Our aim was to examine the genetics of clonal evolution in follicular lymphoma (FL) and to identify genetic alterations associated with disease progression. A total of 100 biopsies from 44 patients diagnosed with t(14;18)-positive FL were examined by array comparative genomic hybridization. In 20 patients the patterns of somatic hypermutations (SHMs) in the variable region of heavy chain gene were additionally analyzed. Gain of chromosome X in male samples was a marker for poor outcome (P < .01). Gains involving chromosome 2, 3q, and 5 were exclusively present in FL biopsies from cases with higher grade transformation and were among the copy number alterations (CNAs) associated with inferior survival. Although we noted a trend for increasing genomic complexity in initial versus late FL samples, the overall frequencies of CNAs in initial and late FL biopsies showed a surprisingly stable pattern through the course of the disease. In 27 of cases the initial samples harbored CNAs that were absent in relapse samples, indicating that tumor cell clones at relapse were not direct descendants of initially dominating clones. The pattern of SHMs confirmed parallel development of tumor cell clones in 14 cases. Our findings support the hypothesis of common progenitor cells in FL.
Introduction
Follicular lymphoma (FL) is second to diffuse large B-cell lymphoma (DLBCL) as the most common non-Hodgkin lymphoma in Western countries. FL affects the middle-aged and elderly population and exhibits a variable disease course with a median survival of 7 to 10 years. Although often initially responsive to chemotherapy or radiotherapy, FL is characterized by relapses and progression to treatment-resistant disease or transformation to higher-grade lymphoma, most commonly DLBCL, with poor treatment outcome. The reported proportions of patients with transformation of FL range from 10% to 70%,1,,–4 depending on the definition of transformation and the observation time.
The translocation t(14;18)(q32;q21), present in approximately 90% of cases, represents the primary oncogenetic event in FL. Leading to overexpression of the antiapoptotic Bcl2, the translocation renders B cells resistant to proapoptotic stimuli in the germinal center. Here, the Bcl2+ B cells may survive to accumulate oncogenetic alterations, leading to lymphomagenesis.5 Cytogenetic aberrations appearing secondary to t(14;18) are mainly chromosomal imbalances with a pattern typical for FL.6 Some of these have been reported as independent prognostic factors in FL, including loss of 1p36, 6q21-26, 9p21, and 17p; gain of 12q, 18; and gain of the X chromosome in men, and some of these have been associated with transformation risk.7,,,,,–13 Although clinical and molecular parameters have been linked to survival and transformation,4 the disease course cannot be reliably predicted. Indeed, it is not clear whether FLs that remain indolent and FLs that transform are biologically distinct. Several studies have compared FL and transformed lymphoma samples to identify molecular mechanisms for transformation.4 Gene expression studies of limited size have shown association of transformation to DLBCL with a germinal centerlike signature and showed changes in the expression of genes controlling the cell cycle and proliferation as well as genes expressed in tumor infiltrating T cells.14,,,,,–20 The presence and activation status of various immune cells in FL have also been linked to prognosis by gene-expression analysis and studies of immune cell markers by immunohistochemistry.21,22 A few studies have addressed the genetic evolution underlying transformation by comparing genomic data from paired samples of FL and DLBCL.20,23,–25 The results indicate that a diverse set of copy number alterations is associated with transformation. Of note, the genetic alterations associated with relapse and clinical progression of FL without histologic transformation have been less studied.
Karyotypic data26,27 and examinations of the variable regions of the B-cell receptor28,,,,,–34 have shown extensive intraclonal heterogeneity in FL B cells. These observations have led to the hypothesis of a common FL progenitor cell giving rise to clonally related, but genetically distinct, subpopulations of tumor cells. This hypothesis has been supported by a study of acquired uniparental disomy in paired FL and DLBCL samples.25
The aim of the present study was to examine the genetics of clonal evolution in FL and to identify genetic alterations associated with transformation of FL or relapse of FL without transformation. We analyzed 100 sequential biopsies from 44 patients with FL, using array comparative genomic hybridization (aCGH). In addition, 49 of the biopsies from 20 of the patients were examined for clonal relationship and evolution by sequencing the rearranged immunoglobulin heavy chain (IGH) genes and analyzing somatic hypermutations (SHMs) in the variable region of IGH (IGVH).
Methods
Patients and biopsies
Samples were selected from the archives of the Pathology Clinic at The Norwegian Radium Hospital, Oslo University Hospital. Informed consent was obtained from all patients in accordance with the Declaration of Helsinki. The study was approved by the regional committee of Oslo, Norway, for research ethics (protocol no. S-05 209, date of approval August 16, 2005).
The selection criteria were the diagnosis of FL according to the World Health Organization classification35 and presence of fresh frozen tissue from multiple samples of FL positive for the translocation t(14;18)(q32;q21). We obtained aCGH results in 100 biopsies from 44 patients collected between 1987 and 2005 (39 patients with aCGH results from 2 or more sequential biopsies and 5 with results from 1 biopsy). The first biopsy in 23 of the patients was obtained at diagnosis, in 5 at progression after watchful waiting, and in 16 at relapse. Histologic review and determination of tumor cell content in the frozen samples was performed by a hematopathologist (J.D.; Table 1). The tumor cell content was greater than 50% (relative to normal immune cells and stroma) in all except 1 sample that showed 40% tumor cells. All but 9 biopsies showed FL, the remaining showed DLBCL. FL with marginal zone differentiation was diagnosed in 15 of the 100 biopsies, and these originated from 10 patients. Two of these samples also had components of DLBCL, and 2 others showed partial FL grade 3a. In total, there were 17 biopsies (from 14 patients) with FL3a and none with FL3b. Most samples were lymph node biopsies. Five samples were from skin (as secondary sites) and 6 from unknown sites. Karyotypic data were available from 89 of the biopsies and confirmed the presence of t(14;18)(q32;q21) in at least 1 biopsy from all 44 cases. All karyotypes were hyperdiploid, except for 4 that were near tetraploid. The karyotypes of the first biopsies from 40 of these patients have been published previously.36
The histology of the 100 biopsies analyzed by aCGH
| Histology/FL grade . | Biopsy 1 . | Biopsy 2 . | Biopsy 3 . | Biopsy 4 . | Total . |
|---|---|---|---|---|---|
| FL grade 1 | 16 | 12 | 2 | 0 | 30 (4 with MZD) |
| FL grade 2 | 21 | 12 | 8 | 2 | 43 (6 with MZD) |
| FL grade 3a | 7 | 9 | 1 | 0 | 17 (2 with MZD) |
| FL (grade unspecified) | 0 | 1 | 0 | 0 | 1 (with MZD) |
| DLBCL | 0 | 5 | 4 | 0 | 9 (2 with FL + MZD) |
| Total | 44 | 39 | 15 | 2 | 100 |
| Histology/FL grade . | Biopsy 1 . | Biopsy 2 . | Biopsy 3 . | Biopsy 4 . | Total . |
|---|---|---|---|---|---|
| FL grade 1 | 16 | 12 | 2 | 0 | 30 (4 with MZD) |
| FL grade 2 | 21 | 12 | 8 | 2 | 43 (6 with MZD) |
| FL grade 3a | 7 | 9 | 1 | 0 | 17 (2 with MZD) |
| FL (grade unspecified) | 0 | 1 | 0 | 0 | 1 (with MZD) |
| DLBCL | 0 | 5 | 4 | 0 | 9 (2 with FL + MZD) |
| Total | 44 | 39 | 15 | 2 | 100 |
aCGH indicates array comparative genomic hybridization; FL, follicular lymphoma; DLBCL, diffuse large B-cell lymphoma; and MZD, follicular lymphoma with marginal zone differentiation.
aCGH and statistical considerations
DNA was isolated from frozen tissue with the use of the EZ1 tissue kit (QIAGEN). DNA copy number alterations (CNAs) were detected by aCGH on in-house arrays containing approximately 4500 Bacterial Artificial Chromosomes/P1-derived Artificial Chromosomes (BAC/PACs) in quadruplicate at an average resolution of 1 megabase. The construction and preparation of the microarrays as well as the procedures for DNA labeling, hybridization, and scanning have been previously described.37 Images were segmented, raw data were filtered, and log2-ratios were calculated in GenePix Pro 6.0. Simple, local normalization was performed in Normalize Suite Version 2.6 BETA.38 Clones belonging to chromosomes 1 to 22 and the X chromosome with known unique chromosomal locations in Ensembl39 were considered for analysis. Clones with missing values in more than 10% of the samples were removed, leaving a total number of 3091 clones for further analysis. For each array, a centering coefficient was calculated, and the normalized aCGH data centered by subtracting this coefficient from every log2-ratio. To determine the centering coefficient, we first estimated the density of the log2-ratios of the array by applying a kernel density estimator with an Epanechnikov kernel and a bandwidth of 0.04. The log-ratio value corresponding to the highest peak in the density was used as the centering coefficient. All subsequent analyses were performed on the centered data. The whole dataset is available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress; accession no. E-TABM-930).
The identification of chromosomal gains and losses was based on fitting piecewise constant curves to the data, using a penalized least square method with the use of the Potts algorithm40 (details are provided in supplemental Document 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Each chromosomal arm was then separated into a set of distinct segments defined by a minimum of 3 consecutive BAC probes. Within each segment the height of the curve was defined by the mean of the probe values. To identify gains and losses, we applied a threshold (log2-ratio = 0.08) to the curves defined by the piecewise constant fits (PCFs). Segments of the PCF curve exceeding the threshold were defined as gains, whereas segments below minus the threshold were defined as losses.
For 47 of the samples, the results were compared with results from chromosomal CGH (cCGH) and karyotypic data to evaluate PCF settings. The aCGH and PCF (with current settings) detected the same recurrent regions of gains and losses as cCGH, and the aCGH results were in agreement with the karyotypes. However, the array-based technique showed higher frequencies of CNAs and allowed more precise delineations of CNAs than the cCGH. In addition, CNAs involving narrow regions were detected by aCGH and PCF and not by cCGH (supplemental Figure 1). The results were also visually evaluated. In a few cases with low tumor cell content, in which standard settings in PCF failed to identify visually observed aberrations, the PCF threshold was lowered (log2-ratio = 0.06). The effect of chromosomal gains or losses on survival was assessed by Cox (proportional hazard) regression. When carrying out such regressions, one depends on a set of variables defined on all biopsies. Because ordinary procedures for identifying aberrations will not provide this, we used an extension to the PCF methods (denoted multi-PCF) in which the least square fit was found simultaneously for all biopsies (supplemental Document 1). The breakpoints were then forced to be equal for all biopsies; thus, the biopsies obtained copy number estimates on the same set of segments. These copy number estimates were then the basis for the Cox regressions. The multi-PCF method will not detect gains or losses present in only 1 to 2 biopsies; however, for regression analysis this is not serious because such rare aberrations will not achieve significance in the analyses. In addition, we checked that the results from the multi-PCF methods provided a fair approximation to results obtained with the use of PCF separately on each biopsy.
When carrying out Cox regression, we used 1-sided tests, assuming that a copy number alteration should give a survival disadvantage (although the assumption is not necessarily true, it seems probable for most cases and may markedly reduce the rate of false discoveries). Moreover, when considering gains, the negative (log2) copy number values were set to zero, similarly positive values were truncated when considering losses. Finally, we compared the number of detected gains and losses to the number expected by chance, with the a priori decision of only reporting results if the number of significant results markedly exceeded the number expected by chance. We additionally entered age as a covariate.
To obtain a simple measure for the level of CNAs in each sample, we defined a CNA index that adds up the areas between the PCF curves and the zero line. That is, the contribution to the index from one segment of constant copy number value corresponds to the absolute value of the curve multiplied by the length of the segment in base pairs. We used multi-PCF to produce the curves because this method is less sensitive to differences in noise levels between samples than ordinary PCF. To check the influence of tumor cell content, we tested the correlation between percentage of tumor cells in each sample and the CNA index. We found only a weak trend for lower CNA index in samples with low tumor cell content and concluded that the CNA index reflects the level of CNAs and can be used as a marker for genomic complexity.
Comparisons of survival in different groups of patients were carried out with the use of Kaplan-Meier curves and log-rank tests. Differences in CNA indexes were tested by variance component models.
Analysis of rearranged IGH genes
DNA was extracted from the frozen samples as described in the previous section. For the detection of IgH rearrangements, a multiplex polymerase chain reaction (PCR) method with the use of a set of primers against the framework 1 region (FR1), FR2, and FR3 of the IgH variable gene segment and joining gene segment was performed as previously described41 (further details about the multiplex PCR and sequencing of PCR products are provided in supplemental Document 2). The obtained sequences were analyzed with the use of the IgBLAST42 and IMGT43 databases. The identity of the clone-specific IGH complementarity 3 regions (CDR3) as well as SHMs in the variable gene segment regions was analyzed. Among the 44 cases, 24 were excluded from analysis because of the lack of a monoclonal PCR product from more than 1 biopsy. One case was excluded because of different CDR3 sequences in the 2 successive biopsies. Genealogic trees were constructed for 19 patients on the basis of SHM patterns from a total of 47 biopsies.
Results
Patient characteristics
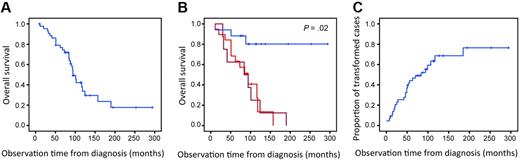
Clinical characteristics of the 44 patients are summarized in Table 2. The median observation time was 88 months (range, 10-294 months). During this time 19 patients (43%) developed histologically verified transformation to DLBCL, of which 9 had fresh frozen tissue showing DLBCL available. Eight additional patients (18%) underwent transformation to higher grade lymphoma, defined by clinical criteria as published by Al-Tourah et al3 (sudden rise in lactate dehydrogenase levels, rapidly growing tumor or new tumor growth at unusual extranodal site, new B symptoms, or hypercalcemia). Thus, the material consisted of 27 cases with transformation and 17 cases without any signs of transformation on the basis of histologic or clinical criteria. Of these 17 patients without transformation, 7 had one biopsy showing FL3a. The patients were treated with various regimens according to standard FL treatment protocols. After year 2000, rituximab was introduced to the treatment of FL. Three of the 44 patients received rituximab as part of the initial treatment, and 20 patients received rituximab as part of the treatment at relapse. Fourteen patients received high-dose chemotherapy with autologous stem cell support. One patient received reduced-intensity conditioning allogeneic stem cell transplantation. The median survival of all patients was 10 years. Patients who showed no signs of transformation during the observation time had significantly better outcome than patients with transformation (P = .02; Figure 1).
Clinical characteristics of the 44 patients
| Patient characteristics . | Value . |
|---|---|
| Year of diagnosis | 1984-2002 |
| Median age, y (range) | 50 (29-71) |
| Sex (M:F) | 2:1 |
| FLIPI score | |
| 0-1, n (%) | 12 (27) |
| 2, n (%) | 17 (38) |
| 3-5, n (%) | 14 (32) |
| Transformation | |
| Histologic (DLBCL), n (%) | 19 (43) |
| Clinical, n (%) | 8 (18) |
| No transformation, n (%) | 17 (39) |
| Median observation time, mo (range) | |
| All 44 patients | 88 (10-294) |
| Patients alive at the end of follow up (n = 22) | 96 (68-294) |
| Dead patients (n = 22) | 68 (10-190) |
| Patients with transformation (n = 27) | 84 (11-190) |
| Patients with no transformation (n=17) | 97 (10-294) |
| Patient characteristics . | Value . |
|---|---|
| Year of diagnosis | 1984-2002 |
| Median age, y (range) | 50 (29-71) |
| Sex (M:F) | 2:1 |
| FLIPI score | |
| 0-1, n (%) | 12 (27) |
| 2, n (%) | 17 (38) |
| 3-5, n (%) | 14 (32) |
| Transformation | |
| Histologic (DLBCL), n (%) | 19 (43) |
| Clinical, n (%) | 8 (18) |
| No transformation, n (%) | 17 (39) |
| Median observation time, mo (range) | |
| All 44 patients | 88 (10-294) |
| Patients alive at the end of follow up (n = 22) | 96 (68-294) |
| Dead patients (n = 22) | 68 (10-190) |
| Patients with transformation (n = 27) | 84 (11-190) |
| Patients with no transformation (n=17) | 97 (10-294) |
FLIPI indicates Follicular Lymphoma International Prognostic Index; and DLCBL, diffuse large B-cell lymphoma.
Overall survival and time to transformation. (A) Overall survival for all 44 patients. (B) Overall survival for patients with transformation by histologic (red; n = 19) and clinical (dark red; n = 8) criteria compared with survival for patients without transformation during the observation time (blue; n = 17; P = .02). (C) Time to transformation for all 44 patients. Patients without signs of transformation (including those who died without transformation) are censored.
Overall survival and time to transformation. (A) Overall survival for all 44 patients. (B) Overall survival for patients with transformation by histologic (red; n = 19) and clinical (dark red; n = 8) criteria compared with survival for patients without transformation during the observation time (blue; n = 17; P = .02). (C) Time to transformation for all 44 patients. Patients without signs of transformation (including those who died without transformation) are censored.
DNA copy number alterations in initial biopsies
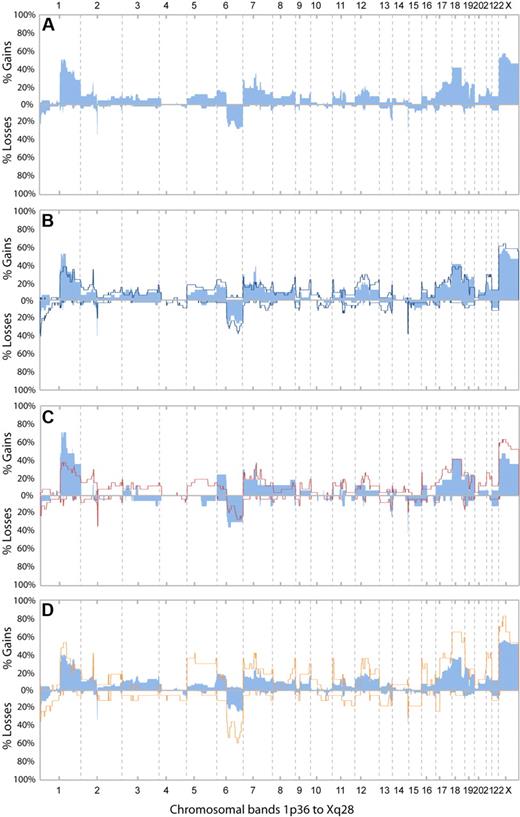
DNA CNAs were detected by aCGH. The frequencies of CNAs in the first available biopsy from all 44 patients are shown in Figure 2A. Twenty-eight biopsies were obtained before any treatment and 16 at relapses. Copy number gains were common, and the most frequent ones included Xp11q21 (57%), 1q22 (50%), and 18p11q21 (41%; a complete list is provided in Table 3). In contrast, only 3 chromosomal regions were involved in losses in more than 15% of cases (1p36, 6q, and 13q34). The rearrangement of immunoglobulin κ light chain and heavy chain genes were seen as frequent narrow losses at 2p11 and 14q32, respectively (Figure 2A). The rearrangement of the λ light chain gene was also seen in several cases but involved only 1 BAC clone and was not scored by the PCF algorithm.
Frequencies of CNAs. (A) Frequencies of copy number alterations (CNAs) in the first available biopsies from all 44 patients. The biopsies were obtained before any treatment in 28 of the patients and at relapse in 16 of the patients. (B) Frequencies of CNAs in the first (light blue bars) and the last (dark blue line) examined biopsies of follicular lymphoma (FL; grade 1, 2, or 3a) from 35 patients. (C) Frequencies of CNAs in the first examined biopsy of FL from 17 cases that did not undergo transformation (light blue bars) and 27 cases that underwent transformation (red lines). (D) Frequencies of CNAs in all biopsies with histologic grades FL1/FL2 (blue bars; n = 74) and in all biopsies with FL grade 3a (orange line; n = 17).
Frequencies of CNAs. (A) Frequencies of copy number alterations (CNAs) in the first available biopsies from all 44 patients. The biopsies were obtained before any treatment in 28 of the patients and at relapse in 16 of the patients. (B) Frequencies of CNAs in the first (light blue bars) and the last (dark blue line) examined biopsies of follicular lymphoma (FL; grade 1, 2, or 3a) from 35 patients. (C) Frequencies of CNAs in the first examined biopsy of FL from 17 cases that did not undergo transformation (light blue bars) and 27 cases that underwent transformation (red lines). (D) Frequencies of CNAs in all biopsies with histologic grades FL1/FL2 (blue bars; n = 74) and in all biopsies with FL grade 3a (orange line; n = 17).
Recurrent CNAs in FL
| Column A (chromosome arm) . | Column B (chromosome band) . | Column C (n = 44), n (%) . |
|---|---|---|
| Gains | ||
| 1q | 1q22 | 22 (50) |
| 2p | 2p16 | 11 (25) |
| 6p | 6p21p25 | 7 (16) |
| 7p | 7p22 | 12 (27) |
| 7q | 7q11 | 16 (36) |
| 8q | 8q24 | 7 (16) |
| 11p | 11p11 | 7 (16) |
| 12q | 12q13 | 9 (21) |
| 16p | 16p13 | 9 (21) |
| 17p | 17p11 | 7 (16) |
| 17q | 17q21 | 12 (27) |
| 18 | 18p11q21 | 18 (41) |
| 19p | 19p13 | 11 (25) |
| 19q | 19q13 | 10 (23) |
| 21 | 21q21 | 8 (18) |
| X | Xp11q21 | 25 (57) |
| Losses | ||
| 1p | 1p36 | 10 (23) |
| 6q | 6q23q25 | 12 (27) |
| 13q | 13q34 | 8 (18) |
| Column A (chromosome arm) . | Column B (chromosome band) . | Column C (n = 44), n (%) . |
|---|---|---|
| Gains | ||
| 1q | 1q22 | 22 (50) |
| 2p | 2p16 | 11 (25) |
| 6p | 6p21p25 | 7 (16) |
| 7p | 7p22 | 12 (27) |
| 7q | 7q11 | 16 (36) |
| 8q | 8q24 | 7 (16) |
| 11p | 11p11 | 7 (16) |
| 12q | 12q13 | 9 (21) |
| 16p | 16p13 | 9 (21) |
| 17p | 17p11 | 7 (16) |
| 17q | 17q21 | 12 (27) |
| 18 | 18p11q21 | 18 (41) |
| 19p | 19p13 | 11 (25) |
| 19q | 19q13 | 10 (23) |
| 21 | 21q21 | 8 (18) |
| X | Xp11q21 | 25 (57) |
| Losses | ||
| 1p | 1p36 | 10 (23) |
| 6q | 6q23q25 | 12 (27) |
| 13q | 13q34 | 8 (18) |
Column A indicates chromosome arms with copy number alterations in more than 15% of the first examined biopsies from all patients (n = 44). Chr 18 and X were the only ones with gain of the whole chromosome in more than 15% of the cases, and a whole chromosome gain was seen in 9 (21%) and 20 (46%) of the cases, respectively. Column B indicates chromosome bands with the highest frequency of gain or loss within the arm specified in column A. Column C indicates the number and frequency of cases harboring gain or loss in the region specified in column B.
DNA CNAs in successive biopsies of FL
To examine the evolution of CNAs in FL, we compared the frequencies of CNAs in the first- and last-examined FL samples from the subgroup of 35 patients with 2 or more FL samples (including FL grade 1-3a). We observed a small increase in overall frequencies of CNAs in the last biopsies of FL (Figure 2B). The frequency of 1p36 loss increased the most, from 23% to 43%. Gains of 1q21q23 and 7q11.23 were exceptions, being more common in the first FL samples, 51% versus 37% and 37% versus 20% respectively. None of the CNAs were specifically associated with either the first or the last biopsies. This finding was consistent when examining clinically or morphologically more homogeneous subgroups of the material (ie, FL samples obtained before and after treatment, first and last FL samples from cases that did not transform, first and last samples with FL grade 1-2 only; supplemental Figure 2A-C). We next studied the level of CNAs in relation to time by comparing the CNA index (as defined in “Methods”) in the first and last FL samples from these 35 patients. In line with the observation of slightly increasing frequencies of several CNAs, a trend for increasing CNA index in the last biopsies was found (not statistically significant), reflecting that chromosomal imbalances tend to become more numerous late in the course of FL. The CNA index in each biopsy is represented by the circle size in Figure 3. We observed considerable variation in the CNA index among different cases and also within each case at different time points. Indeed, as this figure shows, in some relapse samples the CNA index was clearly lower than in preceding samples.
Time distribution of sequential biopsies and representation of the CNA index. The length of the horizontal lines represents the observation time (years) from diagnosis for each patient. Open-ended lines indicate that the patient was alive at the last follow-up, whereas closed lines indicate the time of death. The 17 upper cases marked with blue did not transform. The 27 lower cases marked with red transformed (clinical transformation criteria were applied for cases no. 12, 13, 15, 19, 23, 24, 26, and 27; the rest transformed to diffuse large B-cell lymphoma [DLBCL]). Blue circles represent samples of FL grade 1/FL2, orange FL grade 3a, and red DLBCL. For the transformation cases without aCGH data from a transformed biopsy the red lines mark the transformation time point. The circle sizes correlate to the CNA index for each sample which is a marker for genomic complexity. Of note, in many cases (patient nos. 3, 5, 6, 15, 21, 22, 29, 31, 35, 38, 40, 41, and 43) the CNA index is lower in biopsies taken later in the disease course.
Time distribution of sequential biopsies and representation of the CNA index. The length of the horizontal lines represents the observation time (years) from diagnosis for each patient. Open-ended lines indicate that the patient was alive at the last follow-up, whereas closed lines indicate the time of death. The 17 upper cases marked with blue did not transform. The 27 lower cases marked with red transformed (clinical transformation criteria were applied for cases no. 12, 13, 15, 19, 23, 24, 26, and 27; the rest transformed to diffuse large B-cell lymphoma [DLBCL]). Blue circles represent samples of FL grade 1/FL2, orange FL grade 3a, and red DLBCL. For the transformation cases without aCGH data from a transformed biopsy the red lines mark the transformation time point. The circle sizes correlate to the CNA index for each sample which is a marker for genomic complexity. Of note, in many cases (patient nos. 3, 5, 6, 15, 21, 22, 29, 31, 35, 38, 40, 41, and 43) the CNA index is lower in biopsies taken later in the disease course.
Relation of specific CNAs to disease progression
To examine if any of the CNAs in the first FL biopsies were associated with the risk of higher grade transformation, we assigned the 44 patients into 2 groups, depending on whether they underwent transformation (n = 27) or not (n = 17) during the observation time. Importantly, the observation time was long and comparable for both groups (Table 2). The results are displayed in Figure 2C. Interestingly, gains involving chromosome 2 (other than 2p16), 3q, and 5 occurred exclusively in FL biopsies from a proportion of the patients with transformation. One or 2 of these gained segments were present in 14 of the 27 FL samples (52%) from cases with transformation. Among the 5 transforming cases with gain of 3q, 4 had FL with marginal zone differentiation in the first or in subsequent biopsies. Gains of 12q13 and chromosome 21 were more frequent among the 27 cases that transformed compared with the cases who did not (25% vs 13% and 25% vs 6%, respectively). The following CNAs were more frequent among the cases without transformation: gains of 1q (75% vs 36%) and 6p (25% vs 11%) and loss of 6q14 to q15 (38% vs 17%; Figure 2C).
Comparing the frequencies of CNAs in samples with FL grade 1 to 2 (74 samples from 41 patients) and FL grade 3a (17 samples from 14 patients), we found overrepresentation of gain of chromosome 5/5p. Gains of 2p16, 8q24, 11p15, 16p, and Xp11 and of chromosome 18 and 21 were also more frequent in FL3a, in addition to several losses, including loss of 6q (6q14 and 6q23) and 17p (Figure 2D). Differences in gains of 2p16, 8q24, and 11p15 were especially prominent when comparing paired biopsies of FL1 to FL2 and FL3a from a subgroup of 9 patients (supplemental Figure 2D). The CNA index in FL3a samples was significantly higher than in FL1 to FL2 (P < .001), indicating a higher degree of genomic instability in FL3a.
To examine if CNAs also can be predictors of prognosis in FL, we performed Cox regression analysis to test the effect on survival of all 93 segments defined by multi-PCF. A 1-sided test for negative effect of gains identified 11 significant segments. The number of significant segments of loss did not exceed the expected number of false positive (n = 5). The strongest predictor of unfavorable outcome was gain of the X chromosome in male samples (a high amplitude of the most commonly gained segment of X was associated with increased risk of death; P < .01). The other segments of gain with clearly unfavorable prognostic effect were located on the chromosomal arms 2p (2p24.1ptel), 3q (the whole arm), 5p (the whole arm), 5q (5q23qtel), and 12q (12q12q14.1 and 12q22qtel) (Table 4). Notably, the chromosomal gains found exclusively in FLs that transformed (2p, 3q, and 5; Figure 2C) were among those associated with poor survival. Among all 44 cases, there was a trend for poorer survival with higher CNA index in the initial biopsy (not statistically significant).
Frequencies of gains associated with inferior survival in Cox regression analysis (1-sided test for the negative effect of gains on survival)
| Gains . | No transformation (n = 17), n (%) . | Transformation (n = 27), n (%) . |
|---|---|---|
| 2p24.1ptel | 0 | 4 (15) |
| 3q | 0 | 5 (19)* |
| 5p | 0 | 5 (19) |
| 5q23qtel | 0 | 6 (22)* |
| 12q12q14.1 | 3 (18) | 8 (29)* |
| 12q22qtel | 1 (6) | 3 (11) |
| X† | 6 (60) | 14 (70) |
| Gains . | No transformation (n = 17), n (%) . | Transformation (n = 27), n (%) . |
|---|---|---|
| 2p24.1ptel | 0 | 4 (15) |
| 3q | 0 | 5 (19)* |
| 5p | 0 | 5 (19) |
| 5q23qtel | 0 | 6 (22)* |
| 12q12q14.1 | 3 (18) | 8 (29)* |
| 12q22qtel | 1 (6) | 3 (11) |
| X† | 6 (60) | 14 (70) |
The strongest predictor for unfavorable outcome was gain of the X chromosome in male samples (P < .01). The correlation of copy number alterations with survival was calculated with the log2 ratios of the multi–piecewise constant fits segments.
In 2 of the cases the regions of gain were not overlapping.
A total of 10 males had no transformation, and a total of 20 males had transformation.
Clonal relationship and genomic evolution in successive biopsies of FL
Thirty-five of the 39 cases with 2 or more biopsies harbored at least 1 CNA that persisted in all samples, indicating clonal relationship between the sequential biopsies (Figure 4A-D; supplemental Figure 3A-G). Similarly, when examining the 36 cases from which karyotypic data were available from 2 or more biopsies, we observed structural aberrations in addition to t(14;18)(q32;q21) that persisted in the sequential biopsies from 26 cases. Furthermore, by hierarchical clustering the samples from the same patient tended to cluster together (supplemental Figure 4). To study the clonal relationship between the respective lymphoma samples of the patients, rearranged IGH genes were sequenced, and sequences were compared. At the same time, we analyzed somatic hypermutations in the IGVH to establish a genealogic tree for the respective clones. We obtained monoclonal sequences in 2 or more biopsies from 20 cases. The IGHs in sequential samples originated from the same germline sequences and showed identical CDR3 regions in all serial biopsies except 2 cases. In patient no. 3, the CDR3 sequences were not homologous and may thereby reflect the presence of unrelated tumor cell clones in the 2 examined biopsies. In patient no. 8, the obtained CDR3 sequences were technically of inferior quality, which precluded sequence comparison between biopsies. However, a clonal relationship was probably due to usage of the same IGVH gene and several common SHMs. A clonal relationship between sequential biopsies was probable for all cases on the basis of 1 or more of these methods.
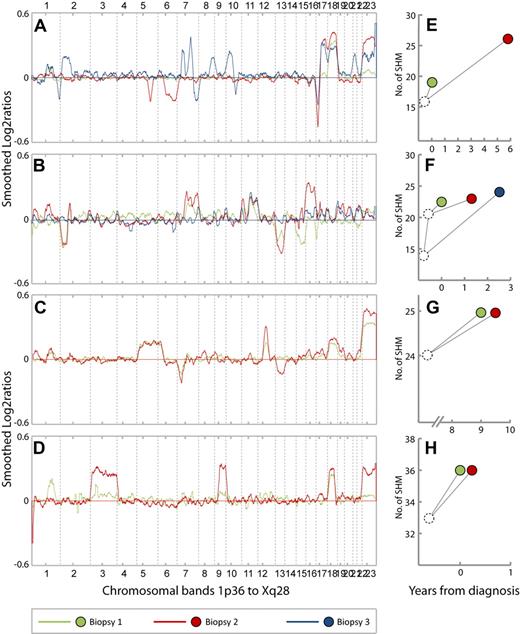
CNA profiles and patterns of SHMs from 4 representative cases. (A-D) CNA profiles from cases 34, 5, 26, and 28, respectively. The chromosomes 1 to 22 and X are aligned along the x-axis from 1p36 to Xq28. Y-axis; smoothed log2 ratios, k = 29. Green, red, and blue lines represent the CNA profiles in biopsy 1, 2, and 3 from each patient, respectively. (E-H) Patterns of somatic hypermutations (SHMs) in samples from cases 34, 5, 26, and 28, respectively. Green, red, and blue dots represent biopsy 1, 2, and 3, respectively. The position of the dots along the x-axis represent the time point at which the biopsies were obtained relative to primary diagnosis (t = 0). Y-axis; number of SHMs. (A) Case 34. Biopsy 1 = FL grade 1 (t = 0). Biopsy 2 = FL grade 2 (t = 5.7). Biopsy 3 = FL grade 3a (t = 6.8). Gain of 18 is present in all 3 biopsies. Losses on 5q and 6q in biopsy 2 are absent in biopsy 3, whereas several new CNAs also appear in biopsy 3. (B) Case 5. Biopsy 1 = FL grade 1 (t = 0). Biopsy 2 = DLBCL (t = 1.2). Biopsy 3 = FL grade 2 (t = 2.4). Gain of 11q is present in all 3 biopsies. Losses of 15 and 17p in biopsy 1 are absent in biopsy 2. Losses of 2p and 13 and gains of 2p, 7q, and 16 in biopsy 2 are absent in biopsy 3. (C) Case 26. Biopsy 1 = FL grade 2 (t = 9). Biopsy 2 = FL grade 3a with marginal zone differentiation (t = 9.4). Loss of 13 appears in biopsy 2. (D) Case 28. Biopsy 1 = FL grade 1 (t = 0). Biopsy 2 = FL grade 1, (t = 0. 1). Gains of chromosome 3 and 9 in biopsy 2 are barely seen in biopsy 1. (E). Case 34. Sixteen mutations are common for biopsies 1 and 2, whereas 3 and 10 additional mutations, respectively, are unique. (F) Case 5. Biopsies 1 and 2 have 21 mutations in common in addition to 1 unique mutation each. Biopsy 3 has 14 mutations in common with the 2 preceding samples in addition to 13 unique mutations. (G) Case 26. Biopsies 1 and 2 have 24 mutations in common and 1 unique mutation each. (H) Case 28. Biopsies 1 and 2 have 33 mutations in common and 3 unique mutations each.
CNA profiles and patterns of SHMs from 4 representative cases. (A-D) CNA profiles from cases 34, 5, 26, and 28, respectively. The chromosomes 1 to 22 and X are aligned along the x-axis from 1p36 to Xq28. Y-axis; smoothed log2 ratios, k = 29. Green, red, and blue lines represent the CNA profiles in biopsy 1, 2, and 3 from each patient, respectively. (E-H) Patterns of somatic hypermutations (SHMs) in samples from cases 34, 5, 26, and 28, respectively. Green, red, and blue dots represent biopsy 1, 2, and 3, respectively. The position of the dots along the x-axis represent the time point at which the biopsies were obtained relative to primary diagnosis (t = 0). Y-axis; number of SHMs. (A) Case 34. Biopsy 1 = FL grade 1 (t = 0). Biopsy 2 = FL grade 2 (t = 5.7). Biopsy 3 = FL grade 3a (t = 6.8). Gain of 18 is present in all 3 biopsies. Losses on 5q and 6q in biopsy 2 are absent in biopsy 3, whereas several new CNAs also appear in biopsy 3. (B) Case 5. Biopsy 1 = FL grade 1 (t = 0). Biopsy 2 = DLBCL (t = 1.2). Biopsy 3 = FL grade 2 (t = 2.4). Gain of 11q is present in all 3 biopsies. Losses of 15 and 17p in biopsy 1 are absent in biopsy 2. Losses of 2p and 13 and gains of 2p, 7q, and 16 in biopsy 2 are absent in biopsy 3. (C) Case 26. Biopsy 1 = FL grade 2 (t = 9). Biopsy 2 = FL grade 3a with marginal zone differentiation (t = 9.4). Loss of 13 appears in biopsy 2. (D) Case 28. Biopsy 1 = FL grade 1 (t = 0). Biopsy 2 = FL grade 1, (t = 0. 1). Gains of chromosome 3 and 9 in biopsy 2 are barely seen in biopsy 1. (E). Case 34. Sixteen mutations are common for biopsies 1 and 2, whereas 3 and 10 additional mutations, respectively, are unique. (F) Case 5. Biopsies 1 and 2 have 21 mutations in common in addition to 1 unique mutation each. Biopsy 3 has 14 mutations in common with the 2 preceding samples in addition to 13 unique mutations. (G) Case 26. Biopsies 1 and 2 have 24 mutations in common and 1 unique mutation each. (H) Case 28. Biopsies 1 and 2 have 33 mutations in common and 3 unique mutations each.
To examine the genomic evolution of tumor cell clones in relapsing and progressing FL more closely, we evaluated the sequence in which CNAs appeared in each of the 39 cases represented by 2 or more biopsies. The CNA profiles from each patient displayed interesting differences during the disease course. Notably, in 27 of 39 cases, 1 or more of CNAs present in initial biopsies were absent in later relapse biopsies and in 24 of these new CNAs also appeared. These observations are shown by representative cases in Figure 4A-B (all 27 cases can be viewed in supplemental Figure 3A-E). In 9 cases, we observed only the appearance of new CNAs in relapse biopsies (Figure 4C; supplemental Figure 3F-G). Three cases (case no. 30, 31, and 41) showed CNA profiles without clear changes.
On the basis of the somatic hypermutations of the IGVH regions from the 19 cases with clonally related tumor samples, we constructed genealogic trees by aligning the IGVH sequences from successive biopsies with germline sequences. Genealogic trees could be constructed for 13 of the 27 cases with early CNAs being absent in later biopsies. The pattern of shared and unique SHMs from 11 cases were compatible with divergent clonal evolution; in 2 cases the SHM patterns were identical in the examined biopsies (supplemental Figure 3A-E). Among the 12 cases displaying the addition of new CNAs or unchanged CNA profiles in relapse biopsies, genealogic trees could be made for 6 cases (supplemental Figure 3F-G). In 3 of these, the SHM patterns were also compatible with divergent clonal evolution. In one case the SHM pattern showed linear evolution, whereas in 2 cases the SHM patterns were identical in the examined biopsies. The genealogic trees from representative cases are displayed in Figure 4E-H. In some cases the changes in CNA profile suggested that different subclones coexisted in biopsies and that the relative proportions of these clones changed during progression. This is indicated by the increased amplitude of CNAs from one biopsy compared with the next, whereas both showed a constant tumor cell content and ploidy (Figure 4D).
Discussion
Secondary genomic aberrations are common in FL and comprise a consistent pattern of recurrent chromosomal imbalances, as shown by classic cytogenetic analysis and CGH analysis.7,,,,,–13,26,27,36,44,,,,,–50 Most studies are based on the analysis of patient biopsies obtained at diagnosis. We have studied sequential samples in a set of patients to elucidate possible patterns in which secondary genetic aberrations are acquired and to identify possible chromosomal regions associated with clinical progression, histologic transformation, and prognosis. By analyzing CNA profiles and patterns of SHMs, we have shown that parallel evolution of tumor cell clones is frequent in FL. Moreover, correlation of CNA frequencies to clinical data indicates that a set of specific CNAs with prognostic effect are present in FL samples from patients with higher grade transformation.
Chromosomal gains involving 1q, 2p, 5q, 6p, 7, 8q24, 12q, 17q, 18, 21, and X and losses of 1p and 6q were common changes in FL samples, as also reported by others.7,,,,,–13,26,27,44,,,,,–50 However, gained segments on 1q, 7q, 9q, 11p, 16p, and 17q; gain of chromosomes 18, 19, 21, and X; and loss of 13q34 appear more frequently in our material than in previous studies. The latter could be due to the selected character of our material as only cases of FL with sequential frozen samples were studied. Notwithstanding, some of these CNAs involve narrow regions, such as gains of 1q21q23, 7p22, 7q11, 8q24, 9q34, 11p15, 11p11, and 16p13 and loss of 13q34, which may not have been detected by the methods used in previous studies. Our study made use of a highly sensitive algorithm (PCF) as evidenced by the detection of rearranged DNA segments in the immunoglobulin κ light chain and heavy chain loci.
Eleven segments of gain were associated with inferior survival in our material; these were located on 2p, 3q, 5p, 5q, 12q, and the X chromosome. The gain of X in FL samples from male patients was the strongest predictor of unfavorable outcome (P < .01), confirming the findings in a previous study.12 By contrast, losses in the initial biopsies had no statistically significant effect on survival when corrected for multiple testing. Thus, our data could not confirm that the losses of 1p36 and 6q23q26 are independent prognostic factors and risk factors for transformation, as previously reported.8,10,11,13,51 The gains of 2p, 3q, 5, and 12q have been associated with aggressive clinical or histopathologic features of FL in previous studies.7,11,–13,20,24,26,27,44 Intriguingly, we observed that gains involving chromosome 2, 3q, or 5 in initial FL biopsies were exclusively present in samples from patients with transformation (14 of 27 cases), in line with the prognostic value associated with gains of these segments. FL with marginal zone differentiation was found in 4 of 5 transforming cases with gain of 3q. This is consistent with previous studies reporting that gain of 3q is frequent in FLs with marginal zone differentiation as well as in nodal marginal zone lymphomas. The association between gain of 3q and FL with marginal zone differentiation suggests that this represents a high-risk variant of FL.52,53 In addition to its prognostic value, the gain of chromosome 5 was more frequent in samples of FL grade 3a than in FL grade 1 to 2, confirming similar findings in 2 previous studies.44,46 In a study comparing cytogenetic data from paired biopsies of FL and DLBCL,20 the gains of 3q and 5p among other CNAs were present in the DLBCL samples and not in the paired FL samples, indicating that these CNAs may also be acquired on transformation to DLBCL. In conclusion; our findings indicate that gains involving 3q and chromosome 5 are associated with the risk of transformation of FL to DLBCL or by clinical criteria. In addition, gain of 3q is associated with marginal zone differentiation in FL and gain of chromosome 5 with morphologic transition to FL grade 3a. An interesting and novel finding is that gain of 2p24.1ptel also may be involved in clinical and histologic transformation.
We noted a trend for increasing genomic complexity measured by comparing the CNA index in initial and late FL samples. Because most samples in these analyses were FL grade 1 to 2 (31 of 35 initial biopsies and 24 of 35 late biopsies), our findings suggest that relapse of FL without histologic progression is associated with few genetic changes. Of interest, comparison of all FL1 to FL2 samples (n = 74) with all FL3a samples (n = 17) showed that several CNAs were more common in FL3a and that the CNA index was significantly higher in FL3a (P < .001), confirming previous findings.44,46 Our findings indicate that a shift in morphology from FL1/FL2 to FL3a is associated with increased genomic instability and acquisition of chromosomal imbalances such as gains involving 5, 8q24, 11p11, 12q, 16, 18, 21, and Xp as well as a diversity of losses, including loss of 6q and 17p. However, despite a trend for increasing genomic complexity with time and grade of FL, the overall CNA frequencies in initial and late FL biopsies showed a surprisingly stable pattern of CNAs characterizing FL throughout the course of the disease. The CNAs that were common in initial biopsies of FL increased slightly in frequency in later relapse biopsies. Loss of 1p36 was the only CNA that appeared markedly more often in the late biopsies. Gain of 1q12q23 and 7q11.23 were more frequent in initial biopsies. This pattern was consistent also when clinically or structurally homogeneous subgroups were examined.
In the majority of the cases with sequential biopsies (27 of 39), 1 or more of the CNAs present in the initial sample were absent in a later sample, and new CNAs were acquired, indicating that tumor cell clones at relapse were not direct descendants of clones dominating in a preceding sample. In the remaining 12 cases with sequential biopsies, the CNA profiles were either unchanged or displayed additional CNAs in the late samples, being compatible with a linear progression from the initial sample to the later relapse. Similar findings were reported from studying karyotypic data from sequential biopsies of FL12,27 and using conventional CGH20 and single nucleotide polymorphism array analysis25 of paired FL and DLBCL samples. A closer examination of the variation in CNA index with time (Figure 3) showed that in several cases the relapse biopsies were actually less complex than the preceding biopsies, indicating that in many cases clones evolve in parallel and with a distinctly different time course in terms of developing genomic instability. In a previous study,20 the mean number of aberrations was higher in DLBCL than in FL, but the opposite was also seen in some cases, consistent with our findings.
To further investigate the question of parallel versus linear clonal evolution in patients with FL, we studied the pattern of SHMs of the rearranged IGVH gene. The analysis of SHMs in sequential biopsies allows the establishment of a genealogic tree for the tumor clones present in the biopsies of each patient. Such a tree clarifies the actual temporal sequence of occurrence of the clones. Of interest, 14 of the 19 cases that could be analyzed for SHMs showed a pattern compatible with an origin of the respective clones in the serial biopsies from a less-mutated common ancestor cell and not directly from each other. These findings are in agreement with those of others.29,32,–34 Taken together, the analysis of CNA profiles and SHM patterns show that parallel evolution of tumor cell clones is common in FL and support the hypothesis of common progenitor cells in FL. The frequent relapse in FL suggests that the hypothetical common progenitor cells escape treatment and give rise to new tumor cell clones that are genetically different from primary clones. The different states of the tumor genome at each relapse seem to be correlated with morphologic and clinical features.
Previous studies have shown that the composition of the tumor microenvironment21,22 in FL has an effect on survival. The factors determining the rate of progression toward a more treatment-resistant phenotype may depend on the interplay between the tumor cells and the tumor-infiltrating immune cells. Whether it is the type of antigen stimuli, the immunologic constitution of the host, or the genetic status of a common progenitor cell in FL that directs this interplay needs to be further investigated.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Ragnhild Lothe, Franclim Ribeiro, and Rolf Skotheim for advice and guidance through aCGH data analysis; Ana Barragan Lid for help with hybridization and scanning of arrays; and Chloe Steen and Abdirashid Warsame for the performance of PCR and sequence analysis of IGVH.
This work was supported by the Faculty of Medicine (University of Oslo), The Norwegian Research Council, and the Norwegian Cancer Society. Microarray services were provided by the Norwegian Microarray Consortium, funded by the FUGE Program of the Norwegian Research Council.
Authorship
Contribution: M.B.E., M.E.H., S.H.K., G.T., and H.V.A. performed experiments; M.B.E., K.L., O.C.L., H.H., E.B.S., and J.D. performed analysis and made figures; L.M.-Z., O.M., H.H., E.B.S, and J.D. designed the research; and M.B.E., K.L., O.C.L., H.H., E.B.S., and J.D. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marianne Brodtkorb Eide, Department of Immunology, Institute for Cancer Research, Oslo University Hospital, Montebello, 0310 Oslo, Norway; e-mail: marianne.brodtkorb.eide@rr-research.no.

![Figure 3. Time distribution of sequential biopsies and representation of the CNA index. The length of the horizontal lines represents the observation time (years) from diagnosis for each patient. Open-ended lines indicate that the patient was alive at the last follow-up, whereas closed lines indicate the time of death. The 17 upper cases marked with blue did not transform. The 27 lower cases marked with red transformed (clinical transformation criteria were applied for cases no. 12, 13, 15, 19, 23, 24, 26, and 27; the rest transformed to diffuse large B-cell lymphoma [DLBCL]). Blue circles represent samples of FL grade 1/FL2, orange FL grade 3a, and red DLBCL. For the transformation cases without aCGH data from a transformed biopsy the red lines mark the transformation time point. The circle sizes correlate to the CNA index for each sample which is a marker for genomic complexity. Of note, in many cases (patient nos. 3, 5, 6, 15, 21, 22, 29, 31, 35, 38, 40, 41, and 43) the CNA index is lower in biopsies taken later in the disease course.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/9/10.1182_blood-2010-03-272278/5/m_zh89991056410003.jpeg?Expires=1769200555&Signature=vUByVTXGybhZQL00OuN0I6XkvNwsUDPcIl3um8W6Yt7dFN0AiszYC0iyv9QruHsIe9ytDxVmPZB3iK5aUu-IBUfATXOS6Ti8FdHocykI91o8gqrnHWfkiicANjjm2YVLCF~wrZTiUAw4k~mqxX5YmaaiNusXUATlj-nMWuH9ZMw~tqvuvrJnM3DdygXsw04EfTTiZcHS4-t5tny0PIoA0qnYtqYOoPgInKQPNmaAsfvvpI8gokint0KNkKkgbtdgpVUteNkljrB04rPEUAwfO1t2kg-mexLfuMNrXOEBWfE-HwYvchZ-KBLZXvsQUNNoMNQVQ~MzgYgelYqAPN4y1A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)