Abstract
A region of the human von Willebrand factor (VWF) gene between −2812 and the end of the first intron (termed vWF2) was previously shown to direct expression in the endothelium of capillaries and a subset of larger blood vessels in the heart and skeletal muscle. Here, our goal was to delineate the DNA sequences responsible for this effect. A series of constructs containing deletions or mutations of vWF2 coupled to LacZ were targeted to the Hprt locus of mice, and the resulting animals were analyzed for reporter gene expression. The findings demonstrate that DNA sequences between −843 and −620 are necessary for expression in capillary but not large vessel endothelium in heart and skeletal muscle. Further, expression of VWF in capillaries and larger vessels of both tissues required the presence of a native or heterologous intron. In vitro assays implicated a role for ERG-binding ETS motif at −56 in mediating basal expression of VWF. In Hprt-targeted mice, mutation of the ETS consensus motif resulted in loss of LacZ expression in the endothelium of the heart and skeletal muscle. Together, these data indicate that distinct DNA modules regulate vascular bed–specific expression of VWF.
Introduction
The von Willebrand factor (VWF) is a high-molecular weight glycoprotein that mediates platelet-vessel wall interactions. VWF is expressed specifically in endothelial cells and megakaryocytes. Its expression in the intact endothelium varies both spatially and temporally. For example, in mice VWF mRNA levels are higher in the lung and heart compared with the liver and kidney, and within a given tissue, VWF expression is generally greater on the venous side of the circulation.1 Expression of VWF may change in pathophysiological conditions. For example, in a mouse model of endotoxemia, VWF mRNA levels were down-regulated in aorta, brain, adipose tissue, testis, thymus, adrenal, skeletal muscle, gut, and liver but increased in the heart and kidney.1 The elucidation of the mechanisms underlying VWF expression may provide important insights into the molecular basis of vascular diversity.
The structural organization of the mouse, human, and bovine VWF promoter-proximal region is closely related.2-4 The first exon (+1 to +246 in human VWF) encodes 5′ untranslated sequence. The second exon contains the ATG translational start site. Exons 1 and 2 are separated by an intron (+247 to +1475 in human VWF). A previous in vitro analysis of the human gene demonstrated that a 734-bp region between −487 and +246 contains information for lineage-specific expression in cultured endothelial cells.5 In standard transgenic mice, this promoter fragment (which we have termed vWF1) retained endothelial cell specificity. However, expression was limited to blood vessels of the brain.6 In contrast, standard transgenic mice carrying a larger region of the VWF gene (between −2182 and the end of the first intron; designated vWF2) directed expression not only in blood vessels of the brain but also in capillaries of the heart and skeletal muscle.7 A similar fragment from the mouse VWF promoter displayed an identical pattern of expression, supporting interspecies similarities in transcriptional control mechanisms.2
To control for copy number and integration site, we targeted a single copy of the vWF2-LacZ cassette to the Hprt locus of mice. Consistent with the results of the standard transgenic mice, the Hprt-targeted vWF2 promoter directed expression in the vasculature of the brain, heart, and skeletal muscle but not in other organs such as aorta, lung, liver, spleen, and kidney.8
The goal of this study was to further delineate the DNA sequences involved in mediating vascular bed–specific expression of the VWF gene. We report that a 224-bp region between −843 and −620 in the upstream promoter is necessary for expression in the capillaries of the heart and skeletal muscle. These DNA sequences are distinct from those that regulate expression in venous endothelium in these 2 organs. Furthermore, we demonstrate that promoter activity in the capillaries and certain larger vessels of the heart and skeletal muscle requires a native or heterologous intron as well as an intact ERG-binding site at −56. These data support a model of gene regulation whereby a constellation of vascular bed–specific DNA regions or modules govern overall expression of VWF in the endothelium. Further, the results underscore the importance of studying gene regulation in the context of the intact endothelium.
Methods
Plasmid constructions
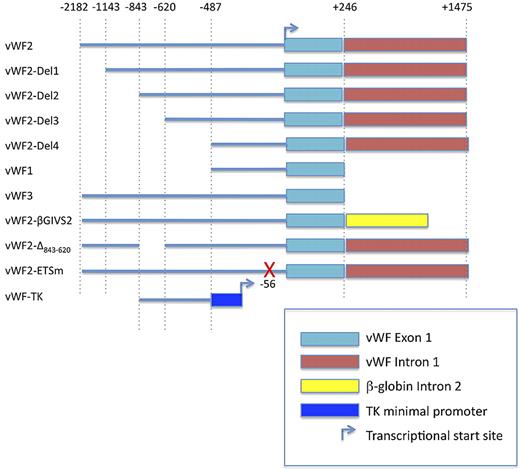
Construction of VWF deletion and mutant promoter constructs used in transient transfection assays and Hprt-locus targeting is detailed in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). The constructs are schematically shown in Figure 1.
Cell culture and transfections
Human umbilical vein endothelial cells (HUVECs) were purchased from Lonza and cultured in endothelial cell medium supplemented with microvascular endothelial cell growth medium-2 (bullet kit; Lonza).
Transient transfection assays
Transient transfections were carried out as described in supplemental Methods.
ChIP assay
Chromatin immunoprecipitation (ChIP) assays were performed using a ChIP assay kit (Upstate Biotechnology Inc.) according to the manufacturer's instructions and as detailed in supplemental Methods.
Transfection with siRNA
HUVECs were transfected with siRNA against ERG as previously described.9
Quantitative reverse transcriptase PCR
Generation and analysis of Hprt-targeted transgenic mice
The generation of Hprt-vWF2-LacZ mice was previously described.8 All animal experiments were approved by the Beth Israel Deaconess Medical Center Institutional Animal Care and Use Committee. To generate the additional Hprt-targeted mice, the targeting vectors were linearized with SalI and transfected into BK4-embryonic stem cells by electroporation. Homologous recombinants were selected in HAT (hypoxanthine-aminopterin-thymidine)–supplemented medium, containing 0.1mM hypoxanthine, 0.0004mM aminopterin, and 0.016mM thymidine (Sigma-Aldrich). HAT-resistant colonies were picked 10 days later for propagation, and a subset of the cells were examined for homologous recombination by LacZ PCR. Targeted embryonic stem cells were injected into C57BL/6-derived blastocysts that were then transplanted into the uteri of recipient Swiss Webster females. Resulting chimeric males were bred with C57BL/6 females to obtain agouti offspring. The agouti female offspring were backcrossed with C57BL/6 males to generate hemizygous male F2 mice. Genotyping was performed by PCR analysis. Whole-mount LacZ staining in adult male tissues was carried out as previously described.6 Smooth muscle actin staining was carried out as described in supplemental Methods.
Bioinformatics
Development and analysis of DNA microarray compendiums and analysis of the human, mouse, and bovine VWF promoters are described in supplemental Methods.
Statistical analysis
Data are expressed as mean ± SE or SD. The statistical significance of differences of the means was determined by 1-way analysis of variance and multiple comparisons by Tukey-Kramer multiple range test.
Results
DNA sequences between −2182 and −487 and/or in the first intron (between +247 and +1475) contain information for expression in vascular beds of the heart and skeletal muscle
Previous studies in standard transgenic mice demonstrated that the vWF1 promoter (between −487 and +246) directed expression in blood vessels of the brain,6 whereas the larger vWF2 promoter fragment (between −2182 and +1475) directed expression not only in the vascular endothelium of the brain but also in the capillary endothelium of the heart and skeletal muscle.7 When targeted as a single-copy transgene to the Hprt locus of mice, vWF2-lacZ demonstrated an identical expression pattern to that of the standard transgenic mice with the exception that it was also active in a subset of larger vessels of the heart and skeletal muscle (supplemental Figure 1).8 In tissue sections of the heart, these larger vessels comprised veins and venules, as defined by their wide diameter and thin wall and low reactivity to antismooth muscle actin antibody (supplemental Figures 2-3). In tissue sections of the diaphragm, the majority of staining in noncapillary endothelial cells also occurred in veins and venules. However, in contrast to the heart, the diaphragm revealed occasional LacZ-positive endothelial cells in arterioles (supplemental Figure 4). LacZ expression was not detected in other organs examined, including the aorta, liver, spleen, lung, and kidney (supplemental Figure 5).
We extended our findings by targeting vWF1 to the Hprt locus. As in standard transgenic mice, vWF1 promoter activity was limited to blood vessels of the brain (supplemental Figure 1). Although the pattern of LacZ expression in the brain was similar between vWF1 and vWF2 lines, overall promoter activity was lower in vWF1 mice. Thus, information for expression in the vascular endothelium of the heart and skeletal muscle is contained within DNA sequences between −487 and −2182 and/or in the first intron (between +247 and +1475).
A region between −843 and −620 contains information for expression in the capillary endothelium of the heart and skeletal muscle
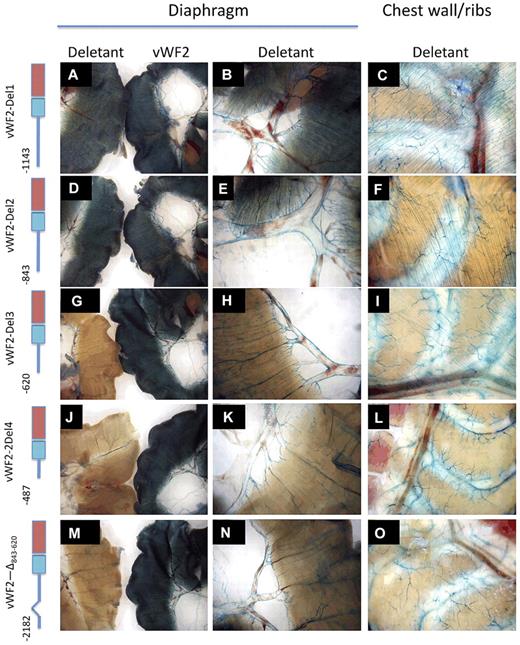
To further delineate the promoter regions responsible for expression in blood vessels of the heart and skeletal muscle, we targeted a series of deletion constructs to the Hprt locus of mice (Figure 1). 5′-deletion variants of vWF2 containing 1143-bp 5′-flanking sequence (vWF2-Del1-lacZ) and 843-bp 5′-flanking sequence (vWF2-Del2-lacZ) retained promoter activity in the endothelium of capillaries and larger vessels in the heart and skeletal muscle, while 5′-deletion constructs containing 620 bp (vWF2-Del3-lacZ) or 487 bp (vWF2-Del4-lacZ) were expressed in a similar subset of macrovessels but not in capillaries of the heart and skeletal muscle (Figure 2 and supplemental Figures 6-7).
Schematic of the human VWF promoter fragments used in transfections and Hprt-targeted mice. TK, thymidine kinase.
Schematic of the human VWF promoter fragments used in transfections and Hprt-targeted mice. TK, thymidine kinase.
LacZ staining of diaphragm and chest wall from Hprt-targeted mice carrying vWF2-lacZ or deletant VWF promoter-lacZ constructs. Diaphragms and chest walls were harvested from 6- to 8-week-old F2 male Hprt-targeted mice carrying vWF2-lacZ or deletant VWF transgenes and processed in parallel for whole-mount staining with X-Gal. In the left-most column (A,D,G,J,M), the diaphragm from the vWF2-lacZ transgenic mouse is on the right, and the diaphragm from the deletant VWF transgenic mouse is on the left. Panels in the second column (B,E,H,K,N) represent higher power images of the deletant VWF transgenic tissues shown A, D, G, J, and M, respectively. Wild-type vWF2-lacZ and all 5 deletant VWF transgenic mice demonstrate LacZ staining in macrovessels of skeletal muscle (diaphragm and intercostal muscle). However, note the loss of microvascular LacZ staining in vWF2-Del3-lacZ, vWF2-Del4-lacZ, and vWF2-Δ843-620-lacZ tissues. Whole-mount tissues were analyzed under a Nikon SMZ-U dissecting microscope, and microphotographs were collected using a Nikon Coolpix 8400 camera.
LacZ staining of diaphragm and chest wall from Hprt-targeted mice carrying vWF2-lacZ or deletant VWF promoter-lacZ constructs. Diaphragms and chest walls were harvested from 6- to 8-week-old F2 male Hprt-targeted mice carrying vWF2-lacZ or deletant VWF transgenes and processed in parallel for whole-mount staining with X-Gal. In the left-most column (A,D,G,J,M), the diaphragm from the vWF2-lacZ transgenic mouse is on the right, and the diaphragm from the deletant VWF transgenic mouse is on the left. Panels in the second column (B,E,H,K,N) represent higher power images of the deletant VWF transgenic tissues shown A, D, G, J, and M, respectively. Wild-type vWF2-lacZ and all 5 deletant VWF transgenic mice demonstrate LacZ staining in macrovessels of skeletal muscle (diaphragm and intercostal muscle). However, note the loss of microvascular LacZ staining in vWF2-Del3-lacZ, vWF2-Del4-lacZ, and vWF2-Δ843-620-lacZ tissues. Whole-mount tissues were analyzed under a Nikon SMZ-U dissecting microscope, and microphotographs were collected using a Nikon Coolpix 8400 camera.
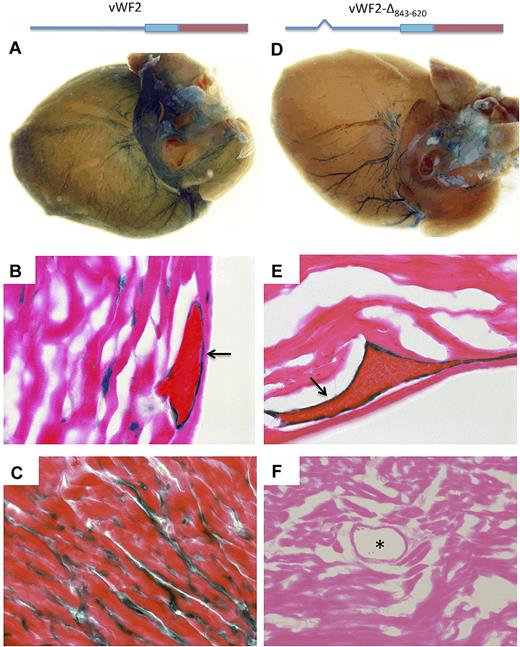
Based on the results of the 5′-deletion analysis, we generated a vWF2 promoter-LacZ cassette containing an internal deletion of DNA sequences between −843 and −620. When targeted to the Hprt locus of mice, the resulting construct (vWF2-Δ843-620-lacZ) also demonstrated an identical expression pattern to that of vWF2-Del3-lacZ, with selective loss of capillary expression in the heart and skeletal muscle (Figures 2–3 and supplemental Figures 5-9). Together, these data indicate that a region between −843 and −620 contains information for expression of VWF in the capillary endothelium of the heart and skeletal muscle, whereas a region between −620 and +1475 confers expression in the veins/venules of the heart and veins/venules and occasional arterioles of skeletal muscle.
LacZ staining of heart whole mounts and tissue sections from vWF2-lacZ and vWF2-Δ843-620-lacZ mice. Hearts were harvested from 6- to 8-week-old F2 male Hprt-targeted mice carrying vWF2-lacZ transgene (A-C) or vWF2-Δ843-620-lacZ transgene (identical to vWF2 except for an internal deletion of DNA sequences between −843 and −620; D-F) and processed in parallel for whole-mount (A,D) or cryosection (B-C,E-F) staining with X-Gal. vWF2-lacZ and vWF2-Δ843-620-lacZ transgenic hearts demonstrate LacZ staining in veins. However, note loss of capillary endothelial staining in the vWF2-Δ843-620-lacZ heart. Whole-mount hearts were analyzed under a Nikon SMZ-U dissecting microscope, and microphotographs were collected using a Nikon Coolpix 8400 camera. Tissue section images were obtained using a ×100 objective (B,E), ×60 objective (C), or ×40 objective (F). Lac-Z-stained sections were counterstained with eosin. Slides were analyzed under a Zeiss Axio Imager upright microscope, and photomicrographs were collected using a Zeiss Axiocam MRc camera and Axiovision 4.6.3 image acquisition software. Arrow, LacZ staining in venous endothelial cells. Asterisk, lumen of an artery.
LacZ staining of heart whole mounts and tissue sections from vWF2-lacZ and vWF2-Δ843-620-lacZ mice. Hearts were harvested from 6- to 8-week-old F2 male Hprt-targeted mice carrying vWF2-lacZ transgene (A-C) or vWF2-Δ843-620-lacZ transgene (identical to vWF2 except for an internal deletion of DNA sequences between −843 and −620; D-F) and processed in parallel for whole-mount (A,D) or cryosection (B-C,E-F) staining with X-Gal. vWF2-lacZ and vWF2-Δ843-620-lacZ transgenic hearts demonstrate LacZ staining in veins. However, note loss of capillary endothelial staining in the vWF2-Δ843-620-lacZ heart. Whole-mount hearts were analyzed under a Nikon SMZ-U dissecting microscope, and microphotographs were collected using a Nikon Coolpix 8400 camera. Tissue section images were obtained using a ×100 objective (B,E), ×60 objective (C), or ×40 objective (F). Lac-Z-stained sections were counterstained with eosin. Slides were analyzed under a Zeiss Axio Imager upright microscope, and photomicrographs were collected using a Zeiss Axiocam MRc camera and Axiovision 4.6.3 image acquisition software. Arrow, LacZ staining in venous endothelial cells. Asterisk, lumen of an artery.
Intronic splicing is necessary for expression in the capillary endothelium of the heart and skeletal muscle
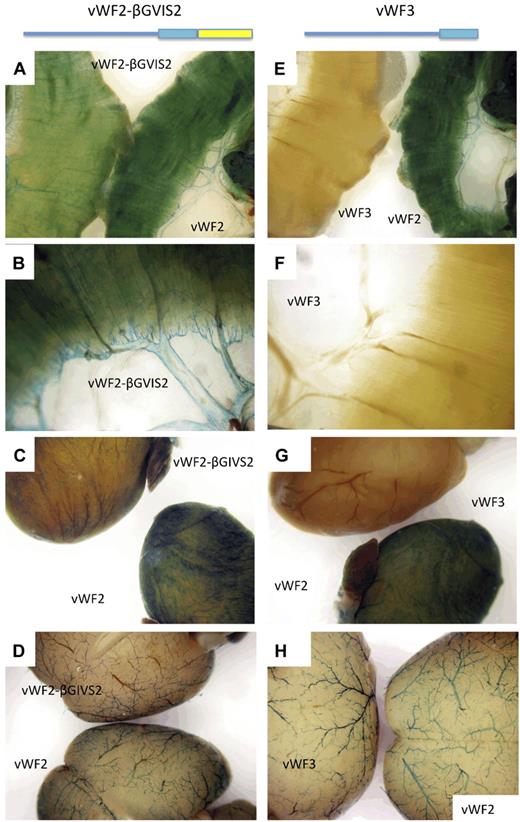
Previous studies have demonstrated a role for intronic enhancers in mediating the expression of other endothelial cell-specific promoters in vivo, including vascular endothelial growth factor receptor 2/fetal liver kinase (FLK)1, TIE-2, GATA2, and endoglin.11-15 Thus, we asked whether the first intron of VWF plays a role in mediating vascular bed–specific expression of the gene. To that end, we generated Hprt-targeted mice with an intron-less version of vWF2 (between −2182 and +246; termed vWF3) coupled to LacZ. Similar to vWF1 (-487 to +246), vWF3 promoter activity was limited to blood vessels of the brain (Figure 4). These findings suggested that the first intron of VWF plays an important role in mediating VWF expression in the heart and skeletal muscle.
LacZ staining of diaphragm, heart and brain from Hprt-targeted mice carrying vWF2-lacZ, vWF3-lacZ, or vWF2-βGIVS2-lacZ. Diaphragms, hearts, and brains were harvested from 6- to 8-week-old F2 male Hprt-targeted mice carrying vWF2-lacZ (vWF sequences between −2182 and +1475), vWF3-lacZ (vWF sequences between −2182 and +246), or vWF2-βGIVS2-lacZ (vWF sequences between −2182 and +246 and intron 2 from the human β-globin gene) and processed in parallel for whole-mount staining with X-Gal. In panels A and E, the diaphragm from the vWF2-lacZ transgenic mouse is on the right, and the diaphragm from the vWF2-βGIVS2-lacZ or vWF3-lacZ mouse is on the left. Panels B and F are higher power images of vWF2-βGIVS2-lacZ and vWF3-lacZ diaphragms, respectively. In panel C, the vWF2-lacZ heart is on the bottom, and the vWF2-βGIVS2-lacZ heart is on the top. In panel G, the vWF2-lacZ heart is on the bottom, and the vWF3-lacZ heart is on the top. In panel D, the vWF2-lacZ brain is on the bottom, and the vWF2-βGIVS2-lacZ brain is on the top. In panel H, the vWF2-lacZ brain is on the right, and the vWF3-lacZ brain is on the left. In panels A through D, vWF2-βGIVS2-lacZ organs reveal a similar pattern (though lower absolute levels) of expression compared with vWF2-lacZ. In E-H, vWF3-lacZ organs show the X-Gal reaction product in brain but not heart or skeletal muscle. Whole-mount tissues were analyzed under a Nikon SMZ-U dissecting microscope, and microphotographs were collected using a Nikon Coolpix 8400 camera.
LacZ staining of diaphragm, heart and brain from Hprt-targeted mice carrying vWF2-lacZ, vWF3-lacZ, or vWF2-βGIVS2-lacZ. Diaphragms, hearts, and brains were harvested from 6- to 8-week-old F2 male Hprt-targeted mice carrying vWF2-lacZ (vWF sequences between −2182 and +1475), vWF3-lacZ (vWF sequences between −2182 and +246), or vWF2-βGIVS2-lacZ (vWF sequences between −2182 and +246 and intron 2 from the human β-globin gene) and processed in parallel for whole-mount staining with X-Gal. In panels A and E, the diaphragm from the vWF2-lacZ transgenic mouse is on the right, and the diaphragm from the vWF2-βGIVS2-lacZ or vWF3-lacZ mouse is on the left. Panels B and F are higher power images of vWF2-βGIVS2-lacZ and vWF3-lacZ diaphragms, respectively. In panel C, the vWF2-lacZ heart is on the bottom, and the vWF2-βGIVS2-lacZ heart is on the top. In panel G, the vWF2-lacZ heart is on the bottom, and the vWF3-lacZ heart is on the top. In panel D, the vWF2-lacZ brain is on the bottom, and the vWF2-βGIVS2-lacZ brain is on the top. In panel H, the vWF2-lacZ brain is on the right, and the vWF3-lacZ brain is on the left. In panels A through D, vWF2-βGIVS2-lacZ organs reveal a similar pattern (though lower absolute levels) of expression compared with vWF2-lacZ. In E-H, vWF3-lacZ organs show the X-Gal reaction product in brain but not heart or skeletal muscle. Whole-mount tissues were analyzed under a Nikon SMZ-U dissecting microscope, and microphotographs were collected using a Nikon Coolpix 8400 camera.
Other studies using standard transgenic mice have shown that intronic splicing per se may result in increased expression of the transgene in nonendothelial cells.16-19 To test the possibility that such a mechanism is involved in mediating VWF expression, we replaced the first intron of VWF with the second intron (intervening sequence 2 [IVS2]) of the human β-globin gene. We chose this particular intron because it has been shown to increase the expression of native and heterologous transgenic promoters by an enhancer-independent mechanism.16,20 The resulting Hprt-targeted mice (termed vWF2-βGIVS2-lacZ) revealed an identical pattern of expression to that of vWF2-lacZ mice (Figure 4 and supplemental Figure 10). These findings suggest that VWF promoter activity in the heart and skeletal muscle is dependent on intronic splicing. The absolute level of vWF2-βGIVS2-lacZ expression was, however, lower compared with vWF2-lacZ. Thus, the native intron of VWF may also contain positive-acting DNA elements.
To determine whether the microvascular and/or macrovascular-specific region of the VWF promoter functions as an autonomous enhancer, a fragment spanning the region between −843 and −487 was coupled to the 5′ end of the thymidine kinase (TK) promoter and LacZ, and the resulting construct (VWF enhancer-TK-lacZ) was targeted to the Hprt locus. TK-lacZ demonstrated high-level expression in cardiomycoytes but not in skeletal muscle. The addition VWF promoter sequences between −843 and −620 did not affect the level or pattern of expression (supplemental Figure 11 shows diaphragm). Thus, this DNA region does not function as an autologous enhancer.
An ERG-binding motif at −56 is necessary for vWF2 promoter activity in vivo
We recently reported that a mutation of a GATA motif at +220 in vWF2 results in moderately reduced promoter activity in the heart, skeletal muscle, and brain of Hprt-targeted mice.10 Thus, it is likely that vascular bed–specific regions of the promoter (such as the one between −843 to −620) require proximal promoter elements for basal activity in their respective vascular beds. To further test this hypothesis, we focused on an ETS motif (TTCC) at −56. A previous study showed that this site binds the ETS factor ERG but not ETS-1/2 and that it positively regulates VWF promoter activity.21 Bioinformatic approaches offer additional insights into the potential role of the −56 ETS site and the ERG transcription factor in regulating VWF expression. For example, cross-species sequence comparisons of the proximal VWF promoter reveal evolutionary conservation of the −56 ETS motif (supplemental Figure 12). As a second strategy, we plotted expression levels of ERG and VWF across 2 compendia of publicly available DNA microarray experiments. The first such compendium is dominated by endothelial cells and smooth muscle cells of heterogeneous origin. It includes transcriptional profiles from 53 purified endothelial cell samples, representing 14 distinct locations, all propagated in identical culture conditions.22 The sample set included endothelial cells from 5 different arteries (aorta, coronary artery, pulmonary artery, iliac artery, and umbilical artery), 2 different veins (umbilical vein and saphenous vein), and 7 different tissues (skin, lung, intestine, uterus myometrium, nasal polyps, bladder, and myocardium).22 These arrays are complemented by 60 profiles of smooth muscle cells representing a similar set of locations and a study of hypoxia and serum starvation of endothelial cells, smooth muscle cells, and 2 lines of epithelial cells (64 further arrays).23,24 We assembled a second compendium of 5532 microarray experiments restricted to human cells, as described in supplemental Methods. As shown in Figure 5A, the correlation between VWF and ERG transcription factor mRNA levels was very high within the endothelial cell population, as well as within all arrays of the compendium representing multiple cell types and tissue samples. Indeed, ERG ranked second among 1081 genes that are annotated by GO with “transcriptional regulatory activity” based on correlation and ranked fourth based on mutual information. Although there is no guarantee that transcription factor mRNA levels reflect their transcriptional activity, these unbiased data strongly suggest that ERG plays an important role in mediating VWF expression.
ERG binds to the −56 ETS motif. (A) VWF expression is shown as a function of ERG expression levels. Results are based on the Brown datasets (left) and our compendium of 5532 arrays (right). Red, endothelial cells; black, nonendothelial cells. (B) Quantitative real-time PCR analysis of β-actin, CD31, ERG, friend leukemia virus integration-1, and VWF in HUVECs treated with 20 ng/mL TNF-α for 0 (untreated control), 4, 12, or 24 hours. For each gene, expression is shown relative to control values. Data are expressed as mean + SD of 3 independent experiments. (C) ChIP assay was performed using HUVECs treated in the absence of presence of TNF-α for 12 hours. DNA was sheared, and the resulting DNA-protein complexes were immunoprecipitated in the absence or presence of antibodies to ERG or control IgG. Real-time PCR analysis was performed using the precipitated DNA fragments and primers for VWF proximal region, which included the −56 ETS site. (D) Transient transfection of vWF2-Luc or vWF2-ETSm-Luc in HUVECs treated in the absence or presence of TNF-α for 4 hours. The results show the mean + SD of luciferase light units (relative to vWF2 in untreated cells) obtained in triplicate from at least 3 independent experiments. *P < .05. (E) HUVECs were either not transfected (NT) or transfected with control siRNA (CTR) or siRNA against ERG (ERG) and assayed for mRNA expression of ERG or VWF by real-time PCR. The results in E show the mean + SD of mRNA expression (relative to control siRNA-transfected cells) obtained in triplicate from 3 independent experiments. *P < .05, relative to control siRNA. (F) Same HUVECs as described in E were assayed for ERG protein expression using Western blot. For loading control, the blot was stripped and reprobed with antibodies against tubulin and against the nuclear marker, proliferating cell nuclear antigen (PCNA). (G) Parallel plates of HUVECs used for mRNA and protein analysis in panels E and F were transiently transfected with vWF2-Luc or vWF2-ETSm-Luc. The results show the mean + SD of luciferase light units (relative to vWF2 in control siRNA-transfected cells) obtained in triplicate from at least 3 independent experiments. *P < .05; **P < .01; n/s, nonsignificant.
ERG binds to the −56 ETS motif. (A) VWF expression is shown as a function of ERG expression levels. Results are based on the Brown datasets (left) and our compendium of 5532 arrays (right). Red, endothelial cells; black, nonendothelial cells. (B) Quantitative real-time PCR analysis of β-actin, CD31, ERG, friend leukemia virus integration-1, and VWF in HUVECs treated with 20 ng/mL TNF-α for 0 (untreated control), 4, 12, or 24 hours. For each gene, expression is shown relative to control values. Data are expressed as mean + SD of 3 independent experiments. (C) ChIP assay was performed using HUVECs treated in the absence of presence of TNF-α for 12 hours. DNA was sheared, and the resulting DNA-protein complexes were immunoprecipitated in the absence or presence of antibodies to ERG or control IgG. Real-time PCR analysis was performed using the precipitated DNA fragments and primers for VWF proximal region, which included the −56 ETS site. (D) Transient transfection of vWF2-Luc or vWF2-ETSm-Luc in HUVECs treated in the absence or presence of TNF-α for 4 hours. The results show the mean + SD of luciferase light units (relative to vWF2 in untreated cells) obtained in triplicate from at least 3 independent experiments. *P < .05. (E) HUVECs were either not transfected (NT) or transfected with control siRNA (CTR) or siRNA against ERG (ERG) and assayed for mRNA expression of ERG or VWF by real-time PCR. The results in E show the mean + SD of mRNA expression (relative to control siRNA-transfected cells) obtained in triplicate from 3 independent experiments. *P < .05, relative to control siRNA. (F) Same HUVECs as described in E were assayed for ERG protein expression using Western blot. For loading control, the blot was stripped and reprobed with antibodies against tubulin and against the nuclear marker, proliferating cell nuclear antigen (PCNA). (G) Parallel plates of HUVECs used for mRNA and protein analysis in panels E and F were transiently transfected with vWF2-Luc or vWF2-ETSm-Luc. The results show the mean + SD of luciferase light units (relative to vWF2 in control siRNA-transfected cells) obtained in triplicate from at least 3 independent experiments. *P < .05; **P < .01; n/s, nonsignificant.
A previous study showed that ERG knockdown in HUVECs or incubation of HUVECs with tumor necrosis factor-α (TNF-α) reduced the expression of several endothelial genes, including VWF.25 We recently demonstrated that TNF-α suppresses ERG expression in endothelial cells.9 To confirm that TNF-α results in concomitant inhibition of ERG and VWF, HUVECs were treated with 20 ng/mL TNF-α for varying times and assayed for mRNA expression using quantitative real-time PCR. As shown in Figure 5B, TNF-α resulted in a time-dependent reduction in ERG and VWF mRNA (46% and 52% of control at 24 hours, respectively) but not CD31, β-actin, or the ETS transcription factor, friend leukemia virus integration (Fli)–1. To assay for ERG binding to the proximal VWF promoter, we performed ChIP assays in HUVECs treated in the absence or presence of 20 ng/mL TNF-α for 12 hours. Native chromatin was immunoprecipitated with antibodies against ERG or control IgG, and the immunoprecipitated fractions were subjected to PCR using VWF exon 1-specific primers. As shown in Figure 5C, the immediate upstream promoter of VWF (which contains the −56 ETS motif) was bound by ERG, and the level of binding was significantly reduced by TNF-α treatment. In transient transfection assays of HUVECs, a mutation of the −56 ETS site (from GGAA to TTAA) resulted in a slight but significant reduction (72.2% of untreated vWF2 control) in basal promoter activity (Figure 5D). TNF-α treatment of transfected HUVECs inhibited the activity of vWF2 (74.4% of untreated vWF2 control) but not vWF2-ETSm (Figure 5D).
To further verify a functional role for ERG binding to the −56 ETS site in mediating VWF promoter activity, we used siRNA against ERG to knock down ERG expression in HUVECs and transfected these cells with wild-type (vWF2) or mutant VWF promoter-reporter gene constructs. ERG knockdown resulted in significant inhibition of ERG mRNA (18% of control) and VWF mRNA (37% of control), as assayed by quantitative real-time PCR (Figure 5E). siRNA-mediated down-regulation of ERG protein was confirmed in Western blot analyses (Figure 5F). In ERG-deficient cells, vWF2 promoter activity was reduced to 45% of control (CTR siRNA) levels (Figure 5G). Consistent with the data shown in Figure 5D, vWF2-ETSm demonstrated lower activity in untreated or control siRNA-treated HUVECs, implicating a functional role for the −56 ETS motif (Figure 5G). vWF2 and vWF2-ETSm demonstrated comparable activity in knockdown cells, suggesting that ERG mediates vWF2 expression via the −56 ETS site (Figure 5G). There was a further small reduction in vWF2-ETSm activity in ERG-deficient cells compared with control cells. Together, these in vitro data strongly suggest that ERG binds to the −56 ETS site and that the resulting DNA-protein interaction positively regulates VWF expression.
Finally, to determine the role of the ERG-binding site in mediating expression of the VWF promoter in vivo, we targeted the Hprt locus with the vWF2 promoter containing a mutation of the −56 ETS motif coupled to LacZ. Analysis of the resulting mice did not reveal detectable expression of β-galactosidase with the exception of occasional positive blood vessels in the hindbrain (Figure 6). Thus, the −56 ETS motif is necessary for vWF2 promoter activity in blood vessels of the heart and skeletal muscle.
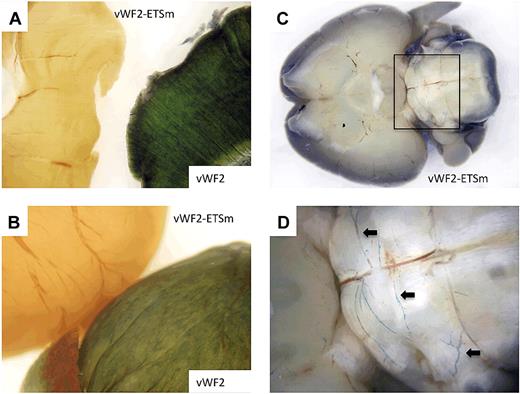
LacZ staining of diaphragm, heart, and brain from Hprt-targeted mice carrying vWF2-lacZ or vWF2-ETSm-lacZ. Diaphragms, hearts, and brains were harvested from 6- to 8-week-old F2 male Hprt-targeted mice carrying vWF2-lacZ or vWF2-ETSm-lacZ and processed in parallel for whole-mount staining with X-Gal. (A) The diaphragm from the vWF2-lacZ transgenic mouse is on the right, and the diaphragm from the vWF2-ETSm-lacZ mouse is on the left. (B) The vWF2-lacZ heart is on the bottom, and the vWF2-ETSm-lacZ heart is on the top. (C-D) The vWF2-ETSm-lacZ brain at low and high power, respectively (panel D is from the inset shown in panel C). Note the lack of detectable LacZ staining in skeletal muscle (diaphragm) and heart. Whole-mount tissues were analyzed under a Nikon SMZ-U dissecting microscope, and microphotographs were collected using a Nikon Coolpix 8400 camera. Arrows, LacZ-positive blood vessels in posterior brain.
LacZ staining of diaphragm, heart, and brain from Hprt-targeted mice carrying vWF2-lacZ or vWF2-ETSm-lacZ. Diaphragms, hearts, and brains were harvested from 6- to 8-week-old F2 male Hprt-targeted mice carrying vWF2-lacZ or vWF2-ETSm-lacZ and processed in parallel for whole-mount staining with X-Gal. (A) The diaphragm from the vWF2-lacZ transgenic mouse is on the right, and the diaphragm from the vWF2-ETSm-lacZ mouse is on the left. (B) The vWF2-lacZ heart is on the bottom, and the vWF2-ETSm-lacZ heart is on the top. (C-D) The vWF2-ETSm-lacZ brain at low and high power, respectively (panel D is from the inset shown in panel C). Note the lack of detectable LacZ staining in skeletal muscle (diaphragm) and heart. Whole-mount tissues were analyzed under a Nikon SMZ-U dissecting microscope, and microphotographs were collected using a Nikon Coolpix 8400 camera. Arrows, LacZ-positive blood vessels in posterior brain.
Discussion
One feature of the endothelium that is often overlooked is its rich diversity of regional and organ-specific phenotypes. Endothelial cell heterogeneity has been described at the level of cell morphology, function, gene expression, and antigen composition. Endothelial cell phenotypes vary between different organs, between different segments of the vascular loop within the same organ, and between neighboring endothelial cells of the same organ and blood vessel type (reviewed by Aird26 ). In vivo proteomic approaches have revealed a striking array of vascular bed–specific phenotypes. For example, antibody and subfractionation strategies have been employed to generate monoclonal antibodies that specifically target the caveolae in one vascular bed or another.27 Others have used phage display peptide libraries to select for peptides that home to specific vascular beds in vivo. These latter studies have uncovered a vascular address system that allows for site-specific targeting of biologically active compounds, for example, to the endothelial lining of tumor blood vessels.28
The molecular basis of endothelial heterogeneity remains poorly defined. One approach to this problem is to study the mechanisms underlying differential gene regulation. Previous studies using standard transgenic mice have shown that most endothelial promoters contain information for expression in a subset of vascular beds in vivo.12,29-36 In Hprt-targeted mice, different fragments of the VWF, FLT-1, endothelial nitric oxide synthase, and ROBO4 promoters have been shown to direct expression in distinct vascular beds.8,37-39 Collectively, these data argue against the existence of a pan-endothelial “master regulator” and suggest that at least some endothelial genes are regulated by vascular bed–specific promoter modules.
The results of the current study provide compelling new evidence for such a mechanism in mediating VWF expression. Specifically, we have shown that DNA sequences between −843 and −620 contain information for expression of VWF in the capillary endothelium of the heart and skeletal muscle. Indeed, the finding that vWF2-Del3-lacZ mice (containing VWF sequences between −620 and +1475) and vWF2-Δ843-620-lacZ mice (containing VWF sequences between −2182 and +1475, but with an internal deletion of −843 to −620) lose expression in the capillaries of the heart and skeletal muscle indicates that the 224-bp region is necessary for site-specific expression. The −843 to −620 region does not appear to function as a classical enhancer because it failed (at least in the context of the larger −843 to −487 region) to confer microvascular expression in the heart and skeletal muscle when coupled to a minimal TK promoter.
Although we have examined a large number of organs in vWF2 and vWF2-Δ843-620-lacZ mice for LacZ expression, the survey is not exhaustive. Thus, we cannot exclude the possibility that the vWF2 promoter is active in other, yet unexplored vascular beds. This limitation does not detract from the main finding in the paper, namely that capillary endothelial expression in the heart and skeletal muscle is selectively lost in the vWF2-Δ843-620-lacZ mice.
The present data raise a fundamental question: what DNA sequences within this region are responsible for mediating promoter activity in such a limited subset of endothelial cells? To identify candidate regulatory elements, we used Multiz Alignment through the University of California Santa Cruz Genome Browser to compare the −843 to −620 region in the human promoter to similar sequences in the mouse and bovine VWF promoters. On the human promoter fragment, 5 TRANSFAC binding matrices were detected by MATCH, 3 of which, ZNF143, TAL1/TCF3 complex, and BCL6, were moderately conserved between all 3 species (supplemental Figure 13). Furthermore, 25 JASPAR matrix binding sites were also identified, 8 of which are conserved: 3 ETS1 sites, SP1, HOXA5, PDX1, PRRX2, and MZF1 (supplemental Figure 13). Given their location (between −843 and −620), each of these evolutionarily conserved sequences is a candidate for site-specific expression of VWF in the microvascular endothelium of heart and skeletal muscle. In transient transfections of HUVECs, the vWF2 and vWF2-Δ843-620 promoters demonstrate comparable activity, and VWF promoter activity is not induced by the addition of conditioned medium from primary rat cardiomycoytes (data not shown). To date, we have been unsuccessful in transiently transfecting primary mouse heart capillary endothelial cells. Thus, in the absence of an appropriate in vitro system, continued efforts to delineate the microvascular bed–specific DNA elements between −843 and −620 will rely on Hprt-locus targeting.
In addition to generating increased protein diversity through alternative splicing, many studies have shown that constitutively spliced introns are required for optimal gene expression. In some cases, the effect is mediated by cis-regulatory elements present in the native intron. Such a mechanism has been implicated for vascular endothelial growth factor receptor 2/FLK-1, TIE-2, GATA2, endoglin, and VWF (the latter involving DNA sequences in intron 51).11-15 In other cases, gene expression enhancement occurs with native or non-native introns alike, suggesting a role for splicing per se. The mechanisms underlying splicing-mediated enhancement of gene expression are poorly understood but may involve increased transcription16,17,40 and/or increased nuclear export of mRNA.41
The finding that the intron-less vWF3 promoter (−2182 to +246) lacked activity in the heart and skeletal muscle pointed to an important role of the promoter proximal intron in mediating expression in these vascular beds. However, the finding that the non-native second intron of the human β-globin gene could largely rescue expression of the −2182 to +246 promoter argues against a critical role for gene-specific regulatory elements in the VWF intron. Previous studies demonstrated that the second intron of the human β-globin gene rescued expression of a heterologous promoter when placed between the promoter and the cDNA but not when placed 3′ of the gene or 5′ of the promoter.16,20 Thus, it seems unlikely that the β-globin intron is augmenting VWF expression by an enhancer-dependent mechanism. Rather, the data point to a requirement for intronic splicing. This is the first study of which we are aware to implicate a role for intronic splicing in the expression of an endothelial gene. More importantly, the observation that vWF3-lacZ expresses in the brain but not the heart or skeletal muscle suggests that the effects of splicing on gene expression vary between vascular beds, and thus may represent a novel mechanism of endothelial cell heterogeneity.
A mutation of the −56 ETS motif in the context of the vWF2 promoter resulted in complete loss of expression in the heart and skeletal muscle and significantly reduced expression in the brain. These findings implicate a critical role for the −56 ETS motif in mediating expression of VWF. Consensus ETS binding motifs have been identified within the promoters of several endothelial-restricted genes, including VWF, FLT-1, TIE1, TIE2, FLK-1, endothelial nitric oxide synthase, and vascular endothelial-cadherin.42-46 Previous studies from our own group and others have demonstrated the functional relevance of ETS motifs in mediating endothelial gene expression of VWF using standard transgenic mice, Hprt-targeted mice, and by introducing a mutation into the endogenous promoter.38,44,45,47,48 Various ETS family members have been implicated in the regulation of endothelial gene expression, including ELF, ETS-1, ETS-2, GABP, and ERG (reviewed by Oettgen49 ). We have recently shown that ERG is enriched in the endothelium.9 A previous study of the VWF promoter demonstrated that the −56 ETS motif binds ERG but not other ETS factors.21 In the current study, we have provided additional evidence for the role of ERG in mediating expression of VWF via the −56 ETS motif. ChIP assays confirm binding of ERG to the proximal promoter region in HUVECs, and transient transfections of ERG-deficient HUVECs with vWF2 and of ERG-competent HUVECs with vWF2-ETSm revealed comparable promoter activity. Collectively, the data suggest that an interaction between ERG and the −56 ETS motif is necessary for VWF promoter activity in the vascular beds of the heart and skeletal muscle. The −56 ETS site may also be necessary for VWF expression in other vascular beds and/or megakaryocytes. To test this hypothesis, we must first identify the promoter regions that direct authentic expression of VWF throughout the vasculature and in megakaryocytes/platelets. An alternative approach would be to mutate the −56 ETS motif in the context of the endogenous mouse VWF locus as previously described for the ROBO4 gene.48
In summary, we have shown that a 224-bp region of the VWF promoter (between −843 and −620) functions in conjunction with an ETS motif and an intron (ie, by splicing) to mediate gene expression in the capillary endothelium of the heart and skeletal muscle.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
This work was supported by National Institutes of Health grant HL076540.
National Institutes of Health
Authorship
Contribution: J.L., L.Y., G.M., E.R., L.J., D.B., K.C.S., Y.O., and T.M. designed and performed experiments and analyzed the data; P.O. designed experiments and analyzed the data; and W.C.A. designed and performed experiments, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: William C. Aird, Beth Israel Deaconess Medical Center, Molecular and Vascular Medicine, RW-663, 330 Brookline Ave, Boston, MA; e-mail: waird@bidmc.harvard.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal