Hereditary hemorrhagic telangiectasia (HHT) is an autosomal dominant genetically inheritable vascular dysplasia caused by mutations in genes encoding receptors of the transforming growth factor-β (TGF-β) family: ENG, encoding endoglin (HHT1), and ACVRL1, encoding activin receptor-like kinase-1 (ALK1; HHT2). Our recent discovery of bone morphogenetic protein 9 (BMP9) as the specific ligand for ALK1 allowed us to reevaluate the functional significance of ACVRL1 mutations. We generated 19 ALK1 mutants reproducing HHT2 mutations (4 were novel mutations) found throughout the protein. We show that all ALK1 mutant proteins were expressed by transfected cells; most of them were present at the cell surface and retained their ability to bind BMP9 (except for the extracellular mutants). However, most were defective in BMP9 signaling. None of the ALK1 mutants had a dominant negative effect on wild-type ALK1 activity. These data demonstrate that mutations of ACVRL1 fit with a functional haploinsufficiency model affecting BMP9 signaling. Our study also identified 4 ACVRL1 mutations (D179A, R386C, R454W, and A482V) that did not alter the BMP9 responses that are polymorphisms and 2 novel mutations that are pathogenic (L381P and I485F). This demonstrates that the analysis of BMP9 responses can be used as a diagnostic tool by geneticists confronted with novel or conflicting ACVRL1 mutations.
Introduction
Hereditary hemorrhagic telangiectasia (HHT, Rendu-Osler disease) is an autosomal dominant genetically inheritable vascular dysplasia. HHT affects approximately 1 in 8000 persons, and approximately 1.2 million people worldwide have HHT. The clinical features of the disorder include recurrent nosebleeds, mucocutaneous telangiectases, and arteriovenous malformations in lung, liver, and central nervous system. HHT is caused by germline mutations in 1 of 2 major genes: ENG, encoding endoglin, and ACVRL1, encoding activin receptor-like kinase type 1 (ALK1), respectively, defining the HHT1 and HHT2 clinical variants.1,2 Mutations have also been identified in MADH4 that encodes Smad4 in a subset of families with combined juvenile polyposis and HHT, and there is evidence for the existence of 2 other HHT-associated genes.3,–5 ACVRL1 and ENG mutations are currently found in 80% to 90% of patients with a definite clinical diagnosis.6 HHT displays wide allelic heterogeneity, and more than 600 mutations have been reported in the HHT mutation database (www.hhtmutation.org). Mutations in ACVRL1 or ENG are distributed over the entire coding sequence. Mutations can also be found in intronic splice sites of both genes. Large deletions or duplications, from single exons to the whole gene, have also been reported.7,–9
The mechanisms by which these mutations cause HHT are not yet clearly understood. The different manifestations vary considerably, even among members of the same family sharing the same mutation; to date, disease severity has not been associated with specific mutations.7,10 HHT1 patients express reduced levels of endoglin.11,12 HHT1 is therefore the result of haploinsufficiency.13 Similarly, haploinsufficiency may account for HHT2, although the abundance of missense mutations affecting exons 7 and 8 suggests abnormal function of certain ALK1 proteins. ALK1 expression has been measured in human umbilical vein endothelial cells (HUVECs) by fluorescence-activated cell sorter (FACS) or metabolic labeling and immunoprecipitation and has been found to be reduced in 3 patients with ACVRL1 missense mutant codons (G48E/A49P, W50C, and S333I) and to be normal in 1 patient with an in-frame deletion (S232del).14 However, the number of patients is too small to definitively conclude that HHT2 is really the result of haploinsufficiency.
Endoglin and ALK1 are components of a transforming growth factor-β (TGF-β) family receptor complex that are primarily expressed on endothelial cells. Endoglin is a type III receptor with no kinase activity that enhances ligand binding to its signaling receptors. ALK1 is 1 of 5 different type I receptors of the TGF-β family that act downstream of the type II receptors and determine the signaling specificity by recruiting receptor-Smads. ALK1 promotes Smad1 and Smad5 phosphorylation. The ALK1 receptor had long been considered an orphan receptor. However, we recently identified bone morphogenetic protein 9 (BMP9) and BMP10 as ligands for ALK1.15 These data were subsequently confirmed by another group.16 Furthermore, we could establish that BMP9, but not BMP10, is present in human plasma and contributes to adult vascular quiescence.17
The discovery of BMP9 as a specific ligand for ALK1 may represent a significant step in understanding HHT as it allows us to reconsider the molecular mechanisms behind the disease pathogenesis.18 The aim of the present study was to evaluate the functional significance of ACVRL1 mutations found in HHT2 patients using BMP9 as a ligand. We generated 19 ALK1 mutants corresponding to 15 previously described mutations and 4 novel mutations. These mutations were distributed throughout different regions of the protein. We found that all the ALK1 mutant proteins were expressed and that most of them were present at the cell surface and retained their ability to bind BMP9 (except for the extracellular mutations). However, most were defective in BMP9 signaling. Furthermore, none of the ALK1 mutant behaved as a dominant negative receptor by inhibiting signaling through the wild-type receptor. This study suggests that HHT2 follows a model of functional haploinsufficiency in which the BMP9 response in heterozygotic patients is decreased by 50% because of the loss of 1 functional allele. We also demonstrated that analysis of the BMP9 response could discriminate pathogenic (L381P, I485F) from polymorphic (D179A, R386C, R454W) ACVRL1 mutations. This further demonstrates that the study of the BMP9 response through the BRE-luciferase reporter gene activity will be a useful tool for molecular geneticists.
Methods
Cell line
NIH-3T3 fibroblasts and COS-7 cells were maintained in Dulbecco modified Eagle medium, 4.5 g/L glucose (Invitrogen) supplemented with 10% fetal bovine serum (Biowest; Abcys).
DNA constructs
ALK1 mutants were generated by polymerase chain reaction through site-directed mutagenesis of the WT-ALK1 plasmid cloned in pcDNA3 (kindly provided to us by Dr C. H. Heldin, Ludwig Institute for Cancer Research, Uppsala, Sweden) using the Stratagene QuikChange kit.
Fifteen mutations found in HHT2 patients throughout the ACVRL1 gene were selected (Figure 1A; Table 1). Fourteen were missense mutations: 3 were in the extracellular domain (C51Y, C77W, and N96D); 1 was located in the intracellular GS (glycine/serine rich) region, a helix-loop-helix structure proximal to the kinase domain (D179A); and 10 were located in the intracellular kinase domain (A347P, R374W, R374Q, R411Q, R411W, R411P, R479Q, R484W, R484Q, and I485F). One was a 1-bp frameshift insertion (c.1112dupG) leading to a premature stop codon (G371fsX391). This mutation is frequent in patients of French descent and has been shown to be the result of a regional founder effect.19 Four other ACVRL1 missense mutations were also selected because they were found in patients with conflicting results with respect to their molecular diagnosis: 3 ACVRL1 mutations (L381P, A482V, and R454W) corresponded to patients who also had a mutation in the ENG gene, whereas the last (R386C) was observed in a patient with 2 mutations located on the same ACVRL1 allele.
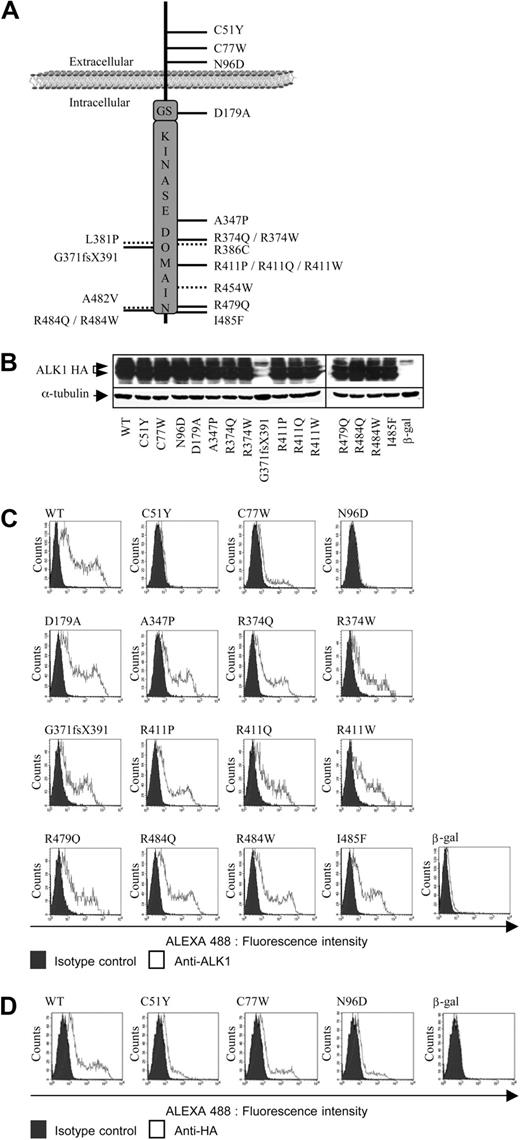
Expression and localization of the different ALK1 mutants. (A) The 19 different mutations are distributed on the map of the ALK1 protein. The mutations indicated with a dotted line are the conflicting mutations. (B) Protein expression of ALK1 mutants. Expression vectors encoding HA-tagged (at their C-terminus), ALK1 (WT or mutants), or β-gal (control) were transiently transfected into NIH-3T3 cells for 24 hours. Cell lysates (20 μg proteins) were resolved by 10% SDS-PAGE and immunoblotted with antibodies directed against HA and α-tubulin (as a loading control). (C-D) Flow cytometry analysis of NIH-3T3 cells transfected with expression vectors encoding HA-tagged (at their N-terminus), ALK1 (WT or mutants), or β-gal (control) for 24 hours. Nonpermeabilized transfected cells were stained for cell surface ALK1 expression (C, anti-ALK1 antibody) or (D, anti-HA antibody).
Expression and localization of the different ALK1 mutants. (A) The 19 different mutations are distributed on the map of the ALK1 protein. The mutations indicated with a dotted line are the conflicting mutations. (B) Protein expression of ALK1 mutants. Expression vectors encoding HA-tagged (at their C-terminus), ALK1 (WT or mutants), or β-gal (control) were transiently transfected into NIH-3T3 cells for 24 hours. Cell lysates (20 μg proteins) were resolved by 10% SDS-PAGE and immunoblotted with antibodies directed against HA and α-tubulin (as a loading control). (C-D) Flow cytometry analysis of NIH-3T3 cells transfected with expression vectors encoding HA-tagged (at their N-terminus), ALK1 (WT or mutants), or β-gal (control) for 24 hours. Nonpermeabilized transfected cells were stained for cell surface ALK1 expression (C, anti-ALK1 antibody) or (D, anti-HA antibody).
Functional significance of ACVRL1 mutations
| Mutation location . | Mutation category . | Nucleotide change . | Amino acid change . | Location of mutation . | Novel mutation . | Conflicting mutation* . | Expression . | Localization . | BMP9 binding . | BMP9 response . |
|---|---|---|---|---|---|---|---|---|---|---|
| Exon 3 | Missense | c.152G → A | p.C51Y | Extracellular | — | — | Yes | Intracellular | No | No |
| Exon 3 | Missense | c.231C → G | p.C77W | Extracellular | — | — | Yes | Partial cell surface | No | No |
| Exon 3 | Missense | c.286A → G | p.N96D | Extracellular | — | — | Yes | Intracellular | No | No |
| Exon 5 | Missense | c.536A → C | p.D179A | GS domain | — | — | Yes | Cell surface | Yes | Yes |
| Exon 7 | Missense | c.1039G → C | p.A347P | Kinase domain | — | — | Yes | Cell surface | Yes | No |
| Exon 8 | Missense | c.1121G → A | p.R374Q | Kinase domain | — | — | Yes | Cell surface | Yes | No |
| Exon 8 | Missense | c.1120C → T | p.R374W | Kinase domain | — | — | Yes | Cell surface | Yes | No |
| Exon 8 | Frameshift | c.1112_1113dupG | p.G371fsX391 | Kinase domain | — | — | Yes | Cell surface | No | No |
| Exon 8 | Missense | c.1142T → C | p.L381P | Kinase domain | Yes | Yes | Yes | Not tested | Not tested | No |
| Exon 8 | Missense | c.1156C → T | p.R386C | Kinase domain | Yes | Yes | Yes | Not tested | Not tested | Yes |
| Exon 8 | Missense | c.1232G → C | p.R411P | Kinase domain | — | — | Yes | Cell surface | Yes | No |
| Exon 8 | Missense | c.1232G → A | p.R411Q | Kinase domain | — | — | Yes | Cell surface | Yes | No |
| Exon 8 | Missense | c.1231C → T | p.R411W | Kinase domain | — | — | Yes | Cell surface | Yes | No |
| Exon 9 | Missense | c.1360C → T | p.R454W | Kinase domain | Yes | Yes | Yes | Not tested | Not tested | Yes |
| Exon 10 | Missense | c.1436G → A | p.R479Q | Kinase domain | — | — | Yes | Cell surface | Yes | No |
| Exon 10 | Missense | c.1445C → T | p.A482V | Kinase domain | — | Yes | Yes | Not tested | Not tested | Yes |
| Exon 10 | Missense | c.1451G → A | p.R484Q | Kinase domain | — | — | Yes | Cell surface | Yes | No |
| Exon 10 | Missense | c.1451C → T | p.R484W | Kinase domain | — | — | Yes | Cell surface | Yes | No |
| Exon 10 | Missense | c.1453A → T | p.I485F | Kinase domain | Yes | — | Yes | Cell surface | Yes | No |
| Mutation location . | Mutation category . | Nucleotide change . | Amino acid change . | Location of mutation . | Novel mutation . | Conflicting mutation* . | Expression . | Localization . | BMP9 binding . | BMP9 response . |
|---|---|---|---|---|---|---|---|---|---|---|
| Exon 3 | Missense | c.152G → A | p.C51Y | Extracellular | — | — | Yes | Intracellular | No | No |
| Exon 3 | Missense | c.231C → G | p.C77W | Extracellular | — | — | Yes | Partial cell surface | No | No |
| Exon 3 | Missense | c.286A → G | p.N96D | Extracellular | — | — | Yes | Intracellular | No | No |
| Exon 5 | Missense | c.536A → C | p.D179A | GS domain | — | — | Yes | Cell surface | Yes | Yes |
| Exon 7 | Missense | c.1039G → C | p.A347P | Kinase domain | — | — | Yes | Cell surface | Yes | No |
| Exon 8 | Missense | c.1121G → A | p.R374Q | Kinase domain | — | — | Yes | Cell surface | Yes | No |
| Exon 8 | Missense | c.1120C → T | p.R374W | Kinase domain | — | — | Yes | Cell surface | Yes | No |
| Exon 8 | Frameshift | c.1112_1113dupG | p.G371fsX391 | Kinase domain | — | — | Yes | Cell surface | No | No |
| Exon 8 | Missense | c.1142T → C | p.L381P | Kinase domain | Yes | Yes | Yes | Not tested | Not tested | No |
| Exon 8 | Missense | c.1156C → T | p.R386C | Kinase domain | Yes | Yes | Yes | Not tested | Not tested | Yes |
| Exon 8 | Missense | c.1232G → C | p.R411P | Kinase domain | — | — | Yes | Cell surface | Yes | No |
| Exon 8 | Missense | c.1232G → A | p.R411Q | Kinase domain | — | — | Yes | Cell surface | Yes | No |
| Exon 8 | Missense | c.1231C → T | p.R411W | Kinase domain | — | — | Yes | Cell surface | Yes | No |
| Exon 9 | Missense | c.1360C → T | p.R454W | Kinase domain | Yes | Yes | Yes | Not tested | Not tested | Yes |
| Exon 10 | Missense | c.1436G → A | p.R479Q | Kinase domain | — | — | Yes | Cell surface | Yes | No |
| Exon 10 | Missense | c.1445C → T | p.A482V | Kinase domain | — | Yes | Yes | Not tested | Not tested | Yes |
| Exon 10 | Missense | c.1451G → A | p.R484Q | Kinase domain | — | — | Yes | Cell surface | Yes | No |
| Exon 10 | Missense | c.1451C → T | p.R484W | Kinase domain | — | — | Yes | Cell surface | Yes | No |
| Exon 10 | Missense | c.1453A → T | p.I485F | Kinase domain | Yes | — | Yes | Cell surface | Yes | No |
Patients with another described mutation in ENG or ACVRL1.
Within these mutations, 4 (L381P, R386C, R454W, and I485F) had not been reported previously in patients with HHT2. The mutations (D179A, R374W, R374Q, L381P, R411W, R411Q, and R484W) have been described in pulmonary artery hypertension (PAH) patients.20,–22
Double ALK1 mutants carrying both a mutation turning the receptor into a constitutively active ALK1 (ALK1ca) form (Q201D) and a mutation found in HHT2 (Q201D/A347P, Q201D/R479Q, and Q201D/R484W) were also generated by site-directed mutagenesis.
All these constructs were HA-tagged at their C-terminus. Some of the mutants were also (His)6-tagged at their C-terminus (WT-ALK1, R479Q, R484Q, and I485F) or HA-tagged at their N-terminus (WT-ALK1, C51Y, C77W, and N96D). A β-gal plasmid cloned in pcDNA3 was used as a control.
Reporter gene constructs
The reporter plasmid pGL3(BRE)2-luc encoding firefly luciferase downstream of a BMP response element23 was kindly provided by Dr P. ten Dijke (Leiden University Medical Center, Leiden, The Netherlands). The pRL-TK-luc plasmid encoding renilla luciferase downstream of the thymidine kinase promoter was purchased from Promega.
DNA transfection and dual luciferase activity assay
NIH-3T3 cells were transfected in Opti-MEM (Invitrogen) using lipofectamine (Invitrogen) with 0.1 μg pGL3(BRE)2-luc, 0.02 μg of pRL-TK-luc, and different doses of plasmids encoding WT-ALK1 or the different mutants. Four hours after transfection, cells were treated with or without recombinant BMP9 (100 pg/mL; R&D Systems) for 15 hours. Firefly and renilla luciferase activities were measured sequentially with the Dual-Luciferase reporter assay (Promega).
Immunoprecipitation and Western blot analysis
NIH-3T3 cells were transfected as described in “DNA transfection and dual luciferase activity assay” with the different ALK1 mutants. At 48 hours later, cells were serum-deprived for 1 hour and then stimulated with BMP9 (100 pg/mL) for 1 hour. Cells were then washed twice with PBS and lysed in radioimmunoprecipitation assay (RIPA) buffer with a cocktail of protease inhibitors (P8340; Sigma-Aldrich). For immunoprecipitation, cells were lysed in 50mM phosphate buffer (pH 8), 300mM NaCl, 5mM imidazole, 0.5% (vol/vol) Triton X-100. Cell lysates were subjected to overnight immunoprecipitation with the anti-His antibody (H1029; Sigma-Aldrich), followed by adsorption to protein G Plus-Agarose (Calbiochem) for 1 hour. Proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE; 10%) and analyzed by immunoblotting with anti-HA (3F10; Roche Diagnostic GmbH), anti-phosphoSmad1/5 (41D10; Cell Signaling Technology), and anti–α-tubulin (generously provided by Dr D. Job, Inserm U366, Grenoble, France) antibodies.
Flow cytometric analysis of ALK1 expression
NIH-3T3 cells were transfected as described in “DNA transfection and dual luciferase activity assay” with the different ALK1 mutants. Twenty-four hours later, cells were detached with diluted trypsin solution (0.000625%). Cells were then labeled for 1 hour at 4°C with polyclonal anti-ALK1 (AF370; R&D Systems) or anti-HA (3F10; Roche Diagnostic GmbH) antibodies or with isotype-matched control IgGs. Labeling was detected with Alexa488-conjugated secondary antibodies (Invitrogen). Labeled cells were analyzed by FACS using a FACScan apparatus and the CellQuest software (BD Biosciences).
[125I]-BMP9 radioreceptor assay
BMP9 was radioiodinated using the chloramine T method.24 COS-7 cells were incubated on ice for 2 hours with radiolabeled BMP9 (36 μCi/μg). After incubation, cells were washed, lysed (NaOH 0.1M), and bound radioactivity was measured in a γ-counter. Nonspecific binding, as determined on β-gal-transfected cells, was deduced from the values of total binding on ALK1-transfected cells.
Results
The ALK1 protein consists of a small extracellular domain (encoded by exons 2, 3, and part of exon 4), a short transmembrane domain (exons 4, and part of 5), and a large intracellular domain including the GS domain (from exon 5) and the serine/threonine kinase domain (exons 5-10). To investigate the effects of HHT2-related ALK1 mutations on BMP9 signaling, we first generated 15 mutants, reproducing human mutations found throughout the ACVRL1 gene in HHT2 patients (Table 1; Figure 1A).
First, we analyzed whether these mutations affect protein expression. To this end, we transfected the ALK1 mutant constructs into NIH-3T3 cells that do not endogenously express ALK1,15 and evaluated mutant proteins by anti-HA immunoblotting. A β-gal-expressing plasmid was used as a control in all experiments. As shown in Figure 1B, all mutants, except 1 (G371fsX391), were expressed at levels comparable with that of wild-type ALK1 (WT). The mutation G371fsX391 could not be detected by Western blotting as this mutation leads to a truncated protein lacking the C-terminal HA tag. To evaluate the expression of this protein and to determine whether ACVRL1 mutations affect ALK1 localization at the plasma membrane, we examined ALK1 cell surface expression using FACS analysis with a polyclonal anti-ALK1 antibody. No ALK1 expression was detected in the absence of ALK1 transfection in accordance with our previous work (β-gal control, Figure 1C).15 As shown in Figure 1C, ALK1 cell surface expression was detected at a similar level as WT-ALK1 in all cases, including the G371fsX391 mutant, except for cells transfected with the mutants within the extracellular domain (C51Y, C77W, and N96D). To determine whether the ALK1 extracellular mutant protein was present at the cell surface or whether the mutations induced conformational changes that hampered epitope recognition, we HA-tagged the extracellular mutants at their N-terminus and measured cell surface expression by FACS analysis using anti-HA antibodies. As shown in Figure 1D, the C77W mutant was partially expressed at the cell surface, whereas the C51Y and N96D mutants were barely expressed at the cell surface.
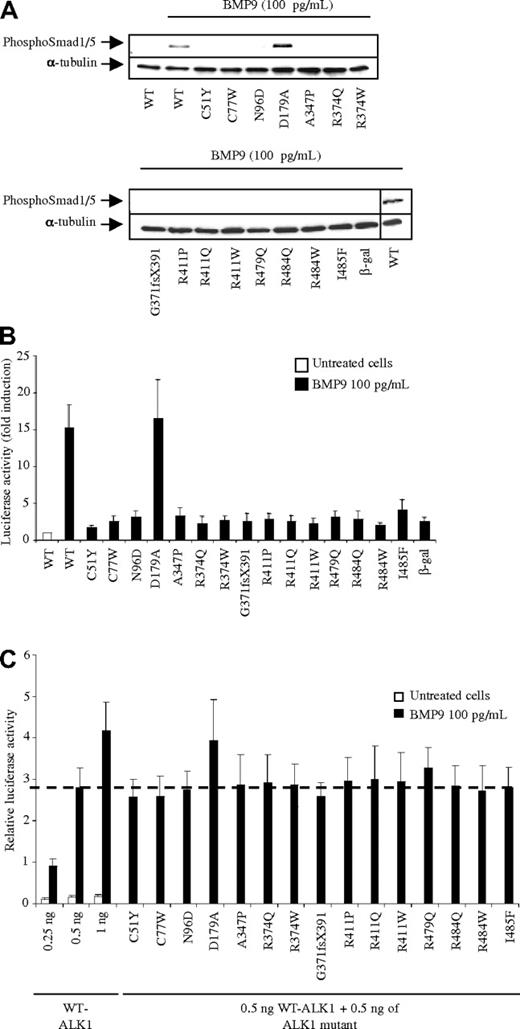
We next evaluated the functional activity of these different ALK1 mutants. First, we investigated the effect of the different mutations on ALK1 activity in response to BMP9 by Western blot analysis of Smad1/5 phosphorylation. For this, 24 hours after transfection, NIH-3T3 cells were serum-deprived for 1 hour and then stimulated for 1 hour with a low dose of BMP9 (100 pg/mL) as we previously demonstrated that, at this dose, BMP9 only binds to ALK1.17 As shown in Figure 2A, the addition of BMP9 strongly induced Smad1/5 phosphorylation in the presence of WT-ALK1 but did not induce Smad phosphorylation in any mutant except D179A, which carries a mutation in the GS box. We also evaluated the activity of these mutants in response to BMP9, using the Smad1/5-responsive transcriptional reporter, pGL3(BRE)2-luciferase.23 The reporter assay was carried out in mutant ALK1/pGL3(BRE)2-luciferase transfected NIH-3T3 cells stimulated with BMP9 (100 pg/mL) for 12 hours, starting 4 hours after tranfection. BMP9 stimulation induced a 15-fold induction of luciferase activity in cells transfected with WT-ALK1 (Figure 2B). Again, none of these mutants responded to BMP9 stimulation, with the exception of D179A, which responded to BMP9 similarly to WT-ALK1 (16-fold induction, Figure 2B).
BMP9 response of ALK1 mutants. (A) Expression vectors encoding WT-ALK1 or the different ALK1 mutants or β-gal (control) were transiently transfected into NIH-3T3 cells. After 24 hours, cells were serum deprived for 1 hour and subsequently treated or not with BMP9 (100 pg/mL) for 1 hour. Cell lysates (20 μg proteins) were resolved by 10% SDS-PAGE and immunoblotted with antibodies against phosphoSmad1/5 or against α-tubulin (as a loading control). (B) NIH-3T3 cells were transiently transfected with pGL3(BRE)2-luc, pRL-TK-luc, and plasmids (0.5 ng) encoding either WT-ALK1 or the different ALK1 mutants or β-gal (control). After 4 hours, cells were treated or not with BMP9 (100 pg/mL) for 15 hours. The luciferase activities were then measured as described in “DNA transfection and dual luciferase activity assay.” The relative firefly luciferase activity was normalized to renilla luciferase activity. Results are expressed as fold induction over the value obtained for each ALK1 mutant in the absence of BMP9. Data are mean ± SD of 3 independent experiments. (C) NIH-3T3 cells were transiently transfected with pGL3(BRE)2-luc, pRL-TK-luc, and different combinations of WT and mutant ALK1: WT-ALK1 alone (0.25, 0.5, or 1 ng) or WT-ALK1 (0.5 ng) and either one of the different mutants ALK1 (0.5 ng). After 4 hours, cells were treated or not with BMP9 (100 pg/mL) for 15 hours. The luciferase activities were then measured as described in “DNA transfection and dual luciferase activity assay.” The relative firefly luciferase activity was normalized to renilla luciferase activity. Data are mean ± SD of 3 independent experiments. The dotted line represents the level of luciferase activity obtained with 0.5 ng of WT-ALK1 stimulated with BMP9.
BMP9 response of ALK1 mutants. (A) Expression vectors encoding WT-ALK1 or the different ALK1 mutants or β-gal (control) were transiently transfected into NIH-3T3 cells. After 24 hours, cells were serum deprived for 1 hour and subsequently treated or not with BMP9 (100 pg/mL) for 1 hour. Cell lysates (20 μg proteins) were resolved by 10% SDS-PAGE and immunoblotted with antibodies against phosphoSmad1/5 or against α-tubulin (as a loading control). (B) NIH-3T3 cells were transiently transfected with pGL3(BRE)2-luc, pRL-TK-luc, and plasmids (0.5 ng) encoding either WT-ALK1 or the different ALK1 mutants or β-gal (control). After 4 hours, cells were treated or not with BMP9 (100 pg/mL) for 15 hours. The luciferase activities were then measured as described in “DNA transfection and dual luciferase activity assay.” The relative firefly luciferase activity was normalized to renilla luciferase activity. Results are expressed as fold induction over the value obtained for each ALK1 mutant in the absence of BMP9. Data are mean ± SD of 3 independent experiments. (C) NIH-3T3 cells were transiently transfected with pGL3(BRE)2-luc, pRL-TK-luc, and different combinations of WT and mutant ALK1: WT-ALK1 alone (0.25, 0.5, or 1 ng) or WT-ALK1 (0.5 ng) and either one of the different mutants ALK1 (0.5 ng). After 4 hours, cells were treated or not with BMP9 (100 pg/mL) for 15 hours. The luciferase activities were then measured as described in “DNA transfection and dual luciferase activity assay.” The relative firefly luciferase activity was normalized to renilla luciferase activity. Data are mean ± SD of 3 independent experiments. The dotted line represents the level of luciferase activity obtained with 0.5 ng of WT-ALK1 stimulated with BMP9.
HHT2-related ALK1 mutations affect only 1 allele of ACVRL1. Potential consequences at the protein level include haploinsufficiency, with the generation of proteins totally or partially lacking receptor activity, or a dominant-negative effect, impairing the function of the normal allele. To determine whether ALK1 mutants behaved as dominant-negatives, we cotransfected equal amounts of WT-ALK1 with each individual ALK1 mutant into NIH-3T3 cells and assayed the response to BMP9. We used ALK1 plasmid amounts (0.25, 0.5, and 1 ng) that would yield cells responsiveness to BMP9 in a linear range (Figure 2C). Then, 0.5 ng of plasmid encoding WT-ALK1 together with 0.5 ng of plasmid encoding the different ALK1 mutants were transfected, after which the response to BMP9 was measured and compared with the response obtained with the WT-ALK1 (0.5 and 1 ng) alone. Addition of the different ALK1 mutants yielded a similar response to that obtained with 0.5 ng of WT-ALK1 (dotted line). This demonstrates that addition of the ALK1 mutants had no dominant inhibitory effect on the BMP9 response. Again, the mutant D179A behaved as the WT-ALK1 (Figure 2C).
Our data demonstrate that all the mutants except 1 (D179A) did not respond to BMP9, although most of them were expressed at the cell surface. We therefore determined whether these mutations could affect BMP9 binding to ALK1. To this end, transfected cells were labeled with [125I]BMP9 for 2 hours at 4°C. We found that the 3 extracellular mutants and the frameshift mutant (G371fsX391) did not bind BMP9, whereas the other mutants bound BMP9 at a similar level to WT-ALK1 (Figure 3A).
BMP9 binding to the different ALK1 mutants and functional analysis of the NANDOR ALK1 mutants. (A) β-gal, WT-ALK1, or the different ALK1 mutants were transiently transfected in COS-7 cells for 48 hours. Cells were then incubated with [125I]BMP9 for 2 hours at 4°C. Cells were then washed, lysed, and the bound radioactivity was counted as described in “[125I]-BMP9 radioreceptor assay.” Data are mean ± SD from 1 representative experiment of 3. (B) NIH-3T3 cells were transiently transfected with pGL3(BRE)2-luc, pRL-TK-luc, and WT-ALK1, or the constitutively active ALK1 mutant (ALK1ca, Q201D) or the different ALK1 double mutants (0.5 ng) or β-gal (control). After 4 hours, cells were treated or not with BMP9 (100 pg/mL) for 15 hours. The luciferase activities were then measured as described in “DNA transfection and dual luciferase activity assay.” The relative firefly luciferase activity was normalized to renilla luciferase activity. Data are mean ± SD of 3 independent experiments. (C) COS-7 cells were cotransfected with expression vectors encoding an HA-tagged and a His-tagged version of either WT-ALK1 or ALK1 mutants for 24 hours. Cell lysates (1-mg proteins) were subjected to immunoprecipitation with anti-His. These lysates were then resolved by 10% SDS-PAGE and immunoblotted with antibodies against HA (IP/IB-HA). In parallel, cell lysates (20 μg proteins) were also resolved by 10% SDS-PAGE and immunoblotted with antibodies against HA (IB-HA).
BMP9 binding to the different ALK1 mutants and functional analysis of the NANDOR ALK1 mutants. (A) β-gal, WT-ALK1, or the different ALK1 mutants were transiently transfected in COS-7 cells for 48 hours. Cells were then incubated with [125I]BMP9 for 2 hours at 4°C. Cells were then washed, lysed, and the bound radioactivity was counted as described in “[125I]-BMP9 radioreceptor assay.” Data are mean ± SD from 1 representative experiment of 3. (B) NIH-3T3 cells were transiently transfected with pGL3(BRE)2-luc, pRL-TK-luc, and WT-ALK1, or the constitutively active ALK1 mutant (ALK1ca, Q201D) or the different ALK1 double mutants (0.5 ng) or β-gal (control). After 4 hours, cells were treated or not with BMP9 (100 pg/mL) for 15 hours. The luciferase activities were then measured as described in “DNA transfection and dual luciferase activity assay.” The relative firefly luciferase activity was normalized to renilla luciferase activity. Data are mean ± SD of 3 independent experiments. (C) COS-7 cells were cotransfected with expression vectors encoding an HA-tagged and a His-tagged version of either WT-ALK1 or ALK1 mutants for 24 hours. Cell lysates (1-mg proteins) were subjected to immunoprecipitation with anti-His. These lysates were then resolved by 10% SDS-PAGE and immunoblotted with antibodies against HA (IP/IB-HA). In parallel, cell lysates (20 μg proteins) were also resolved by 10% SDS-PAGE and immunoblotted with antibodies against HA (IB-HA).
All of the intracellular mutations that do not respond to BMP9 are located in the kinase domain. To determine the molecular mechanisms behind the loss of activity of these different intracellular mutants, we tested their intrinsic kinase activity. However, we were unable to measure an intrinsic kinase activity for ALK1 in absence of activation (data not shown). The mechanisms of ALK1 activation have not been studied in detail and are generally extrapolated from what is known about TGF-β signaling through ALK5. TGF-β initially binds to TβRII, resulting in type I receptor (ALK5) recruitment, and its subsequent phosphorylation by the type II receptor. This transactivation step then leads to activation of the type I receptor. Several ALK5 mutants have been described that are mutated either in the kinase domain (G322D and G261E)25 or in a region, referred to as the NANDOR box (nonactivating non–down-regulating), present at the C-terminal tail of ALK5.26 ALK5 NANDOR mutants have an intact kinase activity but are not transactivated by the type II receptor in response to ligand binding. Interestingly, 4 HHT2-derived ACVRL1 mutations studied here were located in the NANDOR box of the ALK1 protein (R479Q, R484Q, R484W, and I485F). To determine whether these mutations might be involved in the transactivation step, we attempted to evaluate the result of bypassing RII transactivation. For this, we used a constitutively active ALK1 mutant (ALK1ca, Q201D) that is activated in a ligand-independent manner,27 in which we introduced additional mutations. We first tested a double mutant carrying both the Q201D and the A347P mutations. The A347P mutation is located within the catalytic domain and is adjacent to the amino acid that binds Mg2+, an essential cofactor of adenosine triphosphate (ATP) binding. Indeed, this mutation completely abolished the BRE response of ALK1ca (Figure 3B). Surprisingly, double mutants carrying the Q201D mutation and a mutation in the NANDOR box (R479Q and R484W) were also inactive (Figure 3B). Another step that has been shown to be crucial in ALK5 signaling is the formation of homodimeric complexes between 2 type I receptors.25,28,29 To test whether ALK1 NANDOR mutants form homodimers, we performed coimmunoprecipitation studies on cells cotransfected with 2 ALK1 constructs, each carrying a different epitope tag (HA or (His)6). Lysates from cells expressing these differentially tagged receptors were immunoprecipitated with anti-His antibodies. The immunoprecipitates were then subjected to SDS-PAGE and Western blotting using anti-HA antibodies. The results show that the NANDOR mutants tested (R479Q, R484Q, and I485F) coimmunoprecipitate and therefore form homodimers (Figure 3C).
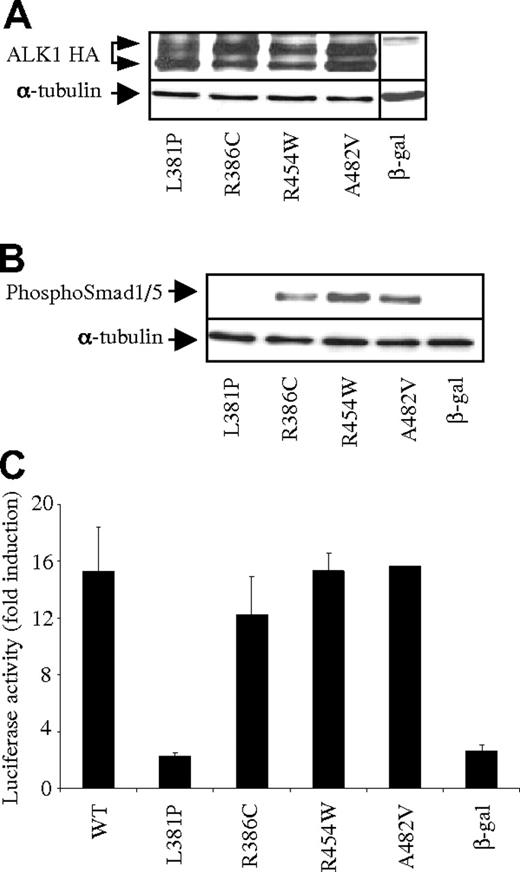
Our data suggested that the study of the BMP9 responses in cells expressing the different ALK1 mutants could be used to assess the functional consequences of missense mutations in ACVRL1. This could be particularly useful in the interpretation of the pathologic significance of novel missense mutations of ACVRL1, especially in complex situations where several mutations (in ACVRL1 or in ACVRL1 and ENG) are found in the same patient. To test this hypothesis, we analyzed 4 ACVRL1 missense mutations (L381P, R386C, R454W, and A482V) identified in HHT patients carrying another mutation in either ACVRL1 or ENG (Service de Génétique Moléculaire et Clinique, Hôpital Edouard Herriot, Lyon, France; Figure 1A dotted lines). We found that these 4 mutant proteins were all expressed (Figure 4A). We next evaluated the functional activity of these different ALK1 mutants in response to BMP9. Three ALK1 mutants responded to BMP9 (R386C, R454W, and A482V) and 1 mutant, L381P, did not respond to BMP9 stimulation either measured in the BRE assay or by Smad1/5 phosphorylation (Figure 4B-C). Functional testing strongly suggested that R386C, R454W, and A482V mutations are rare polymorphisms, whereas the L381P mutation leads to a nonfunctional protein.
Functional analysis of the BMP9 response of conflicting ALK1 mutants. (A) Protein expression of WT-ALK1 or ALK1 mutants. Expression vectors encoding HA-tagged WT-ALK1 or ALK1 mutants or β-gal (control) were transiently transfected in NIH-3T3 cells for 24 hours. Cell lysates (20 μg proteins) were resolved by 10% SDS-PAGE and immunoblotted with antibodies against HA and α-tubulin (as a loading control). (B) Expression vectors encoding WT-ALK1 or the different ALK1 mutants or β-gal (control) were transiently transfected into NIH-3T3 cells. After 24 hours, cells were serum deprived for 1 hour and subsequently treated with BMP9 (100 pg/mL) for 1 hour. Cell lysates (20 μg proteins) were resolved by 10% SDS-PAGE and immunoblotted with antibodies against phosphoSmad1/5 or against α-tubulin (as a loading control). (C) NIH-3T3 cells were transiently transfected with pGL3(BRE)2-luc, pRL-TK-luc, and plasmids (0.5 ng) encoding either WT-ALK1 or the different ALK1 mutants or β-gal (control). After 4 hours, cells were treated (▬) with BMP9 (100 pg/mL) for 15 hours. The luciferase activities were then measured as described in “DNA transfection and dual luciferase activity assay.” The relative firefly luciferase activity was normalized to renilla luciferase activity. Results are expressed as fold induction over the value obtained for each ALK1 mutant in the absence of BMP9. Data are mean ± SD of 3 independent experiments.
Functional analysis of the BMP9 response of conflicting ALK1 mutants. (A) Protein expression of WT-ALK1 or ALK1 mutants. Expression vectors encoding HA-tagged WT-ALK1 or ALK1 mutants or β-gal (control) were transiently transfected in NIH-3T3 cells for 24 hours. Cell lysates (20 μg proteins) were resolved by 10% SDS-PAGE and immunoblotted with antibodies against HA and α-tubulin (as a loading control). (B) Expression vectors encoding WT-ALK1 or the different ALK1 mutants or β-gal (control) were transiently transfected into NIH-3T3 cells. After 24 hours, cells were serum deprived for 1 hour and subsequently treated with BMP9 (100 pg/mL) for 1 hour. Cell lysates (20 μg proteins) were resolved by 10% SDS-PAGE and immunoblotted with antibodies against phosphoSmad1/5 or against α-tubulin (as a loading control). (C) NIH-3T3 cells were transiently transfected with pGL3(BRE)2-luc, pRL-TK-luc, and plasmids (0.5 ng) encoding either WT-ALK1 or the different ALK1 mutants or β-gal (control). After 4 hours, cells were treated (▬) with BMP9 (100 pg/mL) for 15 hours. The luciferase activities were then measured as described in “DNA transfection and dual luciferase activity assay.” The relative firefly luciferase activity was normalized to renilla luciferase activity. Results are expressed as fold induction over the value obtained for each ALK1 mutant in the absence of BMP9. Data are mean ± SD of 3 independent experiments.
Discussion
In the present manuscript, we studied the functional consequences of a series of ACVRL1 mutations (19) found in HHT2 patients affecting different domains of the ALK1 protein. This is the largest study of this type and the first to analyze ALK1 mutants in both the extracellular and intracellular domains. This is also the first study to evaluate the BMP9 response of ALK1 as opposed to the TGF-β response analyzed in previous studies.30,31
We have analyzed the BMP9-response of 19 ALK1 mutations described in HHT2 patients: 15 had already been published, and 4 were novel mutations. Of the 19 studied mutations, we identified 4 (D179A, R386C, R454W, and A482V) that were probably not causing HHT disease but instead represent rare polymorphisms. The first mutation (D179A) has been described in the intracellular GS region, which is phosphorylated by the type II receptor. This is the only mutation reported in this region.21 We found that this mutant was expressed at the cell surface in accordance with previously published data,21 that it could bind BMP9, and that it induces a similar phosphoSmad1/5 response after binding BMP9 to that of WT-ALK1. This patient died, and no parent or surviving relatives were available for further studies. Our data suggest that this mutation is very unlikely to cause the disease. The second mutation, A482V, was studied by 3 different groups. It was first identified in a patient with a gonadotroph tumor; however, the patient and its relatives showed no symptoms of HHT.32 This mutation was then described by Lesca et al in an HHT patient.33 Finally, Letteboer et al also described this mutation; however, it was found with another ACVRL1 (P424L) or ENG (W261R) mutation.34 In line with this result, we reanalyzed DNA from this patient and found a mutation in ENG c.1121_1124delAAGA, which leads to a truncated protein, and so, is very probably the cause of the disease. We can therefore conclude that the mutation A482V is probably a rare polymorphism. The patient carrying the mutation R386C was later found to carry also the mutation R411W in ACVRL1, which has been analyzed in this study and demonstrated to result in a defective BMP9 response. The patient carrying the mutation R454W had an additional mutation in ENG leading to a truncated protein (G296fsX), which is probably the cause of the disease. In this study, we also identified 2 novel ACVRL1 mutations in HHT2 patients (L381P and I485F) that are pathogenic. The mutation L381P has already been described in patients with PAH.20 The HHT2 patient, carrying the mutation L381P, is also carrying a mutation in ENG (V504M). Because this ENG mutation is a missense mutation, it is of major importance for genetic counseling to establish whether it has a pathogenic consequence. Taken together, our data clearly demonstrate that we can use the BMP9 response as a useful diagnostic tool for geneticists confronted with novel or conflicting ACVRL1 mutations both in HHT and PAH.
We have studied 3 extracellular ACVRL1 mutations (C51Y, N96D, and C77W) that were first described by Klaus et al.35 We found that these 3 mutants were expressed, but that they did not reach the cell surface properly, did not bind BMP9, and did not respond to BMP9. Two corresponded to cysteine substitutions (C51Y and C77W); the mutant C51Y disrupts the first disulfide bridge, whereas the mutant C77W disrupts the fourth disulfide bridge. The last mutation corresponded to an asparagine mutated into an aspartic acid (N96D), which adds a negative charge to the protein. This asparagine is conserved in all ALKs, which might indicate that it is of functional importance. None of these amino acids was shown to be directly involved in BMP2/ALK3 binding.36
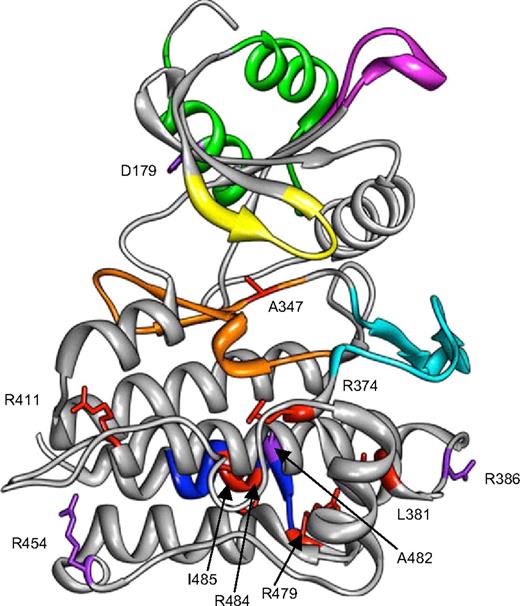
The other mutations were intracellular mutations and involved the large kinase domain. All were expressed at the cell surface, and all bound BMP9, except c.1112dupG (G371fsX391) mutation, which encodes a truncated protein. None of the mutants was functional in response to BMP9. The ALK1 intracellular structure has not been determined yet, but the amino acid sequence alignment of ALK1 and ALK5 reveals an overall similarity of ALK1 with ALK5, allowing us to generate a structural model of the cytoplasmic domain of ALK1 by computer modeling based on the homologous structure of activated ALK537 (Figure 5). The different missense mutations were analyzed for their position in the kinase domain. The A347P mutation is located within the catalytic domain and is adjacent to the amino acid that binds Mg2+, an essential cofactor of ATP binding. Therefore, a change from alanine into proline would clearly inhibit ATP binding. Arginine-411 has been described as mutated to a glutamine, a tryptophan, or a proline residue in HHT2 patients; therefore, we chose to analyze these 3 mutations to determine whether these mutants would differently impair ALK1 activity. We found that none of these mutants was functional in response to BMP9, demonstrating that this amino acid is essential for ALK1 activity and that substitutions with Q, W, or P have similar consequences. The expression of the mutant R411Q was already studied by 2 other groups, and our expression data are in accordance with the work of Gu et al30 but not Harrisson et al21 who found no membranous expression of the R411Q mutant. This last study was performed using GFP-ALK1 proteins, which might lead to mislocalization of the protein. In contrast to Gu et al,30 who found that R411Q retained TGF-β-induced BRE activity, suggesting that reduced activity might be sufficient to cause the disease phenotype, we could not find any residual activity in response to BMP9. We also studied R374W, which is a rather common mutation, shared by patients from different countries and R374Q. We found that these 2 mutants were expressed at the cell surface. R374W had already been shown to be expressed at the cell surface by Gu et al30 and Fernandez et al,39 whereas R374Q was studied by the group of Harrison et al21 who found no membrane expression. As discussed earlier, GFP-ALK1 proteins were used in those studies, which might lead to mislocalization. Further, we demonstrated that none respond to BMP9. R374W had already been shown to be inactive in response to TGF-β by Gu et al30 and Fernandez et al.39 Amino acid 374 is located within the subdomain VIII, which plays an important role in substrate recognition by providing a pocket to accommodate hydrophobic residues. The other intracellular mutations studied here were localized at the C-terminal tail of ALK1, located within the highly conserved −COOH domain, which contains a NANDOR box.26 In ALK5, this NANDOR box appears to be necessary for the regulation of TGF-β signaling through ALK5. This box of 11 amino acids shows 100% identity between the different type I receptors of the TGF-β family. Mutations in this box (R486W and R486Q) have been described in ALK6 causing brachydactyly type A2,40,41 and in ALK5 (R487Q/W/P and R478Q) causing Loeys-Dietz syndrome.42,–44 Four ACVRL1 mutations within this box were analyzed: R479Q, R484Q, R484W, and I485F. Interestingly, the R484W mutation has been identified in a patient with HHT and PAH.22 I485F, which has not been reported earlier, was found in a HHT2 patient who underwent liver transplantation because of severe hepatic damage. These mutants were expressed at the cell surface and could bind BMP9, but they had no functional activity in response to BMP9. It was shown for ALK5 that this box is necessary for type I receptor phosphorylation by the type II receptor in response to ligand binding and that mutation within this box does not modify its kinase activity.25,26 In accordance with these data, it was found that the mutation R486W in ALK6 had a normal kinase activity while it inhibited chondrogenic differentiation.41 Because we were unable to perform successful in vitro ALK1 kinase assays, probably because of its very low intrinsic kinase activity, as previously described,45 we attempted to discriminate transactivation mutants from kinase-dead mutants by bypassing the transactivation step using the constitutively active ALK1ca (Q201D) mutant that has been shown to be ligand-independent.27 We found that mutations in the NANDOR box completely abolished ALK1ca-induced response (Figure 3B). This was unexpected as NANDOR box mutants of ALK5 have been shown to transcomplement kinase-dead mutants of ALK5.26 Another important step that has been shown to be important in ALK5 signaling is the formation of homodimeric complexes between 2 type I receptors.25,28,29 We therefore wondered if a similar mechanism might occur for ALK1 and if some of the ALK1 NANDOR mutants might be nonfunctional because of their inability to homodimerize. To test this hypothesis, we performed coimmunoprecipitation studies on cells cotransfected with 2 ALK1 forms, each carrying a different epitope tag. The results show that the NANDOR mutants could coimmunoprecipitate (Figure 3C), indicating that they form homodimers. Our conclusion is that mutations within the ALK1 NANDOR box result in kinase-dead ALK1 receptor, suggesting that ALK1 does not behave in a similar fashion as ALK5.
Structural model of ALK1 cytoplasmic domain. A structural model of ALK1 intracellular domain was generated by homology modeling to the corresponding domain of ALK5 deduced from its crystal structure.37,38 The different domains of the proteins are indicated in different colors: GS box in green (amino acids 172-201), phosphate binding domain in yellow (amino acids 208-214), Smad binding loop (L45) in magenta (amino acids 263-270), catalytic loop in orange (amino acids 328-348), activation domain in cyan (amino acids 351-375), and NANDOR box in blue (amino acids 479-489). The positions of the 18 missense mutations, overlaid on the modelized structure, are indicated in red for the pathogenic mutations and in purple for the rare polymorphisms.
Structural model of ALK1 cytoplasmic domain. A structural model of ALK1 intracellular domain was generated by homology modeling to the corresponding domain of ALK5 deduced from its crystal structure.37,38 The different domains of the proteins are indicated in different colors: GS box in green (amino acids 172-201), phosphate binding domain in yellow (amino acids 208-214), Smad binding loop (L45) in magenta (amino acids 263-270), catalytic loop in orange (amino acids 328-348), activation domain in cyan (amino acids 351-375), and NANDOR box in blue (amino acids 479-489). The positions of the 18 missense mutations, overlaid on the modelized structure, are indicated in red for the pathogenic mutations and in purple for the rare polymorphisms.
TGF-β signaling requires 2 type I receptors complexed to 2 type II receptors. HHT2 patients have only 1 allele mutated. To mimic this heterozygosity, we transfected WT-ALK1 constructs with the different ALK1 mutants at an equal ratio and compared their level of activation with that of WT-ALK1 alone. We found that ALK1 mutants did not decrease the WT-ALK1 response, indicating that these mutants do not behave as dominant-negative receptors. This conclusion differs from that of Fernandez et al39 and Gu et al30 who found that certain ALK1 mutants inhibited the function of the WT allele. However, in their studies, they used TGF-β1 as an inducer, which requires the presence of ALK5 to activate the BRE promoter.45 Therefore, they did not test WT-ALK1/mutALK1 heterozygotes but, rather, ALK5/WT-ALK1 and ALK5/mutALK1. ALK5 has been shown recently to directly phosphorylate Smad1/5 independently of ALK1,46,47 and this is why some residual activity may still be observed with some ALK1 mutants.30,39 In the present study, we demonstrated that the identification of BMP9 as the specific and physiologic ligand for ALK1 now allows specific evaluation of the functional significance of ACVRL1 mutations.
In conclusion, this work demonstrates that ACVRL1 mutations lead to functional haploinsufficiency of the BMP9 response. This indicates that loss of only half of the ALK1 response is sufficient to lead to disease development. This suggests that one could envision therapies directed either at increasing ALK1 levels or increasing the level of its specific ligand, BMP9, which is known to circulate in blood.15 We also demonstrated that analysis of the BMP9 response could discriminate pathogenic (L381P and I485F) from polymorphic (D179A, R386C, and R454W) ACVRL1 mutations. We therefore propose that analysis of the BMP9 response can be used as a diagnostic tool by geneticists confronted with novel or conflicting ACVRL1 mutations.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr J. LaMarre (University of Guelph, ON) for his review of the manuscript.
This work was supported by Inserm, Commissariat à l'Energie Atomique et aux Energies Alternatives, Association pour la Recherche sur le Cancer, Projet Hospitalier de Recherche Clinique, Hospices Civils de Lyon (grant 27), and Association des Malades de Rendu-Osler. N.R. was supported by the Commissariat à l'Energie Atomique et aux Energies Alternatives (CFR grant).
Authorship
Contribution: N.R., M.B., C.M., and S.B. performed research; G.L. and S.G. contributed to the choice of the ACVRL1 mutations and the discussion; R.P. helped in the modelization of ALK1; and J.-J.F. and S.B. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sabine Bailly, U878, 17 rue des Martyrs, 38054 Grenoble, France; e-mail: sbailly@cea.fr.

![Figure 3. BMP9 binding to the different ALK1 mutants and functional analysis of the NANDOR ALK1 mutants. (A) β-gal, WT-ALK1, or the different ALK1 mutants were transiently transfected in COS-7 cells for 48 hours. Cells were then incubated with [125I]BMP9 for 2 hours at 4°C. Cells were then washed, lysed, and the bound radioactivity was counted as described in “[125I]-BMP9 radioreceptor assay.” Data are mean ± SD from 1 representative experiment of 3. (B) NIH-3T3 cells were transiently transfected with pGL3(BRE)2-luc, pRL-TK-luc, and WT-ALK1, or the constitutively active ALK1 mutant (ALK1ca, Q201D) or the different ALK1 double mutants (0.5 ng) or β-gal (control). After 4 hours, cells were treated or not with BMP9 (100 pg/mL) for 15 hours. The luciferase activities were then measured as described in “DNA transfection and dual luciferase activity assay.” The relative firefly luciferase activity was normalized to renilla luciferase activity. Data are mean ± SD of 3 independent experiments. (C) COS-7 cells were cotransfected with expression vectors encoding an HA-tagged and a His-tagged version of either WT-ALK1 or ALK1 mutants for 24 hours. Cell lysates (1-mg proteins) were subjected to immunoprecipitation with anti-His. These lysates were then resolved by 10% SDS-PAGE and immunoblotted with antibodies against HA (IP/IB-HA). In parallel, cell lysates (20 μg proteins) were also resolved by 10% SDS-PAGE and immunoblotted with antibodies against HA (IB-HA).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/116/9/10.1182_blood-2010-03-276881/5/m_zh89991056270003.jpeg?Expires=1767803104&Signature=rnENI2xbdE50QdDQjWecvGcQD4y-1HtMTFEoGPloJuaF4nl4WmD1avXkSQtfWpj~z7pxUNeSTCxsIHApIG78jARcLO77Kc2yLTqIGxuWCtYI2sEC78yw4cgRTyoSleIA4oIatmQmMiaMiad9t3PzqEwzGnBoDSvwjTRUitSJjxGCOE-V6o4Uvw9m8L7ogloT1~3i9JaJn1XvSznp-RX3xLeea~l3VCBu118AtbtiiZCHpcOqrQZcmZd87XpLDFqOu0D9euisXlPpMW2ujCvJLrEHOOEdO3hm6uAHjUYmzHCktyHMBpId-X2rzz~vRUFv4UTOJLsKzqTsSu5j99xM1A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal