search for potential genes regulating HSC self-renewal and differentiation revealed that members of the homoebox (Hox) gene family of transcription factors acted as potentially important regulators of hematopoiesis.8 Hox genes are DNA-binding transcription factors that were first discovered in Drosophila and act as transcriptional activators or inhibitors.9 They are characterized by a highly conserved 61-amino acid homeobox domain, which forms a helix-turn-helix structure that binds DNA.10 Hox genes are well conserved across species and exhibit a site- and time-specific pattern of expression during embryonic development, which correlates with their relative chromosomal position (spatial and temporal colinearity). Although Hox genes were first shown to be important in embryonic development, subsequent experiments have shown that Hox genes also play lineage-specific roles throughout the life of various somatic tissues, including the hematopoietic system.11 It was observed that retroviral-mediated overexpression of HOXB4 stimulated HSC self-renewal and expansion (1000-fold relative to controls) without impairing normal differentiation or causing the cells to become transformed.12,–14 Retroviral-mediated overexpression of HOXB4 was also shown to promote HSC self-renewal ex vivo. Such an expansion of HSCs without impairing their differentiation had not been previously observed. As a result, HOXB4 function has become a prominent research topic among investigators studying HSC self-renewal because the mechanisms by which HOXB4 mediates HSC self-renewal are largely unknown.
More recent experiments have provided additional evidence for the importance of HOXB4 in the process of early hematopoietic stem cell development. It was demonstrated that, by overexpressing HOXB4 in mouse embryonic stem (mES) cells, these cells could then be differentiated in vitro to obtain mES cell-derived hematopoietic progenitor cells (mES-HPCs) capable of long-term engraftment in lethally irradiated mice.15 This phenomenon had not previously been observed and provided an important breakthrough that has allowed researchers to more effectively study the early processes of hematopoietic development.16,17 Despite these advances clearly showing that HOXB4 is important for early hematopoietic development, there have been very few documented reports identifying direct downstream targets of HOXB4, such as Tnfrsf1b in mES-HPCs and Rap1 and Irx5 in Xenopus embryonic development.17,–19
It is thought that the cellular context in which HOXB4 is expressed greatly determines which genes and pathways are up-regulated and/or down-regulated. This is because HOXB4 is known to form complexes by binding to other proteins, such as PBX1, and it is thought that these types of interactions specify where in the genome HOXB4 binds.20,21 Different cell types presumably have different binding partners for HOXB4; hence, the genes that HOXB4 up-regulates and/or down-regulates are probably different according to cell type. Because HOXB4 promotes HSC self-renewal, it is important to study HOXB4 activity in the most primitive hematopoietic progenitor cell populations.
To identify downstream targets of HOXB4 in primitive hematopoietic cells, we overexpressed HOXB4 in the primitive hematopoietic cell line EML.22 We carried out RNA expression profiling and chromatin immunoprecipitation (ChIP)–chip microarray hybridization experiments using the most primitive subpopulation of EML cells, the KLS (c-Kit+, Lin−, Sca-1+) cells (KLS-EML cells). We observed that 465 gene transcripts were differentially expressed at least 2-fold in KLS-EML cells that overexpressed HOXB4 (KLS-EML-HOXB4 cells) compared with control cells that were transduced with the GFP-expressing vector only (KLS-EML-GFP cells). Differential occupancy of different gene promoters by HOXB4 was also demonstrated in the HOXB4-overexpressing cells, consistent with these genes being direct downstream targets of HOXB4. These genes included lineage-specific genes, such as the B220 gene and the erythroid-specific isoform of hydroxymethylbilane synthase (HMBS),23 that were down-regulated, as well as “stemness-related” genes, such as the CD34 gene, that were up-regulated. Other potentially interesting “stemness-related” genes were also identified that should be the subject of further investigation.
Methods
Cell lines
EML cells were maintained in Iscove modified Dulbecco medium containing 20% heat-inactivated horse serum (Invitrogen) and supplemented with 15% conditioned medium from the BHK/MKL cell line containing rat stem cell factor.22 EPRO cells were derived from EML cells by induction with all-trans retinoic acid (ATRA, 10mM; Sigma-Aldrich) and interleukin-3 (IL-3, 10 ng/mL; PeproTech) for 3 days as previously described.22 EPRO cells were then maintained in Iscove modified Dulbecco medium with 20% horse serum and granulocyte-macrophage colony-stimulating factor (GM-CSF, 10 ng/mL; PeproTech). EPRO cells were differentiated along the neutrophil maturation pathway by addition of ATRA (10mM) to the EPRO growth medium for 96 hours. All growth medium contained 5 U/mL penicillin, 5 μg/mL streptomycin sulfate, and 2mM l-glutamine (Invitrogen).
Retroviral vectors and virus production
MSCV-IRES-GFP and MSCV-HOXB4-IRES-GFP vectors were the same as those used in previous studies of HOXB4 overexpression in primary bone marrow cells and were kindly provided by Dr R. K. Humphries and colleagues (Terry Fox Laboratory).12 Vesicular stomatitis virus glycoprotein (VSV-G) pseudotyped viruses were produced using the GP2-293 cell line.24 pCMV-VSV-G plasmid was cotransfected with the MSCV-based retroviral vectors using the FuGENE 6 Transfection Reagent (Roche Applied Science). The ratio of FuGENE 6 Transfection Reagent (microliters) to DNA (micrograms) used was 3:1. VSV-G pseudotyped viruses were concentrated using the Beckman ultracentrifuge. Using an SW28 rotor, 40 mL of supernatant was concentrated by centrifugation using an autoclaved 1 × 3.5 inch (25 × 89 mm) centrifuge tube for 120 minutes (2 hours) at 4°C at 90 000g.
Lineage depletion
The BD IMag Mouse Hematopoietic Progenitor Cell Enrichment Set (BD Biosciences) was used for lineage depletion experiments. Experiments were done according to the manufacturer's guidelines.
Flow cytometry
For cell surface immunophenotyping experiments of EML and EPRO cells, the following antibodies were used: phycoerythrin (PE)–conjugated anti–c-Kit, PE-anti–Sca-1, PE-anti–Mac-1, PE-anti–Gr-1, PE–anti-B220, and PE–anti-Ter119 (eBioscience). Cells were incubated with the antibodies for 20 minutes at 4°C in antibody dilution solution (phosphate-buffered saline with 2mM ethylenediaminetetraacetic acid, 0.5% bovine serum albumin, 2% rat serum, 2% mouse serum, and 0.1% sodium azide). The cells were analyzed on a FACSCalibur machine (BD Biosciences).
Southern and Western blot analyses
Genomic DNA was extracted using the DNeasy Blood and Tissue Kit (QIAGEN). DNA was then digested with EcoRI (New England Biosciences), which cuts once inside the provirus. Digested DNA was then separated in a 1% agarose gel by electrophoresis, and standard Southern blotting techniques were used as described.25 Membranes were probed with an α-32P-dCTP-labeled GFP cDNA sequence.
Total protein lysate preparation and Western blot analysis were carried out as described.20 Blots were probed using the I12 anti–rat human HOXB4 monoclonal antibody (Developmental Studies Hybridoma Bank, University of Iowa).26 Blots were visualized with the horseradish peroxidase-conjugated goat anti–rat secondary antibody (sc-2006; Santa Cruz Biotechnology)
Microarray analysis
RNA was extracted from 3 biologic replicates of control and HOXB4 overexpressing KLS-EML cells using the RNeasy MinElute Cleanup Kit (QIAGEN). The Mouse genome 430 2.0 GeneChip (> 39 000 transcripts) from Affymetrix was used for the RNA expression profiling experiments (Affymetrix). RNA was processed and hybridized to the array chips by the Keck Microarray Resource Center at the Yale School of Medicine (http://keck.med.yale.edu/microarrays/). Three different probe set algorithms (GC-RMA, dChip, and RMA) were used to generate an expression value for each transcript. Gene transcripts were stringently selected as being differentially expressed at least 2-fold with a Student t test false discovery rate of less than 0.05 only when all 3 algorithms confirmed such a result.27 GeneGO access was provided by the Keck Bioinformatics Resource Center at the Yale School of Medicine (http://keck.med.yale.edu/bioinformatics/), and GeneGO analysis was performed by the Yale Center of Excellence in Molecular Hematology. All real-time polymerase chain reaction (PCR) experiments were done using the corresponding gene TaqMan probes from Applied Biosystems. Array data can be accessed from Gene Expression Omnibus under accession GSE20604.
ChIP-chip
Three clones of EML-HOXB4 cells (4, 11, and 17; 1.5 × 109 to 2 × 109) were lineage depleted to isolate KLS-EML-HOXB4 cells (1.5 × 108 to 2 × 108). A modified version of the ChIP-chip procedure provided by Roche NimbleGen (supplemental Methods; available on the Blood Web site; see the Supplemental Materials link at the top of the online article) was used to prepare the ChIP samples for hybridization to the MM8 Refseq promoter gene chip array from Roche NimbleGen, which contains sequences corresponding to 2 kb 5′ and 0.5 kb 3′ to the transcription start site of annotated genes. Array data can be accessed from Gene Expression Omnibus under accession GSE20604.
Results
HOXB4 overexpression did not affect cell morphology or myeloid differentiation in EML cells
Using MSCV retroviral vectors, we transduced EML cells to overexpress either both HOXB4 and GFP (EML-HOXB4 cells) or only GFP (EML-GFP cells).12 By Western blot analysis of protein lysates from parental EML, EML-GFP, and EML-HOXB4 cells, we observed that HOXB4 was only detected in pooled EML-HOXB4 cells (Figure 1A). These and subsequent experiments, unless otherwise noted, were carried out using cells that were not yet clonally selected, but consisted of the entire GFP-positive cell population obtained after transduction. Microscopic examination of cytospin preparations of EML, EML-GFP, and EML-HOXB4 cells revealed that all 3 cell types had a similar morphology, with the presence of hand mirror-shaped cells, characteristic of EML cells (supplemental Figure 1A).22 All 3 cell types could be differentiated along the myeloid pathway to the promyelocyte stage by culturing the cells in the presence of IL-3, ATRA, and GM-CSF. Such cells are blocked at the promyelocyte stage but can proliferate as a GM-CSF-dependent cell line designated as EPRO (EML-derived promyelocyte) cells, as previously reported.22 Cytospin preparations showed that EPRO cells derived from EML cells (EPRO-EML), EML-GFP cells (EPRO-GFP), and EML-HOXB4 (EPRO-HOXB4) cells all had normal EPRO cell morphology, characterized by the presence of primary granules (supplemental Figure 1B). Western blot analysis showed that HOXB4 protein was still expressed at the EPRO cell stage in EPRO-HOXB4 cells (data not shown). Cell surface marker immunophenotyping showed that EPRO-EML, EPRO-GFP, and EPRO-HOXB4 cells had all acquired characteristics of granulocytic cells with similar patterns of expression of Gr-1 and Mac-1 in a high percentage of the cells (supplemental Figure 2). When EPRO-EML, EPRO-GFP, and EPRO-HOXB4 cells were cultured in the presence of high levels of ATRA (10μM) for 96 hours, the cells differentiated into neutrophils (supplemental Figure 1C). Therefore, overexpression of HOXB4 did not affect myeloid differentiation in EML and EPRO cells, and the only difference that could be observed between cell types was that there was a slight delay in the time required for EML-HOXB4 cells to differentiate to the EPRO stage (data not shown).
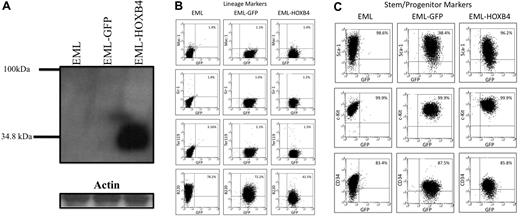
HOXB4 overexpression down-regulates the lymphoid marker B220 in EML cells. (A) Western blot analysis of total cell lysates fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and probed with the I12 monoclonal anti-HOXB4 antibody revealed the presence of HOXB4 only in the lysate of EML cells transduced with retrovirus expressing HOXB4 (EML-HOXB4) and not in control cells. (B) By immunophenotyping of cell surface markers, the percentage of cells that express B220 decreases when HOXB4 is overexpressed in EML cells. All other lineage markers and stem/progenitor cell markers are expressed at similar levels in EML, EML-GFP, and EML-HOXB4 cells.
HOXB4 overexpression down-regulates the lymphoid marker B220 in EML cells. (A) Western blot analysis of total cell lysates fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and probed with the I12 monoclonal anti-HOXB4 antibody revealed the presence of HOXB4 only in the lysate of EML cells transduced with retrovirus expressing HOXB4 (EML-HOXB4) and not in control cells. (B) By immunophenotyping of cell surface markers, the percentage of cells that express B220 decreases when HOXB4 is overexpressed in EML cells. All other lineage markers and stem/progenitor cell markers are expressed at similar levels in EML, EML-GFP, and EML-HOXB4 cells.
HOXB4 down-regulated the expression of the lymphoid marker B220 in EML cells
By flow cytometric analysis, EML-GFP and EML-HOXB4 cells all had similar patterns of expression of lineage markers, such as Ter119, Mac-1, and Gr-1 (Figure 1B). The percentages of cells that expressed c-Kit, Sca-1, and CD34 were also similar between the 3 cell types. However, a striking change in cell surface immunophenotype was observed when HOXB4 was overexpressed in EML cells. The percentage of EML-HOXB4 cells that expressed B220 was much lower than that of EML or EML-GFP cells: approximately 42% for EML-HOXB4 cells compared with approximately 78% for EML cells and approximately 72% for EML-GFP cells.
Analysis of single-cell clones
Single-cell clones were isolated from the total populations of transduced EML-GFP and EML-HOXB4 cells by cell sorting, and 3 single-cell clones were selected from both cell types for use in microarray studies. Each clone isolated had a different retroviral insertion site as indicated by the size of a junctional restriction endonuclease fragment seen on Southern blot analysis (supplemental Figure 3). This helped exclude the possibility that any of the characteristics we observed in the total population of EML-HOXB4 cells was the result of the presence of a dominant cellular clone, the phenotype of which was influenced by its associated retroviral integration site. This approach also provided true biologic triplicates for use in the microarray studies. Cell surface immunophenotyping of the 3 single-cell EML-GFP clones (1, 2, and 14) used for the microarray studies all displayed similar patterns to that of the total unselected population of EML-GFP cells (Table 1). The same was also true for the 3 single-cell EML-HOXB4 clones (4, 11, and 17) that were selected for microarray analysis (Table 1). By Western blot analysis, only the EML-HOXB4 single-cell clones expressed detectable levels of HOXB4 protein (data not shown). As expected from the prior results using cells from the total population of transduced cells, the percentage of cells that expressed B220 was significantly lower in the EML-HOXB4 single-cell clones compared with that in the EML-GFP single-cell clones (Table 1).
The single-cell EML clones selected for microarray analysis studies all exhibit cell-surface marker profiles similar to that of unselected cells in the cell populations from which they were isolated
| Marker . | EML-GFP (unselected) . | EML-GFP 1 . | EML-GFP 2 . | EML-GFP 14 . | EML-HOXB4 (unselected) . | EML-HOXB4 4 . | EML-HOXB4 11 . | EML-HOXB4 17 . |
|---|---|---|---|---|---|---|---|---|
| B220 | 72.2 | 72.9 | 78.3 | 78.4 | 41.5 | 47 | 42.8 | 58.3 |
| Mac-1 | 1.1 | 1.3 | 3.65 | 3 | 1.4 | 4.32 | 2.4 | 4.7 |
| Gr-1 | 1.0 | 1.7 | 4.6 | 0 | 1.2 | 0.16 | 5.4 | 0 |
| Ter119 | 2.1 | 0.3 | 2.06 | 1.6 | 1.5 | 0.3 | 0.8 | 1 |
| c-Kit | 99.9 | 99.8 | 99.8 | 99.4 | 99.9 | 99.1 | 99 | 99.8 |
| Sca-1 | 98.4 | 92.4 | 98 | 93.5 | 96.2 | 94.5 | 90.1 | 96.3 |
| CD34 | 87.5 | 84.3 | 86.1 | 80.4 | 85.8 | 88.2 | 74.9 | 80.1 |
| Marker . | EML-GFP (unselected) . | EML-GFP 1 . | EML-GFP 2 . | EML-GFP 14 . | EML-HOXB4 (unselected) . | EML-HOXB4 4 . | EML-HOXB4 11 . | EML-HOXB4 17 . |
|---|---|---|---|---|---|---|---|---|
| B220 | 72.2 | 72.9 | 78.3 | 78.4 | 41.5 | 47 | 42.8 | 58.3 |
| Mac-1 | 1.1 | 1.3 | 3.65 | 3 | 1.4 | 4.32 | 2.4 | 4.7 |
| Gr-1 | 1.0 | 1.7 | 4.6 | 0 | 1.2 | 0.16 | 5.4 | 0 |
| Ter119 | 2.1 | 0.3 | 2.06 | 1.6 | 1.5 | 0.3 | 0.8 | 1 |
| c-Kit | 99.9 | 99.8 | 99.8 | 99.4 | 99.9 | 99.1 | 99 | 99.8 |
| Sca-1 | 98.4 | 92.4 | 98 | 93.5 | 96.2 | 94.5 | 90.1 | 96.3 |
| CD34 | 87.5 | 84.3 | 86.1 | 80.4 | 85.8 | 88.2 | 74.9 | 80.1 |
Numeric values are expressed as mean percentages from 3 independent experiments.
All of the single-cell EML-GFP and EML-HOXB4 clones used in our microarray studies could be differentiated to the EPRO stage, and their cell surface marker profiles at the EPRO cell stage were all similar to those that were observed in supplemental Figure 2 with the total population of unselected EPRO-GFP and EPRO-HOXB4 cells (data not shown). The cell morphology as assessed by microscopic examination of cytospin preparations was also similar between cloned cells and unselected transduced cells (data not shown).
Lineage-depleted EML-HOXB4 cells contained a higher percentage of CD34+ cells
The 3 EML-GFP single-cell clones (1, 2, and 14) and the 3 EML-HOXB4 single-cell clones (4, 11, and 17) were lineage depleted to isolate the KLS cell fraction for RNA expression profiling experiments. Approximately 10% to 13% of the EML-HOXB4 cells were lineage negative, in contrast to only 2% to 3% of the EML-GFP cells. A striking difference was also observed when the 2 sets of lineage-depleted cells were stained for CD34. A much higher percentage of the lineage-depleted EML-HOXB4 cells stained positively for CD34 (∼ 78%) compared with the lineage-depleted EML-GFP cells (only ∼ 29% CD34+ cells; Figure 2).
CD34 is expressed in a higher percentage of KLS-EML-HOXB4 cells than in KLS-EML-GFP cells. The percentage of cells that express CD34 is much higher in lineage-depleted EML-HOXB4 cells than in lineage-depleted EML-GFP cells, whereas the percentages of cells that express c-Kit and Sca-1 are similar. The cell surface profiles of the numbered lineage-depleted single-cell clones shown are representative of those of the other lineage-depleted single-cell clones that were used for microarray analyses.
CD34 is expressed in a higher percentage of KLS-EML-HOXB4 cells than in KLS-EML-GFP cells. The percentage of cells that express CD34 is much higher in lineage-depleted EML-HOXB4 cells than in lineage-depleted EML-GFP cells, whereas the percentages of cells that express c-Kit and Sca-1 are similar. The cell surface profiles of the numbered lineage-depleted single-cell clones shown are representative of those of the other lineage-depleted single-cell clones that were used for microarray analyses.
RNA expression profiling showed that 465 gene transcripts were differentially expressed when HOXB4 was overexpressed in EML cells
RNA from the 3 KLS-EML-GFP single-cell clones and the 3 KLS-EML-HOXB4 single-cell clones were collected and used for RNA expression profiling experiments. Microarray analysis results showed that 465 gene transcripts were differentially expressed at least 2-fold between KLS-EML-GFP and KLS-EML-HOXB4 cells (supplemental Table 1). Most of these differentially expressed gene transcripts were up-regulated (320 of 465 gene transcripts). Gene transcripts that were observed to be highly up-regulated included Meis1, Dntt, Hlf, Runx2, and CD34. Interestingly, transcripts of 2 genes with relatively unknown function, Latptm4b and Gp49a, were observed to be among the top 5 most highly up-regulated transcripts (Table 2). However, the most striking result was the large number of erythroid-specific gene transcripts that were observed to be strongly down-regulated in KLS-EML-B4 cells. Of the 20 most strongly down-regulated gene transcripts, more than half originated from erythroid-specific genes (Table 3).
Top 20 most highly up-regulated gene transcripts detected by microarray analysis
| Fold difference HOXB4/GFP . | GenBank . | Gene symbol . | Gene title . |
|---|---|---|---|
| 12.345 | NM_008152 | Gpr65 | G-protein coupled receptor 65 |
| 9.82 | AU024771 | Laptm4b | Lysosomal-associated protein transmembrane 4B |
| 9.045 | BB160593 | Dntt | Deoxynucleotidyltransferase, terminal |
| 8.385 | U05264 | Gp49a | Glycoprotein 49 A/leukocyte immunoglobulin-like receptor, subfamily B, member 4 |
| 8.04 | BB744589 | Hlf | Hepatic leukemia factor |
| 7.62 | M94967 | Ptgs2 | Prostaglandin-endoperoxide synthase 2 |
| 7.575 | AW554231 | Cldn12 | Claudin 12 |
| 7.145 | AV058500 | Lzp-s | P lysozyme structural |
| 6.24 | BC027242 | Vav3 | vav 3 oncogene |
| 5.76 | NM_010228 | Flt1 | FMS-like tyrosine kinase 1 |
| 5.64 | AA185889 | Bex6 | Brain expressed gene 6 |
| 4.8 | BG967597 | Pgm5 | Phosphoglucomutase 5 |
| 4.765 | AV172776 | Satb1 | Special AT-rich sequence binding protein 1 |
| 4.725 | AK020608 | 9530053H05Rik | RIKEN cDNA 9530053H05 gene |
| 4.605 | AK003461 | Etv5 | ets variant gene 5 |
| 4.49 | AW547821 | Meis1 | Myeloid ecotropic viral integration site 1 |
| 4.47 | BB515151 | Tcn2 | Transcobalamin 2 |
| 4.465 | BB770873 | Cd200r4 | Cd200 receptor 4 |
| 4.46 | U10551 | Gem | GTP binding protein (gene overexpressed in skeletal muscle) |
| 4.45 | NM_012059 | Sh3d19 | SH3 domain protein D19 |
| Fold difference HOXB4/GFP . | GenBank . | Gene symbol . | Gene title . |
|---|---|---|---|
| 12.345 | NM_008152 | Gpr65 | G-protein coupled receptor 65 |
| 9.82 | AU024771 | Laptm4b | Lysosomal-associated protein transmembrane 4B |
| 9.045 | BB160593 | Dntt | Deoxynucleotidyltransferase, terminal |
| 8.385 | U05264 | Gp49a | Glycoprotein 49 A/leukocyte immunoglobulin-like receptor, subfamily B, member 4 |
| 8.04 | BB744589 | Hlf | Hepatic leukemia factor |
| 7.62 | M94967 | Ptgs2 | Prostaglandin-endoperoxide synthase 2 |
| 7.575 | AW554231 | Cldn12 | Claudin 12 |
| 7.145 | AV058500 | Lzp-s | P lysozyme structural |
| 6.24 | BC027242 | Vav3 | vav 3 oncogene |
| 5.76 | NM_010228 | Flt1 | FMS-like tyrosine kinase 1 |
| 5.64 | AA185889 | Bex6 | Brain expressed gene 6 |
| 4.8 | BG967597 | Pgm5 | Phosphoglucomutase 5 |
| 4.765 | AV172776 | Satb1 | Special AT-rich sequence binding protein 1 |
| 4.725 | AK020608 | 9530053H05Rik | RIKEN cDNA 9530053H05 gene |
| 4.605 | AK003461 | Etv5 | ets variant gene 5 |
| 4.49 | AW547821 | Meis1 | Myeloid ecotropic viral integration site 1 |
| 4.47 | BB515151 | Tcn2 | Transcobalamin 2 |
| 4.465 | BB770873 | Cd200r4 | Cd200 receptor 4 |
| 4.46 | U10551 | Gem | GTP binding protein (gene overexpressed in skeletal muscle) |
| 4.45 | NM_012059 | Sh3d19 | SH3 domain protein D19 |
Top 20 most strongly down-regulated transcripts detected by microarray analysis
| Fold difference HOXB4/GFP . | GenBank . | Gene symbol . | Gene title . |
|---|---|---|---|
| −308.55* | NM_008219 | Hbb-bh1 | Hemoglobin Z, β-like embryonic chain |
| −157.31* | NM_133245 | Eraf | Erythroid-associated factor |
| −78.37* | NM_011269 | Rhag | Rhesus blood group-associated A glycoprotein |
| −76.22* | NM_013848 | Ermap | Erythroblast membrane–associated protein |
| −55.11 | NM_020504 | Cldn13 | Claudin 13 |
| −44.45 | D85596 | Ampd3 | AMP deaminase 3 |
| −36.28 | BC027185 | 2210023G05Rik | RIKEN cDNA 2210023G05 gene |
| −35.31* | NM_010369 | Gypa | Glycophorin A |
| −29.88* | NM_010635 | Klf1 | Kruppel-like factor 1 (erythroid) |
| −29.77* | NM_008218 | Hba-a1 | Hemoglobin α, adult chain 1 |
| −26.57* | BC011223 | Car1 | Carbonic anhydrase 1 |
| −25.79 | BB729271 | — | Transcribed locus |
| −23.46 | AK007743 | Pdia2 | Protein disulfide isomerase associated 2 |
| −22.27* | AF069311 | Rhd | Rh blood group, D antigen |
| −20.86 | NM_008560 | Mc2r | Melanocortin 2 receptor |
| −20.81* | AV156860 | Hbb-y | Hemoglobin Y, β-like embryonic chain |
| −19.85* | NM_032540 | Kel | Kell blood group |
| −19.50 | AV048291 | C1qdc2 | C1q domain containing 2 |
| −17.30 | NM_011452 | Serpinb9b | Serine (or cysteine) peptidase inhibitor, clade B, member 9b |
| −13.80* | BB740660 | LOC630963 | Similar to spectrin α 1 |
| Fold difference HOXB4/GFP . | GenBank . | Gene symbol . | Gene title . |
|---|---|---|---|
| −308.55* | NM_008219 | Hbb-bh1 | Hemoglobin Z, β-like embryonic chain |
| −157.31* | NM_133245 | Eraf | Erythroid-associated factor |
| −78.37* | NM_011269 | Rhag | Rhesus blood group-associated A glycoprotein |
| −76.22* | NM_013848 | Ermap | Erythroblast membrane–associated protein |
| −55.11 | NM_020504 | Cldn13 | Claudin 13 |
| −44.45 | D85596 | Ampd3 | AMP deaminase 3 |
| −36.28 | BC027185 | 2210023G05Rik | RIKEN cDNA 2210023G05 gene |
| −35.31* | NM_010369 | Gypa | Glycophorin A |
| −29.88* | NM_010635 | Klf1 | Kruppel-like factor 1 (erythroid) |
| −29.77* | NM_008218 | Hba-a1 | Hemoglobin α, adult chain 1 |
| −26.57* | BC011223 | Car1 | Carbonic anhydrase 1 |
| −25.79 | BB729271 | — | Transcribed locus |
| −23.46 | AK007743 | Pdia2 | Protein disulfide isomerase associated 2 |
| −22.27* | AF069311 | Rhd | Rh blood group, D antigen |
| −20.86 | NM_008560 | Mc2r | Melanocortin 2 receptor |
| −20.81* | AV156860 | Hbb-y | Hemoglobin Y, β-like embryonic chain |
| −19.85* | NM_032540 | Kel | Kell blood group |
| −19.50 | AV048291 | C1qdc2 | C1q domain containing 2 |
| −17.30 | NM_011452 | Serpinb9b | Serine (or cysteine) peptidase inhibitor, clade B, member 9b |
| −13.80* | BB740660 | LOC630963 | Similar to spectrin α 1 |
Gene transcripts that are erythroid-specific and represent more than half of the top 20 most strongly down-regulated transcripts.
The results of our microarray studies were verified by real-time PCR analysis. Thirteen gene transcripts that were observed to be differentially expressed at least 2-fold were chosen and tested. By our microarray studies, we could not conclude which of the 2 isoforms of the HMBS gene (erythroid-specific or housekeeping isoform) was differentially expressed because the probes for this gene on the array chip did not discriminate between the 2 isoforms.23 Therefore, we carried out separate real-time PCR assays that were specific for each of the 2 isoforms of HMBS. Real-time PCR results confirmed that all 13 gene transcripts (including the 2 isoforms of HMBS) were indeed differentially expressed between KLS-EML-GFP and KLS-EML-HOXB4 cells, as observed in our microarray-based RNA expression profiling experiments (Figure 3). Interestingly, the erythroid-specific isoform of HMBS was down-regulated approximately 28-fold more than the housekeeping isoform of HMBS (62.1-fold vs 2.2-fold down-regulation, respectively, compared with control).
Real-time PCR analysis of selected gene transcripts observed to be differentially expressed in RNA expression profiling experiments. Thirteen gene transcripts that were observed by microarray analysis to be differentially expressed at least 2-fold were chosen and tested. However, by microarray analysis, it was not possible to conclude which of the 2 isoforms of the HMBS gene (erythroid-specific and housekeeping isoforms) was differentially expressed because the probes for this gene on the array chip did not discriminate between the 2 isoforms.23 Therefore, real-time PCR was performed using primers specific for each of the 2 different isoforms of HMBS. Real-time PCR results confirmed that all 13 gene transcripts (including the 2 isoforms of HMBS) were indeed differentially expressed between KLS-EML-GFP and KLS-EML-HOXB4 cells, as observed in our RNA expression profiling experiments. Interestingly, the erythroid-specific isoform of HMBS was observed to be down-regulated approximately 28-fold more than the housekeeping isoform of HMBS (61.2-fold vs 2.2-fold). Values are mean plus or minus SD of triplicate PCR analyses of RNA from 3 different single-cell clones of each cell type.
Real-time PCR analysis of selected gene transcripts observed to be differentially expressed in RNA expression profiling experiments. Thirteen gene transcripts that were observed by microarray analysis to be differentially expressed at least 2-fold were chosen and tested. However, by microarray analysis, it was not possible to conclude which of the 2 isoforms of the HMBS gene (erythroid-specific and housekeeping isoforms) was differentially expressed because the probes for this gene on the array chip did not discriminate between the 2 isoforms.23 Therefore, real-time PCR was performed using primers specific for each of the 2 different isoforms of HMBS. Real-time PCR results confirmed that all 13 gene transcripts (including the 2 isoforms of HMBS) were indeed differentially expressed between KLS-EML-GFP and KLS-EML-HOXB4 cells, as observed in our RNA expression profiling experiments. Interestingly, the erythroid-specific isoform of HMBS was observed to be down-regulated approximately 28-fold more than the housekeeping isoform of HMBS (61.2-fold vs 2.2-fold). Values are mean plus or minus SD of triplicate PCR analyses of RNA from 3 different single-cell clones of each cell type.
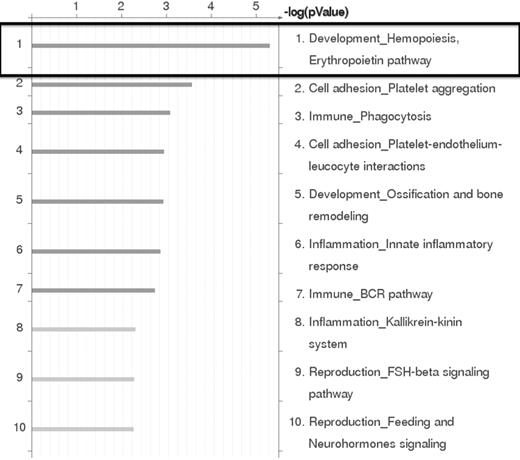
Based on the results of the RNA expression profiling experiments, the genes that were differentially expressed by at least 2-fold between EML-GFP and EML-HOXB4 were subjected to pathway analysis using GeneGo Pathway Analysis Version 5.3.18499 software. This analysis indicated that the erythropoietin pathway in hematopoiesis development was the most significant pathway represented in our results (Figure 4). This result confirmed our findings that erythroid-specific gene transcripts were observed to be strongly down-regulated in KLS-EML-HOXB4 cells.
GeneGO Pathway analysis of microarray results. GeneGo Pathway analysis of the RNA expression profiling results obtained by microarray indicates that the erythropoietin pathway in hematopoiesis development is the most highly affected pathway in KLS-EML cells that overexpress HOXB4.
GeneGO Pathway analysis of microarray results. GeneGo Pathway analysis of the RNA expression profiling results obtained by microarray indicates that the erythropoietin pathway in hematopoiesis development is the most highly affected pathway in KLS-EML cells that overexpress HOXB4.
ChIP-chip analysis revealed that HOXB4 bound to the promoter regions of 1910 genes
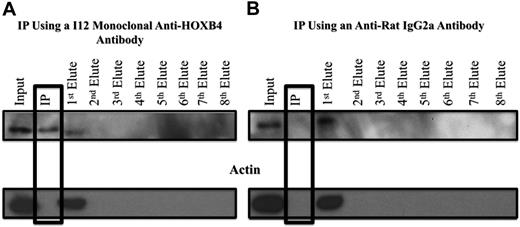
HOXB4 protein was cross-linked with 1% formaldehyde to its DNA binding sites in the 3 KLS-EML-HOXB4 single-cell clones, and the I12 anti-HOXB4 monoclonal antibody was used for ChIP. We observed that, even after cross-linking to its DNA-binding sites, the HOXB4 protein could still be immunoprecipitated with the I12 anti-HOXB4 monoclonal antibody in a specific manner, as indicated by the absence of actin, in the immunoprecipitate (IP) eluate (Figure 5A). Control experiments using an isotype control antirat IgG2a antibody indicated that the HOXB4 protein was specifically immunoprecipitated only when the I12 anti-HOXB4 monoclonal antibody was used (Figure 5B).
Western blot analysis of samples from ChIP experiments, showing specificity of the IP obtained with the I12 anti-HOXB4 monoclonal antibody. (A) Formaldehyde cross-linked chromatin was prepared from clone 4 of KLS-EML-HOXB4 cells, as described in supplemental Methods (ChIP-chip), and incubated with the I12 antihuman HOXB4 rat monoclonal antibody that had been previously bound to magnetic beads (Dynabeads) conjugated to antirat IgG2a antibody. The IP lane represents the material that remained bound to the magnetic beads after repeated washings. Eluates 1 to 8 represent the supernatants from the serial washings of the magnetically bound beads. The HOXB4 protein was specifically immunoprecipitated using the I12 monoclonal anti-HOXB4 antibody, as shown by the presence of the HOXB4 protein and the absence of actin in the IP lane. Some residual HOXB4 protein was observed in the first eluate, indicating that binding was not 100% efficient. (B) Using an antirat IgG2a isotype control antibody, HOXB4 protein is not observed in the IP lane, further documenting the specificity of the I12 monoclonal anti-HOXB4 antibody used for ChIP. Similar results were obtained when chromatin extracts from clones 11 and 17 of KLS-EML-HOXB4 cells were used.
Western blot analysis of samples from ChIP experiments, showing specificity of the IP obtained with the I12 anti-HOXB4 monoclonal antibody. (A) Formaldehyde cross-linked chromatin was prepared from clone 4 of KLS-EML-HOXB4 cells, as described in supplemental Methods (ChIP-chip), and incubated with the I12 antihuman HOXB4 rat monoclonal antibody that had been previously bound to magnetic beads (Dynabeads) conjugated to antirat IgG2a antibody. The IP lane represents the material that remained bound to the magnetic beads after repeated washings. Eluates 1 to 8 represent the supernatants from the serial washings of the magnetically bound beads. The HOXB4 protein was specifically immunoprecipitated using the I12 monoclonal anti-HOXB4 antibody, as shown by the presence of the HOXB4 protein and the absence of actin in the IP lane. Some residual HOXB4 protein was observed in the first eluate, indicating that binding was not 100% efficient. (B) Using an antirat IgG2a isotype control antibody, HOXB4 protein is not observed in the IP lane, further documenting the specificity of the I12 monoclonal anti-HOXB4 antibody used for ChIP. Similar results were obtained when chromatin extracts from clones 11 and 17 of KLS-EML-HOXB4 cells were used.
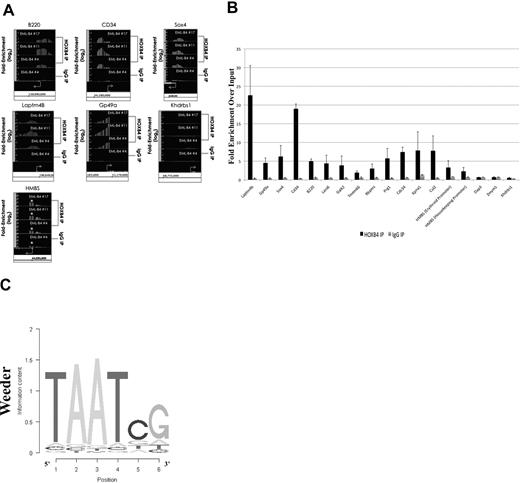
The HOXB4-bound DNA isolated by ChIP using the I12 anti-HOXB4 antibody was hybridized to the MM8 Refseq promoter chip array from Roche NimbleGen. Analysis of the ChIP-chip results using Tilescope software (Version Python Alpha) revealed that the promoter regions of 1910 genes were bound by HOXB4 (supplemental Table 2). Interestingly, the ChIP-chip results indicated that the promoter regions of genes, such as B220, HMBS (promoter regions for both isoforms), CD34, Sox4, and the 2 highly up-regulated genes, Laptm4b and GP49a, were observed to be directly bound by HOXB4 in KLS-EML-HOXB4 cells (Figure 6A). The promoter region for the Khdrbs1 gene was used as a negative control because it was not shown to be differentially expressed in KLS-EML-HOXB4 cells by microarray analysis, and no binding of HOXB4 to its promoter region was detected by ChIP-chip. Seventeen promoter regions that were observed to be enriched at least 2-fold in the IP sample were chosen for validation. Real-time PCR results confirmed that 15 of the 17 promoter regions were indeed enriched in the IP samples compared with the input samples (Figure 6B).
Results of ChIP-chip and their validation by quantitative PCR. (A) ChIP-chip analysis using the I12 anti-HOXB4 monoclonal antibody revealed that there was increased binding to the promoter regions of the B220, CD34, Sox4, Laptm4b, Gp49a, and HMBS genes by the anti-HOXB4 IP, compared with input. In contrast, the control IP obtained using antirat IgG2a antibody beads alone showed no comparable increased binding. By microarray analysis, the Khdrbs1 gene was not shown to be differentially expressed in KLS-EML-HOXB4 cells, and no binding of HOXB4 to its promoter region was detected by ChIP; it is shown as a negative control. *The promoter region for the erythroid-specific isoform of HMBS, which is located within the sequence region of the first intron of the housekeeping isoform of HMBS. Interestingly, there is a greater degree of increased binding of HOXB4 to the promoter region for the erythroid-specific isoform of HMBS than to the promoter region for the housekeeping isoform of HMBS. Arrows represent transcription start sites of the genes. (B) Primers were designed corresponding to the promoter regions of the negative control Khdrbs1 gene and of 17 genes that were observed by ChIP-chip to be enriched at least 2-fold in the IP obtained with the I12 anti-HOXB4 monoclonal antibody. Validation of the enrichment was carried out by quantitative real-time PCR. The PCR results confirmed that 15 of the 17 selected promoter regions were indeed enriched in these IPs compared with the input. Results using the IPs obtained using the control beads conjugated to antirat IgG2a antibody are shown for comparison. Values are mean plus or minus SD of triplicate PCR analyses of ChIP DNA from the 3 different single-cell clones of KLS-EML-HOXB4 cells. (C) Top-scoring DNA-binding motif identified, using the software program Weeder (Version 1.3), in an unbiased analysis of the top 1000 promoter sequences bound by HOXB4 in the ChIP-chip experiments. The known HOX protein binding consensus sequence, TAAT, is found prominently in this motif.
Results of ChIP-chip and their validation by quantitative PCR. (A) ChIP-chip analysis using the I12 anti-HOXB4 monoclonal antibody revealed that there was increased binding to the promoter regions of the B220, CD34, Sox4, Laptm4b, Gp49a, and HMBS genes by the anti-HOXB4 IP, compared with input. In contrast, the control IP obtained using antirat IgG2a antibody beads alone showed no comparable increased binding. By microarray analysis, the Khdrbs1 gene was not shown to be differentially expressed in KLS-EML-HOXB4 cells, and no binding of HOXB4 to its promoter region was detected by ChIP; it is shown as a negative control. *The promoter region for the erythroid-specific isoform of HMBS, which is located within the sequence region of the first intron of the housekeeping isoform of HMBS. Interestingly, there is a greater degree of increased binding of HOXB4 to the promoter region for the erythroid-specific isoform of HMBS than to the promoter region for the housekeeping isoform of HMBS. Arrows represent transcription start sites of the genes. (B) Primers were designed corresponding to the promoter regions of the negative control Khdrbs1 gene and of 17 genes that were observed by ChIP-chip to be enriched at least 2-fold in the IP obtained with the I12 anti-HOXB4 monoclonal antibody. Validation of the enrichment was carried out by quantitative real-time PCR. The PCR results confirmed that 15 of the 17 selected promoter regions were indeed enriched in these IPs compared with the input. Results using the IPs obtained using the control beads conjugated to antirat IgG2a antibody are shown for comparison. Values are mean plus or minus SD of triplicate PCR analyses of ChIP DNA from the 3 different single-cell clones of KLS-EML-HOXB4 cells. (C) Top-scoring DNA-binding motif identified, using the software program Weeder (Version 1.3), in an unbiased analysis of the top 1000 promoter sequences bound by HOXB4 in the ChIP-chip experiments. The known HOX protein binding consensus sequence, TAAT, is found prominently in this motif.
Using the ChIP-chip results, we performed an unbiased search for DNA sequence motifs that could represent HOXB4-DNA interaction sites, using the motif discovery program, Weeder.28 The top-scoring motif discovered using this program was 6 bp in length and contained the HOX protein binding consensus sequence TAAT (Figure 6C).29 This result, together with the real-time PCR results, indicated that the ChIP-chip experiments were valid.
HOXB4 directly regulates 71 of 465 of the differentially expressed genes
The results of the ChIP-chip experiments and those of the RNA expression profiling studies were analyzed to search for genes identified in common by these 2 independent techniques. When the 2 lists were compared with one another, 15.3% (71 of 465) of the differentially expressed genes identified in our RNA expression profiling experiments were also observed to have DNA binding sites occupied by HOXB4 (supplemental Table 3). A majority (54 of 71) of these target genes expressed transcripts that were up-regulated in KLS-EML-HOXB4 cells. CD34, a stem cell antigen, which was observed by flow cytometry to be expressed in a higher percentage of KLS-EML-HOXB4 cells compared with KLS-EML-GFP cells (Figure 2), was present in both lists. Of particular interest are the 2 genes, Laptm4b and Gp49a, which were highly ranked in both lists (Table 4). BloodExpress, a database of gene expression in mouse hematopoietic cells, was used to examine the stage in hematopoietic development at which these 2 genes are expressed.30 This analysis indicated that Laptm4b and Gp49a have both been shown to be preferentially expressed in long-term repopulating HSCs and to be down-regulated in more mature bone marrow cells.31,,–34
Top 20 genes (among 71 genes represented in both ChIP-chip and RNA expression profiling datasets) ranked according to their ChIP-chip score
| Fold difference HOXB4/GFP . | Gene symbol . | Gene title . | ChIP-chip rank (of 1910) . | RNA expression profiling rank (of 465) . |
|---|---|---|---|---|
| 8.39* | Gp49a | Glycoprotein 49 A | 29 | 4 |
| 2.50 | Rab38 | Rab38, member of RAS oncogene family | 68 | 169 |
| −2.53† | 2010301N04Rik | RIKEN cDNA 2010301N04 gene | 88 | 369 |
| 3.65 | Clec4e | C-type lectin domain family 4, member e | 95 | 38 |
| 9.82* | Laptm4b | Lysosomal-associated protein transmembrane 4B | 96 | 2 |
| 2.76 | AI586015 | Expressed sequence AI586015 | 122 | 111 |
| 2.19 | Arhgap15 | Rho GTPase activating protein 15 | 141 | 257 |
| 2.13 | Bmp2k | BMP2 inducible kinase | 162 | 284 |
| −2.14† | Rpl3 | Ribosomal protein L3/similar to 60S ribosomal protein L3 (J1 protein) | 188 | 331 |
| 4.26 | Kazald1 | Kazal-type serine peptidase inhibitor domain 1 | 208 | 24 |
| 3.90 | Chchd6 | Coiled-coil-helix-coiled-coil-helix domain containing 6 | 328 | 32 |
| 2.68 | Zfp521 | Zinc finger protein 521 | 379 | 125 |
| 2.46 | Tm2d2 | TM2 domain containing 2 | 382 | 184 |
| 2.45 | Fcgr2b | Fc receptor, IgG, low affinity IIb | 399 | 187 |
| −2.73† | Rad23a | RAD23a homolog (S cerevisiae) | 440 | 381 |
| 2.17 | Als2 | Amyotrophic lateral sclerosis 2 (juvenile) homolog (human) | 454 | 272 |
| 2.79 | Mfge8 | Milk fat globule-EGF factor 8 protein | 488 | 106 |
| 2.07 | Scap2 | src family associated phosphoprotein 2 | 510 | 309 |
| 2.45 | Rab19 | RAB19, member RAS oncogene family | 533 | 185 |
| 2.64 | 1500041B16Rik | RIKEN cDNA 1500041B16 gene | 586 | 133 |
| Fold difference HOXB4/GFP . | Gene symbol . | Gene title . | ChIP-chip rank (of 1910) . | RNA expression profiling rank (of 465) . |
|---|---|---|---|---|
| 8.39* | Gp49a | Glycoprotein 49 A | 29 | 4 |
| 2.50 | Rab38 | Rab38, member of RAS oncogene family | 68 | 169 |
| −2.53† | 2010301N04Rik | RIKEN cDNA 2010301N04 gene | 88 | 369 |
| 3.65 | Clec4e | C-type lectin domain family 4, member e | 95 | 38 |
| 9.82* | Laptm4b | Lysosomal-associated protein transmembrane 4B | 96 | 2 |
| 2.76 | AI586015 | Expressed sequence AI586015 | 122 | 111 |
| 2.19 | Arhgap15 | Rho GTPase activating protein 15 | 141 | 257 |
| 2.13 | Bmp2k | BMP2 inducible kinase | 162 | 284 |
| −2.14† | Rpl3 | Ribosomal protein L3/similar to 60S ribosomal protein L3 (J1 protein) | 188 | 331 |
| 4.26 | Kazald1 | Kazal-type serine peptidase inhibitor domain 1 | 208 | 24 |
| 3.90 | Chchd6 | Coiled-coil-helix-coiled-coil-helix domain containing 6 | 328 | 32 |
| 2.68 | Zfp521 | Zinc finger protein 521 | 379 | 125 |
| 2.46 | Tm2d2 | TM2 domain containing 2 | 382 | 184 |
| 2.45 | Fcgr2b | Fc receptor, IgG, low affinity IIb | 399 | 187 |
| −2.73† | Rad23a | RAD23a homolog (S cerevisiae) | 440 | 381 |
| 2.17 | Als2 | Amyotrophic lateral sclerosis 2 (juvenile) homolog (human) | 454 | 272 |
| 2.79 | Mfge8 | Milk fat globule-EGF factor 8 protein | 488 | 106 |
| 2.07 | Scap2 | src family associated phosphoprotein 2 | 510 | 309 |
| 2.45 | Rab19 | RAB19, member RAS oncogene family | 533 | 185 |
| 2.64 | 1500041B16Rik | RIKEN cDNA 1500041B16 gene | 586 | 133 |
An RNA expression profile rank of 321 or below are gene transcripts observed to be down-regulated at least 2-fold.
Gp49a and Laptm4b show up as highly ranked in both lists.
Down-regulated genes.
Discussion
Two genome-wide approaches, RNA expression profiling and ChIP-chip, were used to identify downstream target genes of HOXB4 in the multipotent hematopoietic progenitor cell line EML. Comparative analysis of the data from both types of gene chip studies provided novel insights on the function of HOXB4 in these cells. ChIP-chip analysis revealed that there was binding of HOXB4 to the promoter regions of lineage-specific genes, such as the lymphoid gene, B220, and the erythroid-specific isoform of HMBS in KLS-EML-HOXB4 cells (Figure 6). In addition, RNA expression profiling data showed that erythroid-specific genes were significantly down-regulated in KLS-EML-HOXB4 cells (Table 3). Kyba et al observed that HOXB4 overexpression in yolk sac-derived HPCs and mES cell-derived HPCs led to impaired lymphoid engraftment, as evidenced by a low percentage of donor-derived B220+ cells.35 These results, combined with our observation of fewer B220+ EML-HOXB4 cells, suggest that HOXB4 overexpression has a negative regulatory effect on lymphoid development. The finding that the promoter of the B220 gene is directly bound by HOXB4 in KLS-EML-HOXB4 is noteworthy because the KLS-EML-HOXB4 cells had been depleted of B220+ cells before ChIP-chip analysis. This suggests that HOXB4 binds to the promoter of the B220 gene in primitive hematopoietic cells and prevents up-regulation of the gene.
Previous work by Lavelle et al has shown that HOX gene products, such as HOXB2, bind to γ-globin gene-regulatory sequences.36,37 In vitro studies by Kyba et al35 and Pilat et al15 have demonstrated that erythroid cell differentiation is also down-regulated in mES cell-derived HPCs that overexpress HOXB4. These observations are consistent with our observation of marked down-regulation of erythroid-specific gene expression in KLS-EML-HOXB4 cells (Table 3). Despite the large number of erythroid-specific genes down-regulated in these cells, it is noteworthy that, with the exception of the erythroid-specific promoter of HMBS, the promoter regions of these genes were not shown to be bound by HOXB4 in our ChIP-chip experiments. These results suggest that the observed down-regulation of erythroid-specific gene expression was secondary to a phenomenon other than direct binding of HOXB4 to the promoter regions of these genes. This effect could be potentially mediated by the direct effect of HOXB4 on a “master regulator” gene that is not apparent from our studies or by binding of HOXB4 to other (nonpromoter) regulatory sequences of the affected erythroid-specific genes that were not present on the microarrays used in the ChIP-chip analyses. We also performed experiments using EML cells that overexpressed an estrogen receptor-inducible form of HOXB4. After a 4-hour induction of HOXB4 activity in these cells in the presence of tamoxifen and cycloheximide, we did not observe any significant changes in the mRNA levels encoded by the erythroid-specific genes that had been shown to be down-regulated in our RNA profiling experiments using EML cells constitutively overexpressing HOXB4. Thus, these erythroid-specific genes appear to be indirectly regulated by HOXB4. Interestingly, Laptm4b was identified as a directly up-regulated downstream target in these experiments (data not shown).
The effect on myeloid-specific gene expression in KLS-EML-HOXB4 cells could not be investigated thoroughly because of the nature of EML cells that express a dominant negative retinoic acid receptor blocking the myeloid differentiation program in these cells. However, it was possible, by adding ATRA to the culture medium, to overcome this block in KLS-EML-HOXB4 cells and allow them to differentiate to the EPRO cell stage. No differences were observed in the cell surface immunophenotype of EPRO-HOXB4 cells compared with that of control EPRO-GFP cells. Both cell types could also differentiate into mature neutrophils in the presence of ATRA. These results further support the notion that the effect of HOXB4 overexpression in hematopoietic cells does not irreversibly block their ability to differentiate normally.
Milsom et al investigated the effects of HOXB4 overexpression in myeloid differentiation more thoroughly using the IL-3-dependent, hematopoietic progenitor cell line, FDCP-mix.38 They observed that, when FDCP-mix cells were cultured in conditions that promote granulocyte/macrophage differentiation in vitro, cells that overexpressed HOXB4 differentiated with altered kinetics. After 3 days of culture under granulocyte/macrophage differentiation conditions, the percentage of blast cells present in FDCP-mix cells that overexpressed HOXB4 was much higher than in control FDCP-mix cells that overexpressed only GFP. Although FDCP-mix cells that overexpressed HOXB4 did differentiate into mature monocytes and granulocytes, a delay in differentiation was observed, suggesting that HOXB4 might have a negative effect on myeloid-specific gene regulation in the more primitive hematopoietic progenitor cells, but not in the more mature cells. Furthermore, a differentiation arrest was observed in HOXB4-transduced bone marrow cells that undergo in vitro self-renewal.39,40
A genome-wide approach was also used by Schiedlmeier et al to investigate downstream targets of HOXB4 in primitive (KLS) primary hematopoietic stem/progenitor cells.17 When the microarray results from their study were compared with our results, we observed that only 16 of the 156 downstream targets of HOX4 identified by Schiedlmeier et al17 were also shown to be differentially expressed in our RNA expression profiling experiments (supplemental Table 4). Furthermore, only one downstream target of HOXB4, Rasa2, was observed to be a direct downstream target of HOXB4 in both studies. The lack of overlap between the 2 studies can be attributed to the different experimental conditions used to identify the downstream targets. The method used by Schiedlmeier et al17 to identify downstream targets of HOXB4 was an inducible HOXB4/estrogen receptor system in which HOXB4 activity was induced in the presence of tamoxifen and the effects on gene expression analyzed 1 hour and 4 hours after withdrawal of tamoxifen, thereby identifying only relatively early downstream targets, whereas we used a constitutively active version of HOXB4 in our studies. The identification of direct downstream targets of HOXB4 by Schiedlmeier et al17 was based on the differential effect of cycloheximide on gene expression, rather than the use of a genome-wide approach, such as ChIP-chip. In addition, primary cells were used by Schiedlmeier et al,17 whereas we used a cell line where some genes involved in HSC self-renewal may have already been activated before HOXB4 was overexpressed. We think that these important differences in experimental conditions explain the very low degree of overlap between the 2 studies.
Side-by-side comparison of our ChIP-chip and RNA expression profiling datasets provided highly useful information. We identified 2 potential downstream targets (Gp49a and Laptm4b) that were highly ranked in both ChIP-chip and RNA expression profiling datasets (Table 4). These 2 genes have been shown by others to be preferentially expressed in long-term HSCs and down-regulated in more mature bone marrow cells.31 Laptm4b has been shown to be preferentially expressed in long-term HSCs by at least 4 other groups, and the gene is highly conserved across species.31,,–34,41 Laptm4b has also been shown to be up-regulated in various solid tumors (liver, lung, colon, uterus, breast, and ovary) as well as in mouse ES cells.42,43 It is thought to play a dual functional role during tumor progression by promoting tumor cell progression as well as multidrug resistance.44 Gp49a has been shown to be expressed on immature mast cells.31,45 Gp49a is a member of the large family of activating and inhibitory receptors of the immunoglobulin superfamily and is highly homologous to Gp49b (94% homology at nucleotide level).46 It has been suggested that Gp49a can be expressed as a homodimer and act as an activation signal for calcium mobilization, eicosanoid production, and cytokine gene transcription through its incorporation into the glycolipid-enriched membrane fraction. The exact functions of these 2 genes are still relatively unknown. Furthermore, these 2 genes have not yet been studied extensively in hematopoietic cells, and their role in early hematopoietic development remains to be determined.
Deneault et al recently identified novel effectors of HSC activity through an elegant gain-of-function screen that identified 18 nuclear factors that affect HSC function.40 Ten of these nuclear factors were observed to demonstrate HOXB4-like activity when overexpressed in HSCs. Interestingly, Sox4, which we identified as a direct downstream target of HOXB4 and is also up-regulated in HOXB4-overexpressing cells, was one of the genes that demonstrated HOXB4-like activity. These results suggest that Sox4 and HOXB4 might mediate HSC self-renewal through the same pathway.
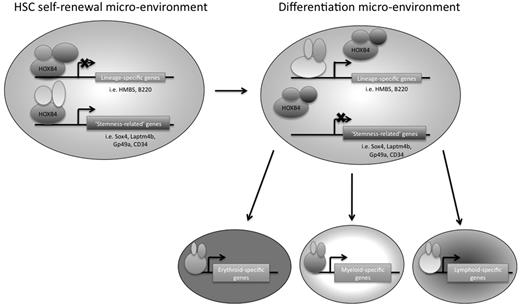
In conclusion, we have used 2 genome-wide approaches to identify downstream targets of HOXB4 in primitive hematopoietic progenitor cells, using the EML cell line as a model system. We think that this is the first study to use ChIP-chip technology to identify potential direct downstream target genes of HOXB4 in primitive hematopoietic progenitor cells (or in any mammalian cell type). HOXB4 overexpression had been previously shown by other groups to have an effect on hematopoietic lineage-specific gene regulation, and our RNA expression profiling data provide additional insights into which specific gene transcripts are affected. We suggest that, in the appropriate HSC microenvironment, HOXB4 might mediate HSC self-renewal by keeping the cells in a primitive state by down-regulating lineage-specific genes and directly up-regulating possible “stemness-related” genes, such as Laptm4b, Gp49a, Sox4, and CD34 (Figure 7). A better understanding of transcriptional regulatory networks in HSCs should provide new insights that could be helpful to the development of future novel stem cell-based therapies.
Model of HOXB4-mediated HSC self-renewal. HOXB4 may promote self-renewal in HSCs by down-regulating the expression of lineage-specific genes and up-regulating “stemness-related” genes. As HSCs encounter microenvironmental changes that are conducive to differentiation, the binding partners of HOXB4 may change, resulting in a reversal of HOXB4 function and/or binding specificity, such that lineage-specific genes that were once down-regulated by HOXB4 directly or indirectly would no longer be down-regulated. In addition, “stemness-related” genes that were once up-regulated by HOXB4 would now be down-regulated as the cells differentiate.
Model of HOXB4-mediated HSC self-renewal. HOXB4 may promote self-renewal in HSCs by down-regulating the expression of lineage-specific genes and up-regulating “stemness-related” genes. As HSCs encounter microenvironmental changes that are conducive to differentiation, the binding partners of HOXB4 may change, resulting in a reversal of HOXB4 function and/or binding specificity, such that lineage-specific genes that were once down-regulated by HOXB4 directly or indirectly would no longer be down-regulated. In addition, “stemness-related” genes that were once up-regulated by HOXB4 would now be down-regulated as the cells differentiate.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Note added in proof:
In this issue of Blood (2010;116(5):711-719), Jiang et al report the results of their studies of downstream targets of induced expression of HOXB4 in primary Lin− bone marrow progenitor cells. One of the targets identified was the gene Hemgn, encoding hemogen, a hematopoietic-specific nuclear protein of unknown function. Retrospective analysis of our ChIP-chip results revealed that the promoter region of Hemgn was bound by HOXB4 in KLS (c-Kit+, Lin−, Sca-1+)—EML cells constitutively overexpressing HOXB4, but the amount of binding was less than that observed in the case of more than 2000 other genes. The results of our microarray analyses revealed that there was no significant increase in the mRNA expression level of Hemgn in HOXB4 overexpressing KLS-EML cells compared with that in control cells. However, it should be noted that differences in the results of these 2 studies could be accounted for by a number of factors, such as the different cellular systems used (primary bone marrow progenitor cells vs an immortalized primitive hematopoietic cell line), as well as differences in the experimental approaches, including study of constitutive versus induced expression of HOXB4 in the target cells and analysis of cells with slightly different cell surface markers.
Acknowledgments
The authors thank Mrs Svetlana Rogulina for skilled technical assistance.
This work was supported in part by the National Institutes of Health (P01-HL63357, B.G.F.; K08-DK064809, H.Z.; T32-HD07149, H.M.L.; T32-HL07262, H.M.L.), Yale Comprehensive Cancer Center (P30-CA016359), and the Yale Center of Excellence in Molecular Hematology (P30-DK72442).
National Institutes of Health
Authorship
Contribution: H.M.L. designed and performed the research, analyzed the data, and wrote the manuscript; H.Z. helped design the research and perform flow cytometric experiments; V.S. and D.P.T. analyzed the gene chip data; and B.G.F. designed the research, analyzed the data, and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bernard G. Forget, Yale University School of Medicine, Departments of Internal Medicine and Genetics, 333 Cedar St, PO Box 208021, New Haven, CT 06520-8021; e-mail: bernard.forget@yale.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal