In a proportion of patients with chronic myeloid leukemia (CML) being treated with dasatinib, we recently observed large granular lymphocyte (LGL) expansions carrying clonal T-cell receptor (TCR) γ/δ gene rearrangements. To assess the prevalence and role of clonal lymphocytes in CML, we collected samples from patients (n = 34) at the time of diagnosis and during imatinib and dasatinib therapies and analyzed lymphocyte clonality with a sensitive polymerase chain reaction–based method of TCR γ and δ genes. Surprisingly, at CML diagnosis, 15 of 18 patients (83%) had a sizeable clonal, BCR-ABL1 negative lymphocyte population, which was uncommon in healthy persons (1 of 12; 8%). The same clone persisted at low levels in most imatinib-treated patients. In contrast, in a distinct population of dasatinib-treated patients, the diagnostic phase clone markedly expanded, resulting in absolute lymphocytosis in blood. Most patients with LGL expansions (90%) had TCR δ rearrangements, which were uncommon in patients without an LGL expansion (10%). The TCR δ clones were confined to γδ+ T- or natural killer–cell compartments and the TCR γ clones to CD4+/CD8+ αβ+ fractions. The functional importance of clonal lymphocytes as a part of leukemia immune surveillance and the putative anergy-reversing role of dasatinib require further evaluation.
Introduction
Chronic myeloid leukemia (CML) is a hematopoietic stem cell disorder characterized by Philadelphia chromosome (Ph) leading to the formation of oncogenic BCR-ABL1 fusion gene and autonomously active tyrosine kinase enzyme. Current therapy for CML is based on tyrosine kinase inhibitor (TKI) treatment with a favorable outcome both in the first-line setting with imatinib (Glivec; Novartis)1,2 and in the second-line setting with dasatinib (Sprycel; Bristol-Myers Squibb)3,4 or nilotinib (Tasigna; Novartis).5 In addition to the intended target kinase, BCR-ABL1, second-generation TKIs (dasatinib in particular) also inhibit a wide variety of other kinases such as SRC and TEC kinases6,–8 known to be key regulators of immune responses. Recent in vitro studies have shown that dasatinib,9,,,–13 imatinib,14,,–17 and nilotinib18 have inhibitory effects on T-cell proliferation and activation. However, the effects of dasatinib were found to be more profound, probably through stronger SRC-kinase inhibition. The drug blocked T-cell proliferation, activation, cell cycle, secretion of various proinflammatory cytokines and interrupted signaling through T-cell receptor (TCR). Dasatinib did not, however, induce apoptosis; therefore, the T-cell viability was unaffected.9,11,12,16,19 In patients, no clinical signs of immunosuppression have been observed during imatinib treatment to date. One earlier study reported a modestly increased incidence of varicella-zoster virus infections during imatinib treatment.20 In the setting after allogeneic stem cell transplantation, imatinib treatment may even have an additive effect to the donor lymphocyte infusions.21 Similarly, in large phase 2/3 clinical studies with dasatinib no increased rates of infections were observed.22,23 However, a recent case report suggested that in some patients with advanced CML, high-dose dasatinib therapy may be associated with opportunistic infections due to Epstein-Barr virus and Pneumocystis jiroveci.24
Recent findings from our and other groups have shown that, in a distinct subgroup of patients with CML and Ph+ acute lymphoblastic leukemia, dasatinib therapy is associated with a chronic monoclonal/oligoclonal large granular lymphocyte (LGL) lymphocytosis in peripheral blood.25,26 Dasatinib-associated lymphocytosis (peak lymphocyte count ranging from 4 to 20 × 109/L) developed at an average of 3 months after the start of treatment and persisted throughout therapy. The expanded lymphocytes had either a CD3+CD8+ effector memory T-cell or a CD3negCD16/CD56+ natural killer (NK)–cell phenotype. Strikingly, the clonal LGL lymphocytosis was associated with exceptional, durable therapy responses in patients with advanced, poor-prognosis Ph+ leukemia.
Monoclonal and oligoclonal T-cell expansions are common during a primary immune response against viral antigens (eg, HIV, cytomegalovirus [CMV]).27,–29 Clonal expansions of cytotoxic cells have also been observed in the context of hematologic malignancies, such as multiple myeloma,30 myelodysplastic syndrome,31 and acute promyelocytic leukemia,32 and have been linked to cancer immune surveillance. Small numbers of clonal and persistent CD4+ and CD8+ cells have been shown to exist in healthy persons and become more common with advanced age. This phenomenon has been attributed to dysregulated systemic immunity in the elderly (immunosenescence).33
Prevalence, detailed molecular background, and clinical implications of clonal lymphocytes in CML are currently unknown, as well as their association to TKI therapy. The aim of this study was to comprehensively analyze lymphocyte clonality and evolution of clones in patients with CML at diagnosis and during TKI therapy. Unexpectedly, clonal lymphocytes were observed in most patients with CML already at diagnosis, they persisted during imatinib therapy at low levels, and expanded during dasatinib therapy. On the basis of the association of clonal LGL expansion to favorable disease outcome, we hypothesize that, by inhibiting distinct off-target kinases in immune effector cells, dasatinib may restore the function of anergic, exhausted leukemia-specific cytotoxic lymphocytes present already at the time of CML diagnosis.
Methods
Study patients and samples
The study population included patients with CML (n = 34) on dasatinib (n = 20) and imatinib (n = 14) therapies. All patients with CML treated with dasatinib from our center (Helsinki University Central Hospital) were included in the study together with 5 additional patients on dasatinib therapy from other centers. Similarly, all patients with CML treated with imatinib from whom we had both diagnostic phase and follow-up samples available were included in the study. Of the 20 patients treated with dasatinib, 10 patients had lymphocyte expansion with LGL morphology during therapy (peak count range, 4-20 × 109/L). The main characteristics of the patients are summarized in Table 1. Blood cell counts, differential analysis of leukocytes, and the presence of antibodies to CMV in the blood (CMV serology) were obtained from routine laboratory tests.
Patient characteristics (n = 34)
| Patient no. . | Diagnosis . | Age, y* . | Sex . | Therapy . | Response . | Duration, mo . | LGL lymphocytosis . | CMV serology . |
|---|---|---|---|---|---|---|---|---|
| 1 | CML BC | 62 | F | Dasatinib | CMR | 16 | Yes | Pos |
| 2 | CML BC | 43 | M | Dasatinib | CMR | 45 | Yes | Pos |
| 3 | CML AP | 74 | F | Dasatinib | CHR | 57† | Yes | Pos |
| 4 | CML CP | 53 | F | Dasatinib | CCgR | 4 | Yes | Pos |
| 5 | CML CP | 65 | F | Dasatinib | MMR | 36† | Yes | Pos |
| 6 | CML CP | 58 | F | Dasatinib | CMR | 48 | Yes | Pos |
| 7 | CML CP | 61 | F | Dasatinib | CCgR | 12† | Yes | ND |
| 8 | CML CP | 52 | M | Dasatinib | CMR | 7 | Yes | Pos |
| 9 | CML CP | 60 | M | Dasatinib | CMR | 39 | Yes | Pos |
| 10 | CML CP | 76 | M | Dasatinib | CMR | 59 | Yes | Pos |
| 11 | CML BC | 68 | F | Dasatinib | PHR | 4 | No | ND |
| 12 | CML CP | 29 | F | Dasatinib | CMR | 58† | No | Pos |
| 13 | CML CP | 64 | F | Dasatinib | MMR | 37† | No | Neg |
| 14 | CML CP | 51 | F | Dasatinib | CMR | 41 | No | ND |
| 15 | CML CP | 27 | M | Dasatinib | MMR | 22 | No | Neg |
| 16 | CML CP | 40 | M | Dasatinib | MMR | 54† | No | Neg |
| 17 | CML CP | 45 | M | Dasatinib | CCgR | 12† | No | Pos |
| 18 | CML CP | 49 | M | Dasatinib | CCgR | 57† | No | Pos |
| 19 | CML CP | 61 | M | Dasatinib | CMR | 25† | No | Neg |
| 20 | CML CP | 78 | M | Dasatinib | CCgR | 4 | No | Pos |
| 21 | CML CP | 22 | F | Imatinib | CMR | 31 | No | Pos |
| 22 | CML CP | 53 | F | Imatinib | CMR | 27 | No | Pos |
| 23 | CML CP | 66 | F | Imatinib | CMR | 48† | No | Pos |
| 24 | CML CP | 36 | M | Imatinib | CCgR | 60† | No | Pos |
| 25 | CML CP | 40 | M | Imatinib | MMR | 49† | No | Neg |
| 26 | CML CP | 43 | M | Imatinib | CCgR | 27† | No | Pos |
| 27 | CML CP | 48 | M | Imatinib | CMR | 52† | No | Pos |
| 28 | CML CP | 49 | M | Imatinib | CMR | 58† | No | ND |
| 29 | CML CP | 49 | M | Imatinib | MMR | 53† | No | ND |
| 30 | CML CP | 52 | M | Imatinib | CCgR | 26† | No | Pos |
| 31 | CML CP | 59 | M | Imatinib | CMR | 16 | No | Neg |
| 32 | CML CP | 58 | M | Imatinib | CMR | 51† | No | Pos |
| 33 | CML CP | 61 | M | Imatinib | MMR | 25† | No | Pos |
| 34 | CML CP | 59 | M | Imatinib | MMR | 63† | No | Pos |
| Patient no. . | Diagnosis . | Age, y* . | Sex . | Therapy . | Response . | Duration, mo . | LGL lymphocytosis . | CMV serology . |
|---|---|---|---|---|---|---|---|---|
| 1 | CML BC | 62 | F | Dasatinib | CMR | 16 | Yes | Pos |
| 2 | CML BC | 43 | M | Dasatinib | CMR | 45 | Yes | Pos |
| 3 | CML AP | 74 | F | Dasatinib | CHR | 57† | Yes | Pos |
| 4 | CML CP | 53 | F | Dasatinib | CCgR | 4 | Yes | Pos |
| 5 | CML CP | 65 | F | Dasatinib | MMR | 36† | Yes | Pos |
| 6 | CML CP | 58 | F | Dasatinib | CMR | 48 | Yes | Pos |
| 7 | CML CP | 61 | F | Dasatinib | CCgR | 12† | Yes | ND |
| 8 | CML CP | 52 | M | Dasatinib | CMR | 7 | Yes | Pos |
| 9 | CML CP | 60 | M | Dasatinib | CMR | 39 | Yes | Pos |
| 10 | CML CP | 76 | M | Dasatinib | CMR | 59 | Yes | Pos |
| 11 | CML BC | 68 | F | Dasatinib | PHR | 4 | No | ND |
| 12 | CML CP | 29 | F | Dasatinib | CMR | 58† | No | Pos |
| 13 | CML CP | 64 | F | Dasatinib | MMR | 37† | No | Neg |
| 14 | CML CP | 51 | F | Dasatinib | CMR | 41 | No | ND |
| 15 | CML CP | 27 | M | Dasatinib | MMR | 22 | No | Neg |
| 16 | CML CP | 40 | M | Dasatinib | MMR | 54† | No | Neg |
| 17 | CML CP | 45 | M | Dasatinib | CCgR | 12† | No | Pos |
| 18 | CML CP | 49 | M | Dasatinib | CCgR | 57† | No | Pos |
| 19 | CML CP | 61 | M | Dasatinib | CMR | 25† | No | Neg |
| 20 | CML CP | 78 | M | Dasatinib | CCgR | 4 | No | Pos |
| 21 | CML CP | 22 | F | Imatinib | CMR | 31 | No | Pos |
| 22 | CML CP | 53 | F | Imatinib | CMR | 27 | No | Pos |
| 23 | CML CP | 66 | F | Imatinib | CMR | 48† | No | Pos |
| 24 | CML CP | 36 | M | Imatinib | CCgR | 60† | No | Pos |
| 25 | CML CP | 40 | M | Imatinib | MMR | 49† | No | Neg |
| 26 | CML CP | 43 | M | Imatinib | CCgR | 27† | No | Pos |
| 27 | CML CP | 48 | M | Imatinib | CMR | 52† | No | Pos |
| 28 | CML CP | 49 | M | Imatinib | CMR | 58† | No | ND |
| 29 | CML CP | 49 | M | Imatinib | MMR | 53† | No | ND |
| 30 | CML CP | 52 | M | Imatinib | CCgR | 26† | No | Pos |
| 31 | CML CP | 59 | M | Imatinib | CMR | 16 | No | Neg |
| 32 | CML CP | 58 | M | Imatinib | CMR | 51† | No | Pos |
| 33 | CML CP | 61 | M | Imatinib | MMR | 25† | No | Pos |
| 34 | CML CP | 59 | M | Imatinib | MMR | 63† | No | Pos |
LGL indicates large granular lymphocyte; CMV, cytomegalovirus; BC, blast crisis; CMR, complete molecular response; AP, accelerated phase; CHR, complete hematologic response; CP, chronic phase; CCgR, complete cytogenetic response; MMR, major molecular response; ND, not done; and PHR, partial hematologic response.
Age at the time of first clonality analysis.
Ongoing therapy.
In addition, 12 healthy volunteers were included as a control group. This group consisted of 6 men (median age, 47 years; range, 28-66 years) and 6 women (median age, 45 years; range, 26-61 years). Provided that the blood counts were normal and with no symptoms of disease, the results from TCR rearrangement assays were not given to the participating volunteers, to prevent unnecessary anxiety from a putative finding of occult clonal lymphocytes with undetermined significance.
The study was conducted in accordance with the principles of the Declaration of Helsinki and was approved by the Helsinki University Central Hospital Ethics Committee. Written informed consents were obtained from all patients and healthy controls, either as part of clinical drug studies or separately.
Separation of mononuclear cells
Peripheral blood (PB) or bone marrow samples were collected from patients at diagnosis and/or during TKI treatment. PB and bone marrow mononuclear cells (MNCs) were separated with Ficoll gradient centrifugation (GE Healthcare). MNCs were stored as cell pellets in −80°C or in liquid nitrogen.
Selection of CD8+ cells
For selection of CD8+ cells, the PB MNCs were labeled with CD8 MicroBeads (Miltenyi Biotec) according to instructions provided by the manufacturer and separated with an AutoMACS cell sorter (Miltenyi Biotec).
Fluorescence-activated cell sorting
Approximately 10 million PB MNCs were stained with antibodies against the following antigens: CD45, CD3, CD8, CD16/CD56. Gates were set on live and CD45+ cells. Different lymphocyte populations were gated and sorted as follows: CD3+CD8neg (CD4+ T cells), CD3+CD8+ (CD8+ T cells), CD3negCD16/56+ (NK cells), and CD3negCD16/56neg (B cells) with FACSAria (Becton Dickinson). TCR αβ+ and TCR γδ+ T cells were stained with antibodies against the following antigens: CD45, CD3, TCR αβ, and TCR γδ. Lymphocyte populations were gated and sorted as the following: CD45+ CD3+ TCR αβ+, CD45+ CD3+ TCR αβ+, and CD45+ CD3neg. All antibodies were from Becton Dickinson. Purity of the sorted fractions was confirmed with flow cytometry to be close to 100%. Sorting strategy and purity of sorted cell fractions are shown in Figure 1.
Strategy of FACS of different lymphocyte populations and purity of sorted populations. (A-G) FACS of a patient with T-cell LGL lymphocytosis (patient no. 2; Table 1) and (H-J) of a patient with NK-cell lymphocytosis (patient no. 6; Table 1). To sort lymphocyte subpopulations, PB MNCs were stained with antibodies against the following antigens: CD45, CD3, CD8, CD16/CD56, TCR αβ, and TCR γδ. Gates were first set on live and CD45+ cells (gray). Then gating of NK- and T-cell populations were done from CD16/CD56 versus CD3 scatter (A,H). CD3+ T cells were further sorted into CD8neg and CD8+ populations (B) and into TCRαβ+ and TCRγδ+ T cells (E). Purity of respective sorted populations is presented (C,D,F,G,I,J).
Strategy of FACS of different lymphocyte populations and purity of sorted populations. (A-G) FACS of a patient with T-cell LGL lymphocytosis (patient no. 2; Table 1) and (H-J) of a patient with NK-cell lymphocytosis (patient no. 6; Table 1). To sort lymphocyte subpopulations, PB MNCs were stained with antibodies against the following antigens: CD45, CD3, CD8, CD16/CD56, TCR αβ, and TCR γδ. Gates were first set on live and CD45+ cells (gray). Then gating of NK- and T-cell populations were done from CD16/CD56 versus CD3 scatter (A,H). CD3+ T cells were further sorted into CD8neg and CD8+ populations (B) and into TCRαβ+ and TCRγδ+ T cells (E). Purity of respective sorted populations is presented (C,D,F,G,I,J).
DNA extraction
Genomic DNA was isolated from fresh, frozen, or sorted MNCs by Blood DNA isolation Kit (MO BIO) or Genomic DNA from Tissue; NucleoSpin Tissue XS (Machery-Nagel). DNA concentration and purity were measured with NanoDrop (Thermo Scientific). DNA was stored at −20°C.
Fluorescence in situ hybridization
Fluorescence in situ hybridization (FISH) analyses were performed on sorted PB CD4+, CD8+, NK, and B cells from 2 patient in diagnostic phase (patients 7 and 17; Table 1). Analyses were done on interphase cells prepared on cytospin slides with the use of locus-specific dual-color, dual-fusion BCR-ABL probe mixture (Vysis). Hybridizations were performed according to the manufacturer's instructions.
Detection of TCR γ and δ gene rearrangements by polymerase chain reaction
TCR γ and δ gene rearrangements were studied by polymerase chain reaction (PCR) analysis with the use of 12 primer pairs for γ gene, and 6 primer pairs for δ genes allowing detection of most of the known TCR γ and δ gene rearrangements. The analysis was conducted according to the BIOMED-1 PCR protocol.34 PCR was performed in the PTC-200 Peltier Thermal Cycler (Bio-Rad).
After PCR amplification of the TCR δ and γ genes, the clonality was confirmed by heteroduplex analysis. PCR products were denaturated at 95°C for 5 minutes, followed by a rapid random renaturation at 4°C for 1 hour. PCR products were separated by electrophoresis on a commercial 5% Criterion polyacrylamide gel (Bio-Rad) in 1× [Tris (tris(hydroxymethyl) aminomethane) borate EDTA (ethylenediaminetetraacetic acid)] buffer. Gels were stained with EtBr and visualized with ultraviolet illumination. Positive findings were cut from the gel and if necessary reamplified with the same set of primers as were used before sequencing. Lower detection limit of the assay is 1% to 5% of clonal cells among polyclonal lymphocytes.
Sequencing of the junctions in TCR γ and δ gene rearrangements
Clonal products were sequenced with BigDye Version 1.1 Cycle Sequencing kit (Applied Biosystems).
Sequencing was performed in the ABI PRISM 310 Genetic Analyzer and sequences were analyzed with the use of the IMGT Database35 (http://www.imgt.org/) and the Blast search.36
Unequivocal sequencing and junction region analysis were obtained for all clonal products, except for 4 clonal TCR γ products that presented as single clonal products on polyacrylamide gel electrophoresis gels, but sequencing disclosed a chromatogram pattern in the junction region compatible with a biallelic product. Determination of functionality for these TCR γ rearrangements thus was not possible.
Design of individual allele-specific oligonucleotide primers and quantification of clones with real-time quantitative PCR
Patient-specific allele-specific oligonucleotide (ASO) primers for real-time quantitative PCR (RQ-PCR) were designed with Primer Express software (Applied Biosystems). For each patient, if possible, 2 targets (TCR γ and TCR δ) were selected for ASO-PCR purposes. ASO primers were designed to match the hypervariable junction regions of the sequenced TCR δ and TCR γ gene rearrangements. Forward ASO primers were used in TaqMan RQ-PCR analysis in conjunction with a consensus reverse primer and a TaqMan probe selected according to the D or J gene present in the rearrangement (reverse primer and probe sequences available from the authors on request). DNA was amplified in triplicate reactions with the use of TaqMan Universal PCR Mastermix (Applied Biosystems) and the individual ASO primer with a consensus primer and a TaqMan probe. RQ-PCR was performed in the 7500 Real Time PCR system (Applied Biosystems), and the thermocycling conditions consisted of an initial step at 50°C for 2 minutes and denaturation step at 95°C for 10 minutes, followed by 50 cycles at 95°C for 15 seconds and at 60°C for 60 seconds.
Standard curves were prepared by diluting a patient DNA sample in polyclonal DNA from healthy donors. A sample with good quality and high amount of DNA, and with confirmed presence of the clonal rearrangement, was used as a reference sample. The ratio of the specific clonally rearranged DNA relative to albumin DNA as measured by RQ-PCR was defined to be 1.0 in the reference samples. The reference sample dilution series for the RQ-PCR standard curve was prepared by 10-fold dilutions. Thus, the amount of DNA with the specific clonal rearrangement relative to the amount of albumin DNA could be measured in all follow-up samples with the use of the reference sample as standard. The measurement scales were completely individual for each clone and for each patient. The purpose of these measurements was to detect relative changes in serial measurements.
The sensitivity of each allele-specific RQ-PCR analysis was estimated from the reference sample dilution series. Sensitivity was defined to be the lowest dilution still giving specific amplification. When determined this way the sensitivity estimate depends on 3 factors: (1) quality of DNA in the reference sample, (2) efficiency of each allele-specific RQ-PCR reaction, and (3) the unknown proportion of clonal cells representing that specific clone in the reference sample. With standard DNA extraction and standard rules for primer design the factors (1) and (2) remain similar for different allele-specific PCR targets. Therefore, a high sensitivity would suggest a high proportion of clonally rearranged cells in the reference sample, whereas a poor sensitivity suggests a low proportion of cells with specific clonal rearrangement in the reference sample.
Results
BCR-ABL1–negative lymphocyte clones are common in patients with CML at diagnosis
Clonality of the lymphocytes was determined by PCR and gel analysis with the use of 12 primer pairs for the TCR γ gene rearrangements and 6 primer pairs for TCR δ gene rearrangements. If a positive product was observed in the gel (Figure 2), it was confirmed by sequencing. Of 12 healthy controls studied, only one female had a sequencing-confirmed clonal TCR γ gene rearrangement. This healthy person was asymptomatic and had normal blood counts. No clonal TCR δ gene rearrangements were detected in any healthy volunteers. In contrast, 15 of 18 (83%) patients with chronic phase CML showed clonal TCR rearrangements at the time of diagnosis. Ten patients had clonal rearrangements only in the TCR γ gene, whereas 5 patients had rearrangements both in TCR γ and δ genes, with the latter being a minority (18% of all rearrangements detected) (Table 2). Most patients displayed more than one clonal TCR rearrangement. The sensitivity of the assay is 1% to 5% of clonal cells, indicating that a significant proportion of diagnostic phase lymphocytes are clonal in untreated patients with CML. In 3 patients no clonal rearrangements could be detected at diagnosis with the set of primers used in this study.
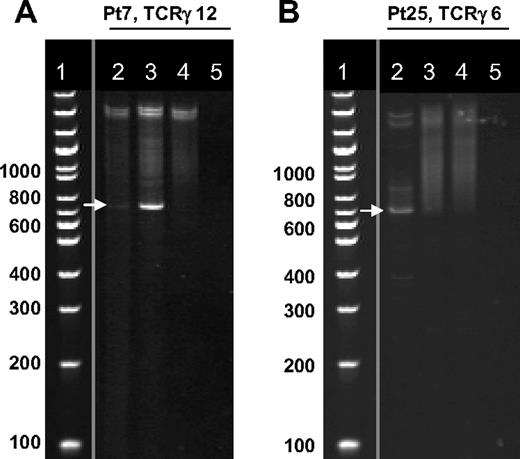
TCRγ rearrangements at the diagnosis and during TKI therapy. Clonality of the lymphocytes was determined by PCR and gel analysis with the use of 12 primer pairs for the TCR γ gene rearrangements and 6 primer pairs for TCR δ gene rearrangements. After PCR amplification and heteroduplex treatment, the PCR products were separated by electrophoresis on a 5% Criterion gel. Gels were stained with EtBr and visualized with ultraviolet illumination. (A) Dasatinib-treated patient with LGL lymphocytosis showing clonal PCR products (white arrow) of the primer pair TCRγ 12 at diagnosis and during dasatinib therapy. Lane 2 indicates at diagnosis; lane 3, 6 months after start of dasatinib therapy during lymphocytosis; lane 4, pool of healthy controls showing polyclonal smear; lane 5, water control. (B) Imatinib-treated patient without lymphocytosis showing clonal PCR product (white arrow) of the primer pair TCRγ 6 at diagnosis. After 12 months of imatinib therapy, no clonal band was visible. Lane 2 indicates at diagnosis; lane 3, 12 months after start of imatinib therapy; lane 4, pool of healthy controls showing polyclonal smear; lane 5, water control. Positive findings were confirmed with sequencing. Vertical lines have been inserted to indicate a repositioned gel lane (DNA ladder).
TCRγ rearrangements at the diagnosis and during TKI therapy. Clonality of the lymphocytes was determined by PCR and gel analysis with the use of 12 primer pairs for the TCR γ gene rearrangements and 6 primer pairs for TCR δ gene rearrangements. After PCR amplification and heteroduplex treatment, the PCR products were separated by electrophoresis on a 5% Criterion gel. Gels were stained with EtBr and visualized with ultraviolet illumination. (A) Dasatinib-treated patient with LGL lymphocytosis showing clonal PCR products (white arrow) of the primer pair TCRγ 12 at diagnosis and during dasatinib therapy. Lane 2 indicates at diagnosis; lane 3, 6 months after start of dasatinib therapy during lymphocytosis; lane 4, pool of healthy controls showing polyclonal smear; lane 5, water control. (B) Imatinib-treated patient without lymphocytosis showing clonal PCR product (white arrow) of the primer pair TCRγ 6 at diagnosis. After 12 months of imatinib therapy, no clonal band was visible. Lane 2 indicates at diagnosis; lane 3, 12 months after start of imatinib therapy; lane 4, pool of healthy controls showing polyclonal smear; lane 5, water control. Positive findings were confirmed with sequencing. Vertical lines have been inserted to indicate a repositioned gel lane (DNA ladder).
Clonal TCR rearrangements in patients with CML at diagnosis and during follow-up samples (n = 18)
| Patient no. . | Date, ddmmmyy . | TCR δ primer pairs . | TCR γ primer pairs . | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | 9 . | 10 . | 11 . | 12 . | ||
| 5 | |||||||||||||||||||
| Diagnosis | 03MAR05 | X* | X* | ||||||||||||||||
| Dasatinib | 11FEB08 | X* | X* | X | |||||||||||||||
| 7 | |||||||||||||||||||
| Diagnosis | 31MAR09 | X* | X | X | X* | ||||||||||||||
| Dasatinib | 29SEP09 | X* | X* | ||||||||||||||||
| 15 | |||||||||||||||||||
| Diagnosis | 18AUG05 | ||||||||||||||||||
| Dasatinib | 24JUL08 | ||||||||||||||||||
| 17 | |||||||||||||||||||
| Diagnosis | 30MAR09 | X | |||||||||||||||||
| Dasatinib | 29SEP09 | X | X | ||||||||||||||||
| 21 | |||||||||||||||||||
| Diagnosis | 12MAR07 | X* | |||||||||||||||||
| Imatinib | 14JAN08 | X* | |||||||||||||||||
| 22 | |||||||||||||||||||
| Diagnosis | 15NOV06 | X | X | X* | |||||||||||||||
| Imatinib | 15OCT07 | X* | |||||||||||||||||
| 23 | |||||||||||||||||||
| Diagnosis | 04OCT05 | X | |||||||||||||||||
| Imatinib | 01MAR06 | ||||||||||||||||||
| 24 | |||||||||||||||||||
| Diagnosis | 03FEB06 | ||||||||||||||||||
| Imatinib | 13SEP06 | ||||||||||||||||||
| 25 | |||||||||||||||||||
| Diagnosis | 26JAN06 | X | X | ||||||||||||||||
| Imatinib | 01FEB07 | ||||||||||||||||||
| 26 | |||||||||||||||||||
| Diagnosis | 04DEC07 | ||||||||||||||||||
| Imatinib | 20NOV08 | ||||||||||||||||||
| 27 | |||||||||||||||||||
| Diagnosis | 13OCT05 | X | |||||||||||||||||
| Imatinib | 12FEB07 | X | |||||||||||||||||
| 28 | |||||||||||||||||||
| Diagnosis | 24MAY05 | X | |||||||||||||||||
| Imatinib | 04OCT07 | X | |||||||||||||||||
| 29 | |||||||||||||||||||
| Diagnosis | 20JUL05 | X | |||||||||||||||||
| Imatinib | 13MAR07 | ||||||||||||||||||
| 30 | |||||||||||||||||||
| Diagnosis | 11JAN07 | X | X* | ||||||||||||||||
| Imatinib | 17APR08 | X* | X | X | |||||||||||||||
| 31 | |||||||||||||||||||
| Diagnosis | 14FEB07 | X* | |||||||||||||||||
| Imatinib | 07JAN08 | X* | |||||||||||||||||
| 32 | |||||||||||||||||||
| Diagnosis | 14SEP05 | X* | X | ||||||||||||||||
| Imatinib | 20FEB07 | X* | |||||||||||||||||
| 33 | |||||||||||||||||||
| Diagnosis | 19FEB07 | X | X | X* | |||||||||||||||
| Imatinib | 30JUN08 | X* | |||||||||||||||||
| 34 | |||||||||||||||||||
| Diagnosis | 28SEP04 | X | X | X* | |||||||||||||||
| Imatinib | 14SEP05 | X* | |||||||||||||||||
| Patient no. . | Date, ddmmmyy . | TCR δ primer pairs . | TCR γ primer pairs . | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | 9 . | 10 . | 11 . | 12 . | ||
| 5 | |||||||||||||||||||
| Diagnosis | 03MAR05 | X* | X* | ||||||||||||||||
| Dasatinib | 11FEB08 | X* | X* | X | |||||||||||||||
| 7 | |||||||||||||||||||
| Diagnosis | 31MAR09 | X* | X | X | X* | ||||||||||||||
| Dasatinib | 29SEP09 | X* | X* | ||||||||||||||||
| 15 | |||||||||||||||||||
| Diagnosis | 18AUG05 | ||||||||||||||||||
| Dasatinib | 24JUL08 | ||||||||||||||||||
| 17 | |||||||||||||||||||
| Diagnosis | 30MAR09 | X | |||||||||||||||||
| Dasatinib | 29SEP09 | X | X | ||||||||||||||||
| 21 | |||||||||||||||||||
| Diagnosis | 12MAR07 | X* | |||||||||||||||||
| Imatinib | 14JAN08 | X* | |||||||||||||||||
| 22 | |||||||||||||||||||
| Diagnosis | 15NOV06 | X | X | X* | |||||||||||||||
| Imatinib | 15OCT07 | X* | |||||||||||||||||
| 23 | |||||||||||||||||||
| Diagnosis | 04OCT05 | X | |||||||||||||||||
| Imatinib | 01MAR06 | ||||||||||||||||||
| 24 | |||||||||||||||||||
| Diagnosis | 03FEB06 | ||||||||||||||||||
| Imatinib | 13SEP06 | ||||||||||||||||||
| 25 | |||||||||||||||||||
| Diagnosis | 26JAN06 | X | X | ||||||||||||||||
| Imatinib | 01FEB07 | ||||||||||||||||||
| 26 | |||||||||||||||||||
| Diagnosis | 04DEC07 | ||||||||||||||||||
| Imatinib | 20NOV08 | ||||||||||||||||||
| 27 | |||||||||||||||||||
| Diagnosis | 13OCT05 | X | |||||||||||||||||
| Imatinib | 12FEB07 | X | |||||||||||||||||
| 28 | |||||||||||||||||||
| Diagnosis | 24MAY05 | X | |||||||||||||||||
| Imatinib | 04OCT07 | X | |||||||||||||||||
| 29 | |||||||||||||||||||
| Diagnosis | 20JUL05 | X | |||||||||||||||||
| Imatinib | 13MAR07 | ||||||||||||||||||
| 30 | |||||||||||||||||||
| Diagnosis | 11JAN07 | X | X* | ||||||||||||||||
| Imatinib | 17APR08 | X* | X | X | |||||||||||||||
| 31 | |||||||||||||||||||
| Diagnosis | 14FEB07 | X* | |||||||||||||||||
| Imatinib | 07JAN08 | X* | |||||||||||||||||
| 32 | |||||||||||||||||||
| Diagnosis | 14SEP05 | X* | X | ||||||||||||||||
| Imatinib | 20FEB07 | X* | |||||||||||||||||
| 33 | |||||||||||||||||||
| Diagnosis | 19FEB07 | X | X | X* | |||||||||||||||
| Imatinib | 30JUN08 | X* | |||||||||||||||||
| 34 | |||||||||||||||||||
| Diagnosis | 28SEP04 | X | X | X* | |||||||||||||||
| Imatinib | 14SEP05 | X* | |||||||||||||||||
Clonality of the lymphocytes was determined by PCR and gel analysis with the use of 12 primer pairs for the TCR γ gene rearrangements and 6 primer pairs for TCR δ gene rearrangements. Positive clonal PCR products were confirmed with the sequencing and are marked in the columns as X.
Identical rearrangements at the diagnosis and follow-up sample.
Lymphocyte clones detected at diagnosis persist during TKI therapy
From 15 patients with CML, who had clonal lymphocytes at diagnosis, a follow-up sample during dasatinib or imatinib therapy was available for analysis (Table 2). TCR γ and δ gene rearrangements were searched from the follow-up samples with the use of a similar approach as from the diagnostic phase samples (PCR and gel analysis followed by confirmation of positive PCR products with sequencing). All 3 patients taking dasatinib and 10 of 12 patients taking imatinib (83%) who had clonal lymphocytes at diagnosis also had confirmed clonal TCR γ/δ rearrangements during TKI therapy (Figure 2; Table 2). In 9 of 13 cases (69%) the clone found in the follow-up sample was identical with the diagnostic phase clone (Table 2). No clonal δ gene rearrangements could be detected during imatinib therapy. None of the patients taking imatinib with lymphocyte clones had increased absolute lymphocyte counts during therapy.
In 3 patients without any confirmed clonal TCR γ/δ rearrangements at diagnosis (patients 15, 24, and 26; Table 2), no clonal products were found in samples during TKI treatment either.
Analysis of TCR γ and TCR δ gene rearrangements in patients treated with dasatinib patients
In our previous study, we showed that in a proportion of patients treated with dasatinib, an expansion of T or NK cells occurs during the therapy.26 In patients with T-cell expansions, we detected clonal TCR γ/δ gene rearrangements. We now used a more sensitive PCR-heteroduplex method to determine the prevalence of clonal lymphocytes in patients with (n = 10) and without lymphocytosis (n = 10) treated with dasatinib. Clonal lymphocytes were found in all patients who developed LGL lymphocytosis during dasatinib therapy (LGLpos) (Table 3). This included also patients with expansion of NK cells (n = 6). The frequency of clonal TCR γ/δ gene rearrangements in patients taking dasatinib without LGL lymphocytosis (LGLneg) was 70% (Table 3), similar to that of patients taking imatinib (64%; Table 2).
Clonal TCR rearrangements in patients treated with dasatinib (n = 20)
| Patient no. . | Immunophenotype of LGL cells . | TCR δ primer pairs . | TCR γ primer pairs . | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | 9 . | 10 . | 11 . | 12 . | ||
| 1 | T and NK cell | X | X | ||||||||||||||||
| 2 | T cell | X | X | X | X | X | X | ||||||||||||
| 3 | T cell | X | X | ||||||||||||||||
| 4 | NK cell | X | X | X | |||||||||||||||
| 5 | T and NK cell | X | X | X | |||||||||||||||
| 6 | NK cell | X | |||||||||||||||||
| 7 | T and NK cell | X | X | ||||||||||||||||
| 8 | T cell | X | X | ||||||||||||||||
| 9 | T cell | X | X | X | |||||||||||||||
| 10 | NK cell | X | X | ||||||||||||||||
| 11 | X | X | X | ||||||||||||||||
| 12 | X | ||||||||||||||||||
| 13 | X | ||||||||||||||||||
| 14 | X | X | X | ||||||||||||||||
| 15 | |||||||||||||||||||
| 16 | |||||||||||||||||||
| 17 | X | X | |||||||||||||||||
| 18 | X | X | |||||||||||||||||
| 19 | |||||||||||||||||||
| 20 | X | ||||||||||||||||||
| Patient no. . | Immunophenotype of LGL cells . | TCR δ primer pairs . | TCR γ primer pairs . | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 1 . | 2 . | 3 . | 4 . | 5 . | 6 . | 7 . | 8 . | 9 . | 10 . | 11 . | 12 . | ||
| 1 | T and NK cell | X | X | ||||||||||||||||
| 2 | T cell | X | X | X | X | X | X | ||||||||||||
| 3 | T cell | X | X | ||||||||||||||||
| 4 | NK cell | X | X | X | |||||||||||||||
| 5 | T and NK cell | X | X | X | |||||||||||||||
| 6 | NK cell | X | |||||||||||||||||
| 7 | T and NK cell | X | X | ||||||||||||||||
| 8 | T cell | X | X | ||||||||||||||||
| 9 | T cell | X | X | X | |||||||||||||||
| 10 | NK cell | X | X | ||||||||||||||||
| 11 | X | X | X | ||||||||||||||||
| 12 | X | ||||||||||||||||||
| 13 | X | ||||||||||||||||||
| 14 | X | X | X | ||||||||||||||||
| 15 | |||||||||||||||||||
| 16 | |||||||||||||||||||
| 17 | X | X | |||||||||||||||||
| 18 | X | X | |||||||||||||||||
| 19 | |||||||||||||||||||
| 20 | X | ||||||||||||||||||
Patients 1 to 10 had persistent lymphocytosis with large granular lymphocyte (LGL) morphology during dasatinib treatment. In patients 11 to 20 lymphocyte counts were normal during therapy. Clonality of the lymphocytes was determined by PCR and gel analysis with the use of 12 primer pairs for the TCR γ gene rearrangements and 6 primer pairs for TCR δ gene rearrangements. Positive clonal PCR products were confirmed with the sequencing and are marked in the columns as X.
Clonal rearrangements in TCR δ gene were found in most patients with LGLpos (90%), whereas only 1 patients with LGLneg had clonal TCR δ gene rearrangement (10%). No clear difference in the frequency of TCR γ gene rearrangements (90% vs 70%) was detected.
Correlation between absolute lymphocyte count and relative proportion of lymphocyte clone
In patients taking dasatinib with LGL lymphocytosis, the lymphocyte counts fluctuated markedly during the therapy. To quantify the amount of lymphocyte clone in correlation to the lymphocyte count, we designed ASO-primers for 4 LGLpos and 2 LGLneg patients. Estimation of the amount of the existing clone was determined by designating one follow-up sample with a reference value of 1 and by calculating other follow-up samples in relation to this reference sample. This method allowed quantitative comparison of the clone in different time points but not between different clones. The median follow-up time during dasatinib treatment was 21 months (range, 8-41 months).
Quantitative follow-up of LGLpos patients showed that the expansion of 1 to 2 predominant lymphocyte clones accounted for LGL lymphocytosis (Figure 3) in most cases. In addition, a few minor clones were often present. Although the method did not allow to determine the absolute amount of the clone from the total lymphocytes, the sensitivity of the assay in some patients with LGL lymphocytosis (1:10 000) suggested that the majority of the analyzed cells belonged to this clone. In patients without LGL lymphocytosis, the amount of clone varied slightly but did account for the variations in the absolute lymphocyte count, suggesting that it was only a minor subpopulation of all lymphocytes (data not shown).
Quantitative follow-up of lymphocyte clone with individual ASO primers. Patient-specific primers were designed to recognize clonal TCR γ and TCR δ gene rearrangements and were used to estimate the amount of clone in various time points before and during dasatinib therapy. The estimation of the amount of the existing clone was determined by designating one follow-up sample with a reference value of 1, and calculating other follow-up samples in relation to this reference sample. Figures on the left side show the estimated amount of clone in the blood at different time points (reference level 1 is marked with dashed black line). The figure on the right side presents the absolute lymphocyte counts at the same time points (upper limit of normal lymphocyte count is marked with dashed black line). (A) Patient 1 (Tables 1,4). This patient had a clonal rearrangement in TCR γ gene that was detected in CD8+αβ+ T cells, whereas the clonal rearrangement in TCR δ gene was found in NK cells. Follow-up of these 2 clones was done before (day −103, on imatinib) and after start of dasatinib therapy (days 56, 117, and 138). (B) Patient 5 (Tables 1,4). This patient had a clonal rearrangement in TCR γ gene detected in CD4+αβ+ T cells, whereas the clonal rearrangement in TCR δ gene was found in CD4+γδ+ T cells. Follow-up samples were available from the time of diagnosis (day −739) and during imatinib (day −187) and dasatinib (days 29, 239, 336, and 351) therapy.
Quantitative follow-up of lymphocyte clone with individual ASO primers. Patient-specific primers were designed to recognize clonal TCR γ and TCR δ gene rearrangements and were used to estimate the amount of clone in various time points before and during dasatinib therapy. The estimation of the amount of the existing clone was determined by designating one follow-up sample with a reference value of 1, and calculating other follow-up samples in relation to this reference sample. Figures on the left side show the estimated amount of clone in the blood at different time points (reference level 1 is marked with dashed black line). The figure on the right side presents the absolute lymphocyte counts at the same time points (upper limit of normal lymphocyte count is marked with dashed black line). (A) Patient 1 (Tables 1,4). This patient had a clonal rearrangement in TCR γ gene that was detected in CD8+αβ+ T cells, whereas the clonal rearrangement in TCR δ gene was found in NK cells. Follow-up of these 2 clones was done before (day −103, on imatinib) and after start of dasatinib therapy (days 56, 117, and 138). (B) Patient 5 (Tables 1,4). This patient had a clonal rearrangement in TCR γ gene detected in CD4+αβ+ T cells, whereas the clonal rearrangement in TCR δ gene was found in CD4+γδ+ T cells. Follow-up samples were available from the time of diagnosis (day −739) and during imatinib (day −187) and dasatinib (days 29, 239, 336, and 351) therapy.
Clonal lymphocyte population existed before dasatinib therapy
DNA samples before the start of dasatinib therapy were available from 5 patients with LGL lymphocytosis. Intriguingly, quantitative follow-up of lymphocyte clones by PCR showed that in all patients the observed clones existed at low levels already before the start of dasatinib therapy during imatinib treatment, but no lymphocyte expansions were then seen. In addition, from 2 patients diagnostic phase samples were available, and the analysis with the patient-specific ASO primers confirmed that the same clone, which expanded during dasatinib treatment, was already present at diagnosis. In patient no. 5, the percentage of the clone was high in the diagnostic phase sample and then subsequently decreased during imatinib therapy. After initiation of dasatinib treatment, the amount of the clone increased again together with increases in the lymphocyte count (Figure 3B).
One patient with persistent NK-cell lymphocytosis during dasatinib therapy discontinued the treatment because of adverse effects (patient 4; Table 1). In the blood sample after 4 years of discontinuation, the earlier expanded NK clone (Table 4) was still present when analyzed with the patient specific primers. However, lymphocyte count and distribution had normalized after discontinuation of dasatinib.
Quantitative follow-up and FACS sorting of lymphocytes with individual allele-specific oligonucleotide (ASO) primer
| Patient no. . | LGL lymphocytosis . | Gene . | Quantitative follow-up . | Lymphocyte population . | Cell-surface TCR expression . |
|---|---|---|---|---|---|
| 1 | Yes | Incomplete TCRD | Yes | NK cell | NA |
| 1 | Yes | Complete TCRG | Yes | CD8+ T cell | TCR αβ+ |
| 2 | Yes | Incomplete TCRD | Yes | CD8+ T cell | ND |
| 2 | Yes | Complete TCRD | Yes | CD8+ T cell | TCR γδ+ |
| 2 | Yes | Complete TCRG | Yes | CD4+ T cell | TCR αβ+ |
| 2 | Yes | Complete TCRG | Yes | CD4+ T cell | ND |
| 4 | Yes | Incomplete TCRD | No | NK cell | NA |
| 5 | Yes | Incomplete TCRD | Yes | CD4+ T cell | TCR γδ+ |
| 5 | Yes | Complete TCRG | Yes | CD4+ T cell | TCR αβ+ |
| 6 | Yes | Incomplete TCRD | No | NK cell | NA |
| 7 | Yes | Incomplete TCRD | No | NK cell | NA |
| 8 | Yes | Complete TCRG | Yes | CD8+ T cell | ND |
| 10 | Yes | Incomplete TCRD | No | ND | ND |
| 10 | Yes | Complete TCRG | No | CD8+ T cell | ND |
| 12 | No | Complete TCRG | Yes | ND | ND |
| 17 | No | Complete TCRG | No | CD8+ T cell | ND |
| 18 | No | Complete TCRG | Yes | CD8+ T cell | ND |
| 18 | No | Complete TCRG | Yes | CD8+ T cell | ND |
| Patient no. . | LGL lymphocytosis . | Gene . | Quantitative follow-up . | Lymphocyte population . | Cell-surface TCR expression . |
|---|---|---|---|---|---|
| 1 | Yes | Incomplete TCRD | Yes | NK cell | NA |
| 1 | Yes | Complete TCRG | Yes | CD8+ T cell | TCR αβ+ |
| 2 | Yes | Incomplete TCRD | Yes | CD8+ T cell | ND |
| 2 | Yes | Complete TCRD | Yes | CD8+ T cell | TCR γδ+ |
| 2 | Yes | Complete TCRG | Yes | CD4+ T cell | TCR αβ+ |
| 2 | Yes | Complete TCRG | Yes | CD4+ T cell | ND |
| 4 | Yes | Incomplete TCRD | No | NK cell | NA |
| 5 | Yes | Incomplete TCRD | Yes | CD4+ T cell | TCR γδ+ |
| 5 | Yes | Complete TCRG | Yes | CD4+ T cell | TCR αβ+ |
| 6 | Yes | Incomplete TCRD | No | NK cell | NA |
| 7 | Yes | Incomplete TCRD | No | NK cell | NA |
| 8 | Yes | Complete TCRG | Yes | CD8+ T cell | ND |
| 10 | Yes | Incomplete TCRD | No | ND | ND |
| 10 | Yes | Complete TCRG | No | CD8+ T cell | ND |
| 12 | No | Complete TCRG | Yes | ND | ND |
| 17 | No | Complete TCRG | No | CD8+ T cell | ND |
| 18 | No | Complete TCRG | Yes | CD8+ T cell | ND |
| 18 | No | Complete TCRG | Yes | CD8+ T cell | ND |
To quantify the amount of lymphocyte clone in correlation with the lymphocyte count and to identify the lymphocyte fraction where clonal cells reside, patient-specific ASO primers were designed for 11 patients. Patients 1 to 10 had persistent lymphocytosis with large granular lymphocyte (LGL) morphology during dasatinib treatment. In patients 12, 17, and 18, lymphocyte counts were normal during the therapy. No sorting was done from patient 12 because of small number of cells.
NK cell indicates CD3neg CD16/56+; CD8+ T cell, CD3+CD8+; CD4+ T cell, CD3+CD8neg; ND, not done; and NA, not applicable.
Clonal TCR γ and δ gene rearrangements were found in distinct lymphocyte subclasses
Analysis of clonal TCR γ and δ gene rearrangements from the magnetic bead–sorted CD8+ and CD8neg cells showed that clones can be detected in both cell populations. To further elucidate the specific cell populations, samples underwent fluorescence-activated cell sorting (FACS) into pure CD4+, CD8+, NK- and B-cell fractions (Figure 1). Sorting and further analysis were performed from 8 LGLpos and 2 LGLneg patients (Table 4).
Analysis with patient-specific ASO primers showed that most patients had clonal rearrangements in the CD8+ cell population, including the 2 LGLneg patients (Table 4). Strikingly in 4 of 6 patients with LGL lymphocytosis who had a clonal TCR δ gene rearrangement, the clonal lymphocyte population resided in the NK-cell fraction. Other fractions from the same samples (CD4+, CD8+, and B cells) were negative for TCR δ gene rearrangements, confirming the specificity. Two other patients with TCR δ gene rearrangements had clonal cells in the γδ+ T-cell population (patient 5 in CD4+ γδ+ T-cell fraction and patient 2 in CD8+ γδ+ T-cell fraction). Thus, clonal TCR δ gene rearrangements were detected only in NK cells and in γδ+ T cells. However, no TCR δ protein was expressed on the NK-cell surface, as assessed by flow cytometry. Sequence alignments of clonal TCR δ gene rearrangements are presented in Table 5.
Sequences of clonal TCR δ-gene rearrangements detected in LGLpos patients
| Patient no. . | Clonal cell population . | Vδ2/Vδ3 . | N1 . | Dδ1 . | N2 . | Dδ2 . | N3 . | Dδ3 . | N4 . | Jδ . | Function of rearrangement and junction region amino acids (complete and productive rearrangements) . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Germline | Vδ2 tgtgcctgtgacacc Vδ3 ctactgtgcctttag | gaaatagt | ccttcctac | actgggggatacg | acaccgataaactcatc | ||||||
| 1 | NK cell | Vδ2 tgtgcctgt | attt | actgggggatacg | Incomplete (no J gene); unproductive | ||||||
| 2 | CD8+ | Vδ2 tgtgcctgtgacacc | gaagac | ctgggggata | aggctc | caccgataaactcatc | Unproductive (stop codons, out-of-frame junction) | ||||
| 2 | TCR γδ+ CD8+ | Vδ3 ctactgtgccttta | atag | cgt | ttcctac | ggggtttg | gggggata | atgg | cgataaactcatc | Productive CAFNSVSYGVWGDNGDKLIF | |
| 3 | NA | Vδ2 tgtgcctgtgacacc | gtatt | actgggggatacg | cggcg | caccgataaactcatc | Productive CACDTVLLGDTRRTDKLIF | ||||
| 4 | NK cell | Vδ2 tgtgcctgtgac | ttcttcccttttg | ctgggggatacg | Incomplete (no J gene); unproductive | ||||||
| 5 | TCR γδ+ CD4+ | ccttccta | actggagggg | accgataaactcatc | Incomplete (no V gene); unproductive | ||||||
| 6 | NK cell | Vδ2 tgtgcctgtgacacc | gtgt | tgggggatacg | Incomplete (no J gene); unproductive | ||||||
| 7 | NK cell | Vδ2 tgtgcctgtgacac | gggct | actgggggatacg | Incomplete (no J gene); unproductive | ||||||
| 9 | NA | Vδ2 tgtgcctgt | tctgtacccgt | actgggggatacg | Incomplete (no J gene); unproductive | ||||||
| 10 | NA | Vδ2 tgtgcctgtgac | cgtt | actgggggatacg | Incomplete (no J gene); unproductive |
| Patient no. . | Clonal cell population . | Vδ2/Vδ3 . | N1 . | Dδ1 . | N2 . | Dδ2 . | N3 . | Dδ3 . | N4 . | Jδ . | Function of rearrangement and junction region amino acids (complete and productive rearrangements) . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Germline | Vδ2 tgtgcctgtgacacc Vδ3 ctactgtgcctttag | gaaatagt | ccttcctac | actgggggatacg | acaccgataaactcatc | ||||||
| 1 | NK cell | Vδ2 tgtgcctgt | attt | actgggggatacg | Incomplete (no J gene); unproductive | ||||||
| 2 | CD8+ | Vδ2 tgtgcctgtgacacc | gaagac | ctgggggata | aggctc | caccgataaactcatc | Unproductive (stop codons, out-of-frame junction) | ||||
| 2 | TCR γδ+ CD8+ | Vδ3 ctactgtgccttta | atag | cgt | ttcctac | ggggtttg | gggggata | atgg | cgataaactcatc | Productive CAFNSVSYGVWGDNGDKLIF | |
| 3 | NA | Vδ2 tgtgcctgtgacacc | gtatt | actgggggatacg | cggcg | caccgataaactcatc | Productive CACDTVLLGDTRRTDKLIF | ||||
| 4 | NK cell | Vδ2 tgtgcctgtgac | ttcttcccttttg | ctgggggatacg | Incomplete (no J gene); unproductive | ||||||
| 5 | TCR γδ+ CD4+ | ccttccta | actggagggg | accgataaactcatc | Incomplete (no V gene); unproductive | ||||||
| 6 | NK cell | Vδ2 tgtgcctgtgacacc | gtgt | tgggggatacg | Incomplete (no J gene); unproductive | ||||||
| 7 | NK cell | Vδ2 tgtgcctgtgacac | gggct | actgggggatacg | Incomplete (no J gene); unproductive | ||||||
| 9 | NA | Vδ2 tgtgcctgt | tctgtacccgt | actgggggatacg | Incomplete (no J gene); unproductive | ||||||
| 10 | NA | Vδ2 tgtgcctgtgac | cgtt | actgggggatacg | Incomplete (no J gene); unproductive |
The table represents detected TCR δ rearrangements. Sequences of the junction region (VDJ) are aligned.
Clonal lymphocytes are BCR-ABL1 negative
To determine whether the clonal lymphocytes belonged to the Ph+ leukemic clone, FISH with a BCR-ABL1 fusion gene probe was performed from the diagnostic phase samples from 2 patients with CML (patients 7 and 17; Table 1). From these patients CD4+, CD8+, NK-, and B-cell populations underwent FACS, and the cell populations were studied separately with FISH. In the first patient, 1% to 2% of cells were Ph+ in CD4+, CD8+, and NK-cell fractions. In B-cell fraction up to 9% of cells were Ph+. In the second patient, all studied lymphocyte populations were negative for the Ph chromosome. This is in accordance with an earlier publication by Takahashi et al37 who have shown that in untreated CML patients T and NK cells are usually negative for the Ph chromosome.
Furthermore, during dasatinib and imatinib therapy, 59% (17 of 29) of the patients with clonal TCR γ/δ rearrangements were negative for BCR-ABL1 transcripts in blood by RQ-PCR, and in 5 patients also high-resolution FISH analysis was performed (1000 cells counted) and no Ph+ cells could be detected. Thus, it can be concluded that the detected clonal lymphocytes did not belong to the malignant Ph+ clone.
CMV seropositivity and clonal lymphocytosis
Of 23 patients who had clonal TCR rearrangements and CMV serology data available, 21 (91%) were positive for CMV immunoglobulin G (IgG; Table 1). Only 2 of 6 patients without clonal TCR rearrangement had a positive CMV IgG result. Patients with LGL lymphocytosis during dasatinib treatment were all CMV IgG positive. Only 50% (4 of 8) of LGLneg patients were CMV IgG positive, and none of the CMV-negative patients had clonal TCR rearrangements. However, 2 patients taking imatinib, who were CMV IgG negative, had clonal TCR rearrangements (patients 25 and 31; Tables 1–2).
Discussion
According to the tumor immune surveillance hypothesis, expansion of occult malignant clones is prevented by the concerted action of innate and adaptive immune system. The development of clinical disease, such as CML, requires an escape from immune recognition through a multifaceted process of immunoediting whereby tumor-reactive cytotoxic cells are either deleted or rendered anergic.38 In accord, PR-1 leukemia antigen reactive CD8+ T cells have been isolated from patients with CML but were shown to be dysfunctional because of a selective deletion of high-avidity clones by apoptosis.39,–41 Recently, in a biologically relevant CML mouse model, leukemia-specific T cells were shown to be exhausted, maintained only limited cytotoxicity, and expressed high levels of the inhibitory receptor PD-1. Similar overexpression of PD-1 was observed in CD8+ cells from patients with CML.42 However, because CML is particularly responsive to immunomodulatory therapies, such as interferon α and donor lymphocyte infusions, it is probable that immunologic defense mechanisms have not been permanently compromised.
Recent data from our group and other groups have suggested that dasatinib has immunostimulatory effects in the form of persistent monoclonal/oligoclonal LGL lymphocytosis in a distinct proportion of patients.25,26 The striking association between LGL expansion and excellent therapy response raised the possibility of TKI-mediated induction of cellular immunity against leukemic antigens. Because the phenotype of the expanding cytotoxic lymphocytes (CD45RO+CD27negCD62Lneg) was in accordance with a preexisting long-lived effector memory cell, we wanted to examine the prevalence of clonal lymphocytes before the start of dasatinib therapy and at the time of CML diagnosis. Unexpectedly, we found lymphocytes with clonal TCR rearrangements in most (83%) patients with CML at diagnosis. Because the sensitivity of the assay is 1% to 5%, a significant proportion of lymphocytes were clonal in untreated patients with CML. The diagnostic phase lymphocyte clones persisted at low levels during imatinib therapy, but they markedly expanded in a proportion of dasatinib-treated patients. These findings strengthen the hypothesis that dasatinib-associated LGL lymphoproliferation is an expansion of preexisting T-memory cell clones present already at the time of diagnosis.
Rearrangements in the gene segment that code for variable TCR regions are responsible for the enormous diversity of antigen-specific receptors in T cells. These receptors are generated during differentiation of T cells by rearrangements of variable (V), diversity (D) (not found in TCR γ gene) and junction (J) gene segments for each lymphocyte. In each person there can be more than 107 different T-cell clones, which constitute a unique and diverse lymphocyte repertoire. Therefore, significant expansions of single-lymphocyte clones are uncommon in healthy persons. Expanded clonotypes of CD4+ or CD8+ cells can be detected with a high frequency only in older persons and are almost exclusively confined to the CD45RO+ memory subset.43,44 Clonal expansions have not been related to particular Vβ families, but are composed of several TCR Vβ families that vary from person to person, suggesting that a multiplicity of antigenic stimulations are involved in the selection of the expanded clones.43 Similarly, in our material, in only 1 of 12 healthy controls a clonal TCR γ gene rearrangement could be found. The high frequency of clonal lymphocytes at CML diagnosis was not related to the older age of patients, because the median age was 51 years in the diagnosis phase patient group compared with 46 years in healthy controls. However, a potential association was observed between positive CMV IgG serology and clonal TCR rearrangements because more than 90% of patients with clonal TCR rearrangements were CMV IgG positive, whereas only 2 of 6 patients without clonal TCR rearrangements were CMV positive. In our previous publication we also showed that in patients who developed LGL lymphocytosis during dasatinib therapy, more than 40% had CMV reactivation during the treatment. Additional studies are warranted to address the role of CMV in this setting.
Rearrangement of TCR genes is considered to be restricted to T cells. However, incomplete TCR δ gene rearrangements have been described to occur in 16% of alleles in B lymphocytes of healthy controls, indicating that the TCR genes are not exclusively rearranged in T cells.45 Furthermore, Fronkova et al46 demonstrated that TCR δ gene rearrangements can occur also in NK cells. This is well in accordance with our results because we detected clonal TCR δ gene rearrangements in the NK-cell fraction of patients with LGL lymphocytosis with NK-cell phenotype. None of the patients had clonal TCR γ gene rearrangement in NK cells. Note that the analysis of TCR gene rearrangements was informative in selected patients with NK-cell phenotype, because NK-cell clonality is notoriously difficult to assess. In addition, this strengthens the view that dasatinib-associated LGL lymphocytosis in patients with NK-cell phenotype is also a clonal or oligoclonal phenomenon similar as in patients with T-cell phenotype.
T cells expressing the α/β chains on the cell surface rearrange first γ and δ genes and thereafter α and β genes, which leads to the formation of the functional protein on the cell surface. TCR δ gene is spliced out during TCR α gene rearrangement. More than 90% of T cells in healthy persons express α/β chains on the surface, and γδ+ T cells are the minority. In a subgroup of patients we observed clonal TCR δ gene rearrangements also among T-cell fraction. Further sorting of T cells into αβ+ and γδ+ T cells showed that clonal cells showing TCR δ gene rearrangements were confined to the γδ+ T-cell fraction. Thus, dasatinib patients with LGL lymphocytosis had more often clonal TCR δ gene rearrangements than patients without lymphocytosis (90% vs 10%, respectively), suggesting that patients with LGL lymphocytosis have an expansion of γδ+ T-cell or NK-cell population. The importance and function of NK and γδ+ T cells in cancer immunosurveillance have been unclear until recently.47,48 These cells of the innate immune system are important effectors in the control of hematologic malignancies and can eliminate minimal residual disease in CML.49,50
To our knowledge, this is the first report on clonal cytotoxic lymphocytes in patients with CML at the time of diagnosis and during TKI therapy. Because the lymphocytes did not belong to the malignant Ph+ clone, their role as a part of anergic immune surveillance machinery requires further studies. Clonal lymphocytes not belonging to the leukemic clone have been described in other hematologic malignancies (acute promyelocytic leukemia, myelodysplastic syndrome) as well.32,51 In our previous report on patients with advanced-phase CML and Ph+ acute lymphoblastic leukemia being treated with dasatinib, clonal LGL lymphocytosis was associated with excellent treatment responses. Similarly, in patients with multiple myeloma, the presence of clonal T cells was related to superior prognosis.52 In accord, long-lasting LGL expansions with clonal TCR γ/δ gene rearrangements have been described in a subset of patients receiving an allogeneic stem cell transplant, which were associated with better overall survival, suggesting a graft-versus-leukemia activity.53,54 We, therefore, speculate that dasatinib treatment reverts the tolerance mechanism and restores the function of unresponsive, exhausted leukemia-specific cytotoxic cells. Our previous results showed that patients with LGL lymphocytosis being treated with dasatinib have a decreased amount of regulatory T cells.26 This observation was confirmed in the current study population (data not shown) and could be one factor facilitating the restoration of T-cell function.
In conclusion, clonal lymphocytes were frequently present in patients with CML at diagnosis and persisted at low levels during TKI therapy. In a distinct subgroup of dasatinib-treated patients, clonal cells markedly expanded during therapy. Expanding clones were not only confined to CD8+ cytotoxic cells but also resided in NK- and γδ+ T-cell fractions. These cells may participate in the elimination of the residual CML cells, because previous studies have linked clonal expansions with excellent, long-lasting therapy responses in dasatinib-treated patients with advanced phase leukemia. The isolation of epitopes and detailed functional analyses of clonal, CML-associated lymphocytes are ongoing.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Drs J. Vakkila and P. Arstila for valuable comments on the paper. We also thank the personnel at the Hematology Research Unit Helsinki and the Laboratory of Molecular Genetics at Turku University Central Hospital for their expert clinical and technical assistance.
This work was supported by the Finnish special governmental subsidy for health sciences, research, and training and by the Finnish Cancer Societies, Emil Aaltonen Foundation, Academy of Finland, Finnish Medical Foundation, Blood Disease Foundation, Biomedicum Helsinki Foundation, Finnish Association of Hematology, Gyllenberg Foundation, and K. A. Johansson Foundation.
Authorship
Contribution: A.K. performed the laboratory analyses; A.K., K.P., and S.M. designed the study, analyzed the data, and wrote the paper; V.J. and V.K. supervised the molecular studies and participated in the data analysis; M.E. and L.S. provided patient samples for analysis and participated in the data analysis; and R.S. participated in the design of the study and analysis of the data.
Conflict-of-interest disclosure: K.P. has received research funding and honoraria from Novartis and Bristol-Myers Squibb. M.E., R.S., and S.M. have received honoraria from Bristol-Myers Squibb. The remaining authors declare no competing financial interests.
Correspondence: Satu Mustjoki, Hematology Research Unit, Biomedicum Helsinki, PO Box 700, FIN-00029 HUCH, Helsinki, Finland; e-mail: satu.mustjoki@helsinki.fi.