Abstract
Macrophages infected with HIV-1 sustain viral replication for long periods of time, functioning as viral reservoirs. Therefore, recognition of factors that maintain macrophage survival and influence HIV-1 replication is critical to understanding the mechanisms that regulate the HIV-1–replicative cycle. Because HIV-1–infected macrophages release the nerve growth factor (NGF), and NGF neutralization reduces viral production, we further analyzed how this molecule affects HIV-1 replication. In the present study, we show that NGF stimulates HIV-1 replication in primary macrophages by signaling through its high-affinity receptor Tropomyosin-related Kinase A (TrKA), and with the involvement of reticular calcium, protein kinase C, extracellular signal-regulated kinase, p38 kinase, and nuclear factor-κB. NGF-induced enhancement of HIV-1 replication occurred during the late events of the HIV-1–replicative cycle, with a concomitant increase in viral transcription and production. In addition, NGF reduced the synthesis of the cellular HIV-1 restriction factor APOBEC3G and also overrode its interferon-γ–induced up-regulation, allowing the production of a well-fitted virus. Because NGF-TrKA signaling is a crucial event for macrophage survival, it is possible that NGF-induced HIV-1 replication plays a role in the maintenance of HIV-1 reservoirs. Our study may contribute to the understanding of the immunopathogenesis of HIV-1 infection and provide insights about approaches aimed at limiting viral replication in HIV-1 reservoirs.
Introduction
HIV-1, the etiologic agent of AIDS, persistently replicates in lymphoid tissues, leading to a profound immunosuppression that can culminate in higher susceptibility to opportunistic infections, tumors, and central nervous system degeneration.1 HIV-1 infects and replicates in CD4+ T lymphocytes and in cells of the monocyte/macrophage lineage using the CD4 molecule and the chemokine receptors CXCR4 or CCR5 to penetrate into host cells.1 During the HIV-1–replicative cycle, viral RNA undergoes reverse transcription, which is accomplished by the action of the viral enzyme reverse transcriptase (RT), and the resulting cDNA can be further integrated in the host cell genome. After long terminal repeat (LTR) activation, HIV-1 transcripts and proteins are produced, new progeny virions are assembled, and, in macrophages, virions are released after maturation in multivesicular bodies.2 Macrophages play a central role in HIV-1 pathogenesis, from the primary infection to the establishment of HIV-1 reservoirs in the gut-associated lymphoid tissue.2
A variety of conditions have been associated with increased viral load and viral persistence in HIV-1–infected persons, including coinfections3-5 and immune activation.6 In this context, cytokines and other endogenous soluble factors can modulate the viral replicative cycle,7,8 and several studies support the influence of these molecules over both the course of the disease and HIV-1 biology.7-9 Pro-inflammatory cytokines, such as tumor necrosis factor-α (TNF-α) and interleukin-6 (IL-6), up-regulate HIV-1 replication, whereas others with anti-inflammatory properties, such as type 1 interferon (IFN), IL-10, and IL-27, may diminish viral replication.8,10 It has been found that some cytokines interfere with HIV-1 replication by modulating cellular restriction factors to the virus. For example, production and activation of the proteins of the APOBEC family are regulated by IL-2, IL-7, IL-15, IL-27, and IFN-α.11
Nerve growth factor (NGF) is a member of the family of neurotrophins that also includes the brain-derived neurotrophic factor (BDNF), neurotrophin-3 (NT-3), and NT-4/5.12,13 All neurotrophins mediate their effects by binding to 2 different receptors: tropomyosin-related kinase (TrK) receptors and p75 neurotrophin receptor (p75NTR). Neurotrophins bind preferentially to specific TrK receptors: NGF to TrKA, BDNF and NT-4 to TrKB, and NT-3 to TrKC. However, some promiscuity may exist, because NT-3 can also bind to TrKA and TrKB receptors. All neurotrophins bind to p75NTR with equal affinity, and the precursors of the neurotrophins, the pro-neurotrophins (such as proNGF), are high-affinity ligands for p75NTR in complex with the coreceptor sortilin.14
The effects of neurotrophins are not restricted to the cells of the nervous system; immune cells and other peripheral tissues are also sensitive to stimulation by these mediators, functioning as key mediators of the neuro-endocrine-immune axis.12,13 For example, NGF induces its own release from CD4+ T cells15 and, during cellular stress, NGF induces an increase of corticosteroid serum levels.16 The production of NGF is up-regulated by several inflammatory mediators, such as IL-1, IL-6, TNF-α, and transforming growth factor-β,17-20 which can stimulate HIV-1 replication in vitro. NGF also participates in many other biologic processes. For example, it stimulates the growth and maturation of myeloid cells,21 the proliferation of B and T cells, and the differentiation of B cells into plasma cells.22,23 More importantly in the context of this study, NGF is vital for the survival and activation of monocytes/macrophages.24,25
It has been demonstrated that NGF is produced by HIV-1–infected cells and can probably support HIV-1 replication for long periods of time via TrK receptors.26 Based on these findings, we investigated whether NGF could enhance HIV-1 replication in vitro. We show here that NGF increases HIV-1 replication in primary monocyte–derived macrophages by engaging its classic TrKA signaling and down-regulating the synthesis of the HIV-1 natural restriction factor APOBEC3G (A3G).
Methods
HIV-1 isolates, reagents, and enzyme-linked immunosorbent assay kits
Assays of cell infection were performed with the monocytotropic, CCR5-dependent isolate HIV-1Ba-L, which was donated to us by the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health (Bethesda, MD). This isolate was expanded in phytohemagglutinin-activated peripheral blood mononuclear cells (PBMCs) from healthy donors, as described elsewhere.27 Recombinant human NGF, BDNF, and proNGF were obtained from PeproTech. The pharmacologic modulators used in this study (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) were purchased from Calbiochem.
Cells
Human monocyte–derived macrophages were obtained from PBMCs that had been isolated by density gradient centrifugation (Histopaque; Sigma-Aldrich) from buffy coat preparations of blood from healthy donors, through adherence onto plastic plates. Briefly, 1.5-2.0 × 106 PBMCs were plated onto 48-well plates (Nalge Nunc) in Dulbecco modified Eagle medium (DMEM; LGC Biotecnologia) containing 10% human serum (Sigma-Aldrich) and penicillin-streptomycin (Sigma-Aldrich). Cells were maintained at 37°C in 5% CO2 for 6-7 days for monocyte differentiation into macrophages. Nonadherent cells were washed out, and the remaining macrophage layer was maintained in DMEM with 10% human serum. Macrophage purity was > 90%, as determined by flow cytometric analysis (FACScan; Becton Dickinson) using anti-CD3 (Pharmingen) and anti-CD16 (Southern Biotech) monoclonal antibodies. For some assays (see below), macrophages were prepared in 25-cm2 plastic culture bottles following the same protocol but dispensing 40 × 106 PBMCs/5 mL of medium/bottle.
HIV-1 infection
Macrophages were infected with the HIV-1 R5-isolate Ba-L by exposing them overnight to viral suspensions containing 5-10 ng/mL of p24 antigen (p24 Ag). Noninternalized viruses were then removed by washing, and cell monolayers were replenished with fresh medium. HIV-1 replication was evaluated in cell-culture supernatants by a commercial enzyme-linked immunosorbent assay (ELISA) kit according to the manufacturer's instructions (ZeptoMetrix).
Effect of the neurotrophins on HIV-1 replication
HIV-1–infected macrophages were treated with NGF, BDNF, or proNGF immediately after cell infection. Cells were maintained in culture for different times, and HIV-1 replication was measured as described in the previous paragraph. Alternatively, to measure the effect of NGF on HIV-1 replication in the presence of different pharmacologic inhibitors, HIV-1–infected macrophages were treated with the appropriate inhibitor 15 minutes before NGF addition (10 ng/mL) at 13-14 days after infection. Virus replication was measured 48-72 hours after NGF addition.
Adsorption and penetration inhibition assays
Virus entry assays were performed as described previously with minor changes.28 Macrophages in 24-well plates were infected with 100 ng/mL of p24 Ag, in the absence or presence of NGF (10 ng/mL) at either 4°C, a permissive temperature for virus adsorption but not internalization, or 37°C, a permissive temperature for both processes. After 3 hours of incubation with virus inoculum, the cells infected at 4°C were washed and lysed with 0.1 mL of phosphate-buffered saline containing 0.5% Triton X-100. For evaluation of penetration, after a 3-hour incubation at 4°C, cells were washed to remove unbound virus and the temperature was raised to 37°C in the presence of NGF (10 ng/mL) for 2 hours. Cells were then lysed as above. Viruses able to adsorb or penetrate were measured by p24 ELISA in these lysates. For comparison, this assay was also carried out with soluble CD4 (sCD4) at 1 μg/mL.
Evaluation of viral and A3G mRNA synthesis
Macrophages adhered in 25-cm2 bottles were infected with HIV-1, and treated with NGF (10 ng/mL) or TNF-α (10 ng/mL). After 2 days, cells were lysed with TRIzol (Invitrogen) and RNA was extracted according to the manufacturer's instructions. Total RNA (5 μL) was incubated with Superscript III with Taq platinum (Invitrogen) in a 25-μL reaction volume and with appropriate primers (300nM), as described previously.29 The quantification of transcript abundance in HIV-1–infected cells treated or not with NGF or TNF-α was determined using the ΔΔCt method. The untreated HIV-1–infected macrophages were used as a reference sample (calibrator) and β-actin as the internal control.
For detection of human A3G and APOBEC3F (A3F), uninfected macrophages were left untreated (mock treated) or treated with NGF or IFN-γ for 24 hours. Cells were then lysed with TRIzol and real-time RT-PCR (qRT-PCR) was performed as described previously.30 Transcript abundance in mock-treated macrophages and in those treated with NGF or IFN-γ were also determined using the ΔΔCt method. The mock-treated macrophages were used as a reference sample (calibrator) and β-actin as the internal control.
All reactions were run in duplicate using the Applied Biosystems Prism 7500 Sequence Detection System. Delta-Ct values from the calibrator and from experimental groups were measured by subtracting Ct values from target versus housekeeping transcripts. Delta-delta-Ct was the difference between calibrators and experimental ΔCts. We determined that our application efficiency was near 100% (0.988) by serial logarithmic dilutions of our transcripts. Thus, the -fold change was assumed to be 2ΔΔCt, considering significant a -fold change more than 5 times the control levels.
Western blot
We performed immunoblotting assays to determine whether NGF could affect A3G expression. In brief, human macrophages in 25-cm2 bottles were treated with NGF (10 ng/mL), IFN-γ (10 ng/mL), or a combination of both molecules at different concentrations for 24 hours at 37°C. Cellular proteins were then extracted using buffer A (0.2% bromophenol blue; 0.5% β-mercaptoethanol; 1M Tris-HCl, pH 6.8; 10% sodium dodecyl sulfate; 1% glycerol) and a 20-μg aliquot of the extracted proteins was separated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride filters. Filters were then blocked and incubated with rabbit polyclonal anti-A3G or anti-GAPDH antibody (Abcam) for 1 hour. Specific reactive proteins were detected with the enhanced chemiluminescence method using a goat anti–rabbit immunoglobulin antibody linked to horseradish peroxidase (Santa Cruz Biotechnology). To evaluate activation of the signaling proteins involved in NGF-mediated enhancement of HIV-1 replication, macrophages were treated with different pharmacologic inhibitors 15 minutes before the addition of NGF (10 ng/mL). After 48 hours, cells were lysed and proteins immunoblotted with anti–TrKA-p, anti-TrKA, anti–protein kinase C-p (anti–PKC-p), anti-PKC, anti–extracellular related kinase-p (anti–ERK-p), anti-ERK, anti–p38K-p, or anti-p38K antibodies (Santa Cruz Biotechnology). We also performed densitometry analysis of the blots using EagleSight software (Version 3.21; Stratagene).
NGF effects on HIV-1 cDNA synthesis and genome integration
Macrophages cultured in 25-cm2 bottles were infected with HIV-1 (10 ng/mL of p24 Ag), and 24 hours later total DNA was extracted using a QIAamp DNA kit (QIAGEN) according to the manufacturer's instructions. PCR assays were performed to amplify total (LTR-LTR) or integrated DNA (LTR-Alu) HIV-1 DNA, as described previously.31 The amplified products were run in 2% agarose. We also performed densitometry analysis of the agarose gel bands using EagleSight. Alternatively, absolute real-time PCR (qPCR) assays were performed to quantify HIV-1–integrated DNA in HIV-1–infected cells treated with NGF. Macrophage DNA was extracted using a QIAamp DNA blood mini kit (QIAGEN), according to the manufacturer's instructions, and then DNA was quantified based on the A260-nm value measured with a Nanodrop 1000 instrument (Thermo Fisher Scientific). To standardize the experiments, DNA was diluted in diethylpyrocarbonate-treated water to test the equivalent to 150 000 cells. DNA was amplified by PCR using LTR-Alu primers.31 The generated amplicon (2 μL) was used as a template for the nested qPCR with 1× SYBR Green Master Mix (Applied Biosystems) and 250nM primers (Alu: 5′-TCCCAGCTACTCGGGAGGCTGAGG-3′ and SC-D-O-A-D: 5′-AGTCACACAACAGACGGGCACACAC-3′) in 25 μL. The copy numbers of proviral HIV-1 DNA were estimated by reference to standard curves of the ACH-2 cell line (ranging from 1-10 000 copies/reaction) under the conditions described previously.32
Functional A3G assays
To investigate whether NGF treatment could interfere with A3G enzymatic activity, we performed sequencing analysis of HIV-1 proviral genomes from cells exposed to this neurotrophin. Macrophages seeded in 25-cm2 bottles were infected with HIV-1 and exposed to NGF (10 ng/mL), IFN-α (10 ng/mL), or a combination of both molecules for 48 to 72 hours after infection. Cells were then lysed and the LTR region of proviral DNA was sequenced, as described in the supplemental materials.
Statistical analysis
All results presented in this study were prepared using Excel 2007 for Windows software (Microsoft). Statistical analysis calculations were performed using the same software, and the comparisons between means were considered significantly different when the P value was less than .05 using the Student t test.
Results
Neurotrophins enhance HIV-1 production in human primary macrophages
It has been shown that NGF release is increased in HIV-1–infected macrophages,26 and that NGF neutralization reduces viral replication and leads to macrophage death.26 These results suggested that NGF could play a relevant role in maintaining macrophages as HIV-1 reservoirs. Based on these findings, we investigated whether NGF—and other neurotrophins as well—would enhance HIV-1 production in primary macrophages. We treated HIV-1–infected human primary monocyte–derived macrophages with NGF, BDNF, or proNGF, and observed a stimulation of virus replication after neurotrophin treatment (Figure 1A). The peak of neurotrophin-induced enhancement of HIV-1 production occurred at 10 ng/mL, and a suboptimal enhancement was achieved at 1 ng/mL. Conversely, a higher neurotrophin concentration (100 ng/mL) induced a smaller but positive effect of these molecules on HIV-1 replication, suggesting receptor desensitization33 (Figure 1A). Comparing the activity of the tested neurotrophins, we observed that NGF, BDNF, and proNGF were equally potent on their effect on amplifying HIV-1 production (Figure 1A); therefore, we performed the next assays using NGF at 10 ng/mL, a concentration range that is commonly used in studies evaluating NGF function.21,23,34 We found the commonly observed donor-to-donor variation in the amplitude of HIV-1 replication, as well as in the magnitude of NGF-induced enhancement of HIV-1 replication, when HIV-1–infected macrophages were treated with NGF (supplemental Figure 1A). The maximum effect of NGF on HIV-1 replication was observed at 14 days, plateauing thereafter (Figure 1B). Similar to what was found by Samah et al,34 when we pretreated macrophages with NGF, no stimulation of HIV-1 replication was observed (supplemental Figure 1B), suggesting that the action of NGF requires the presence of HIV-1 within the cells. When analyzing NGF production by HIV-1–infected macrophages, we detected low NGF levels (supplemental Figure 1C), similar to those found by Samah et al34 and lower than those described Garaci et al26 This discrepancy could have been because of the different sources used for the ELISA assays, such as the commercial kits used by us and Samah et al (both from the same manufacturer) and in-house ELISA.17,26 The basal production of NGF by HIV-1–infected cells was very low and therefore not critical to the observed increment of HIV-1 replication.
NGF, proNGF, and BDNF enhance HIV-1 replication. (A) Macrophages were infected by an R5-tropic HIV-1 isolate (Ba-L), treated only once with different concentrations of neurotrophins as indicated, and virus replication was measured in the culture supernatants by an HIV-1 p24 ELISA 14 days after infection. (B) HIV-1 production was also measured at different time points after infection and addition of NGF (10 ng/mL). Data represent means ± SEM of 6 independent experiments. Virus production in the positive control (HIV-1–infected cells cultured only with medium): 105 ± 27 ng/mL of p24 Ag. *P < .05; **P < .01.
NGF, proNGF, and BDNF enhance HIV-1 replication. (A) Macrophages were infected by an R5-tropic HIV-1 isolate (Ba-L), treated only once with different concentrations of neurotrophins as indicated, and virus replication was measured in the culture supernatants by an HIV-1 p24 ELISA 14 days after infection. (B) HIV-1 production was also measured at different time points after infection and addition of NGF (10 ng/mL). Data represent means ± SEM of 6 independent experiments. Virus production in the positive control (HIV-1–infected cells cultured only with medium): 105 ± 27 ng/mL of p24 Ag. *P < .05; **P < .01.
Cell signaling triggered by NGF in human macrophages
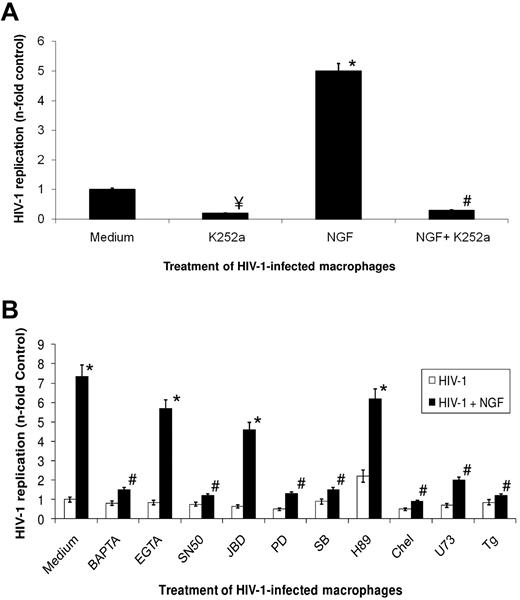
HIV-1 infection in macrophages up-regulates TrKA and down-regulates p75 receptors, which are the high- and low-affinity receptors for NGF, respectively.14 These receptors may also regulate cell survival.26 We next investigated whether NGF-induced enhancement of HIV-1 replication in macrophages would be dependent on TrKA classic signaling.35 Initially, macrophages were infected with HIV-1, and 13-14 days after infection, cells were treated 10 minutes before NGF addition with pharmacologic inhibitors of different cellular pathways classically involved in NGF signaling.35 Virus production was measured 48 hours after the addition of pharmacologic inhibitors. This period of time was chosen because when we treated 13-day-old HIV-1–infected macrophages with NGF, we observed a small but significant enhancement of virus production within 24 hours (data now shown), reaching a peak of stimulation of HIV-1 replication 2 days after NGF addition (Figure 2). The concentrations of the tested pharmacologic inhibitor (supplemental Table 1) were chosen to be virtually ineffective at inhibiting HIV-1 replication “per se” and to be without cellular toxicity (Figure 2B). We show in Figure 2A that K252a, an inhibitor of TrKA catalytic activity, inhibited both basal levels and NGF-enhanced HIV-1 replication, suggesting that this receptor plays an important role in HIV-1 production in macrophages. To investigate downstream events triggered by NGF-TrKA signaling that would be involved in the stimulation of HIV-1 replication, we inhibited different cellular pathways and measured HIV-1 production. Our results (Figure 2B) showed that NGF-induced enhancement of HIV-1 replication was prevented by 1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid-acetoxymethyl (BAPTA-AM), SN50, PD98059, SB230963, chelerythrine chloride, U73122, and thapsigargin, suggesting, respectively, the involvement of intracellular calcium, nuclear factor-κB (NF-κB) translocation, ERK, p38 kinase, PKC, phospholipase C-γ, and calcium from the endoplasmic reticulum on NGF-TrKA effect on HIV-1 replication. On the other hand, EGTA, JBD, and H89 were not able to prevent NGF-induced stimulation of HIV-1 replication, suggesting that extracellular calcium, Janus kinase, and protein kinase A might not be required for the observed phenomenon (Figure 2A).
NGF-mediated signaling involved in the enhancement of HIV-1 production. Macrophages were infected with an R5-tropic HIV-1 isolate (Ba-L), and 13 to 14 days after infection cells were treated with K252a (A) or the indicated pharmacologic modulators (B) 15 minutes before NGF (10 ng/mL) addition. These cells were kept in culture for an additional 2 days, when virus replication was measured in the culture supernatant by an HIV-1 p24 ELISA. Data represent means ± SEM of 4 independent experiments. Virus production in the positive control (HIV-infected cells cultured only with medium): 232 ± 51 of ng/mL p24 Ag. ¥P < .05 for HIV-1–infected cells treated or not with the pharmacologic inhibitors; *P < .05 for HIV-1–infected cells treated or not with NGF; #P < .05 for HIV-1–infected and NGF-boosted cells treated or not with the pharmacologic inhibitors.
NGF-mediated signaling involved in the enhancement of HIV-1 production. Macrophages were infected with an R5-tropic HIV-1 isolate (Ba-L), and 13 to 14 days after infection cells were treated with K252a (A) or the indicated pharmacologic modulators (B) 15 minutes before NGF (10 ng/mL) addition. These cells were kept in culture for an additional 2 days, when virus replication was measured in the culture supernatant by an HIV-1 p24 ELISA. Data represent means ± SEM of 4 independent experiments. Virus production in the positive control (HIV-infected cells cultured only with medium): 232 ± 51 of ng/mL p24 Ag. ¥P < .05 for HIV-1–infected cells treated or not with the pharmacologic inhibitors; *P < .05 for HIV-1–infected cells treated or not with NGF; #P < .05 for HIV-1–infected and NGF-boosted cells treated or not with the pharmacologic inhibitors.
To gain insight into how these pathways are correlated with each other, we performed immunoblotting assays for some of the signaling proteins (supplemental Figure 2). We noticed that the addition of K252a inhibited TrKA, PKC, ERK, and p38 kinase phosphorylation, indicating a TrKA upstream role (supplemental Figure 2A-D). Chelerythrine chloride and BAPTA-AM treatments prevented PKC, ERK, and P38 kinase phosphorylation (supplemental Figure 2B-D). SB230963 and PD98059 treatments only affected p38 kinase and ERK, respectively (supplemental Figure 2C-D). As expected, SN50 did not affect the phosphorylation of any of these proteins (supplemental Figure 2).
Our results suggest that NGF-TrKA signaling triggers phospholipase C-γ, which might lead to reticular calcium mobilization and therefore PKC signaling. PKC seems to be a convergent point for activation of ERK, p38 kinase, and NF-κB.
NGF effects on HIV-1–replicative cycle
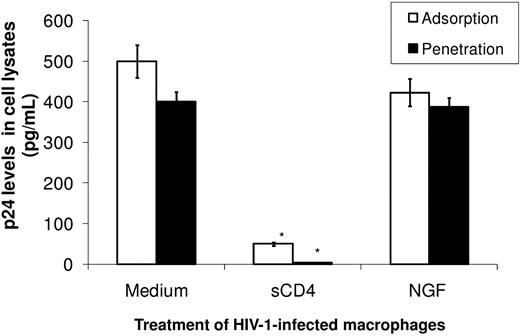
Because NGF triggers multiple signaling pathways in HIV-1–infected human macrophages, we investigated whether this neurotrophin could influence the HIV-1 life cycle. To analyze HIV-1 adsorption, we infected and treated macrophages with NGF at 4°C, a condition during which only virus adsorption occurs. Cells were then washed and lysed, and the amount of p24 Ag derived from the virus adsorbed on the plasma membrane was measured. To evaluate virus penetration, we performed the same assay, but cells were also kept for 1 hour at 37°C to allow virus penetration before the measurement of p24 Ag levels. We found no significant modulation of HIV-1 in NGF-treated cells compared with control cells (Figure 3). For comparison, we used sCD4 as a positive control for virus adsorption/penetration, which, as expected, prevented HIV-1 infection in macrophages (Figure 3).
NGF does not affect HIV-1 adsorption and penetration. Macrophages were exposed to an R5-tropic HIV-1 isolate (Ba-L) and treated with NGF (10 ng/mL) or sCD4 (1 μg/mL) for 3 hours at 4°C. The inoculum was then removed, cells were washed and lysed, and the adsorbed HIV-1 p24 Ag was measured with an HIV-1 p24 ELISA (adsorption). Alternatively, after the incubation at 4°C, the temperature was raised to 37°C for an additional 2 hours and cells were then washed and lysed to measure internalized HIV-1 p24 Ag as above (penetration). Data represent mean ± SEM of 3 independent experiments.
NGF does not affect HIV-1 adsorption and penetration. Macrophages were exposed to an R5-tropic HIV-1 isolate (Ba-L) and treated with NGF (10 ng/mL) or sCD4 (1 μg/mL) for 3 hours at 4°C. The inoculum was then removed, cells were washed and lysed, and the adsorbed HIV-1 p24 Ag was measured with an HIV-1 p24 ELISA (adsorption). Alternatively, after the incubation at 4°C, the temperature was raised to 37°C for an additional 2 hours and cells were then washed and lysed to measure internalized HIV-1 p24 Ag as above (penetration). Data represent mean ± SEM of 3 independent experiments.
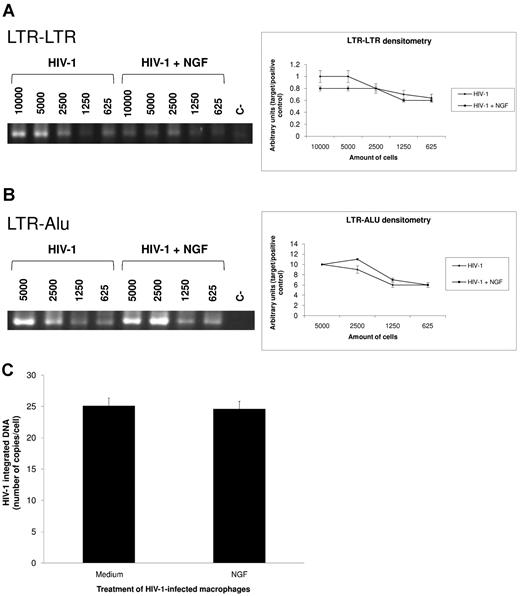
To analyze the primary steps of the HIV-1 life cycle that would be increased after NGF addition, we evaluated whether NGF would enhance HIV-1 cDNA synthesis, integration, and transcription. The neurotrophin was added 24-48 hours after infection instead of 13 days afterward to avoid multiple rounds of cell infection. We measured HIV-1 cDNA synthesis by performing PCR directly toward the 2 LTR segments (LTR-LTR). Proviral HIV-1 DNA was also measured with PCR assays toward LTR-Alu sequences (LTR-Alu) and by qPCR. We did not detect a significant difference in the levels of HIV-1 cDNA (Figure 4A) or virus integration (Figure 4B-C) between NGF-treated HIV-1–infected macrophages and controls.
NGF does not influence HIV-I cDNA synthesis and integration. Macrophages were infected by an R5-tropic HIV-1 isolate (Ba-L) and treated with NGF (10 ng/mL) for 24 hours. Total DNA was then extracted using the QIAamp DNA kit and PCRs were performed directly toward the 2 LTR segments (LTR-LTR) (A) and to LTR and Alu sequences (LTR-Alu) (B). The left panel displays a representative gel with the PCR products for both targets (LTR-LTR and LTR-Alu) in HIV-1–infected cells treated or not with NGF. The numbers shown in this panel represent the number of cells from which total DNA was extracted and used as the template for PCR amplifications. The right panel shows the band densitometry from 4 independent experiments (mean ± SEM). Absolute quantification of provirus DNA was performed by qPCR assays normalized by serial 10-fold dilution of the ACH-2 cell line (C). Bars represent means ± SEM of 2 independent experiments run in triplicate.
NGF does not influence HIV-I cDNA synthesis and integration. Macrophages were infected by an R5-tropic HIV-1 isolate (Ba-L) and treated with NGF (10 ng/mL) for 24 hours. Total DNA was then extracted using the QIAamp DNA kit and PCRs were performed directly toward the 2 LTR segments (LTR-LTR) (A) and to LTR and Alu sequences (LTR-Alu) (B). The left panel displays a representative gel with the PCR products for both targets (LTR-LTR and LTR-Alu) in HIV-1–infected cells treated or not with NGF. The numbers shown in this panel represent the number of cells from which total DNA was extracted and used as the template for PCR amplifications. The right panel shows the band densitometry from 4 independent experiments (mean ± SEM). Absolute quantification of provirus DNA was performed by qPCR assays normalized by serial 10-fold dilution of the ACH-2 cell line (C). Bars represent means ± SEM of 2 independent experiments run in triplicate.
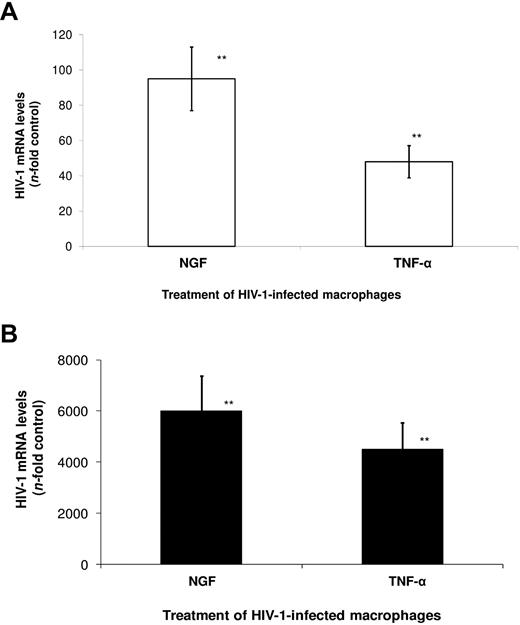
We next evaluated whether NGF could influence viral transcription by measuring levels of multiply spliced (MS) and full-length (FL) HIV-1 transcripts, through qRT-PCR. We observed that NGF enhanced HIV-1 transcription of MS (Figure 5A) and FL (Figure 5B) mRNAs 95- and 6000-fold over control, respectively. For comparison, the classic stimulator of HIV-1 transcription, TNF-α, induced HIV-1 gene expression by 45-fold (MS) and 4200-fold (FL) over control. These results suggest that NGF stimulates HIV-1 replication through activation of its transcription.
NGF enhances HIV-1 transcription in macrophages. Macrophages were infected by an R5-tropic HIV-1 isolate (Ba-L) and treated with NGF (10 ng/mL) or TNF-α (10 ng/mL) for 48 hours. Total RNA was then extracted using the QIAamp RNA kit, and qRT-PCR for the detection of MS (A) and FL (B) HIV-1 transcripts were performed as described previously.29 Data represent means ± SEM of 3 independent experiments. **P < .01.
NGF enhances HIV-1 transcription in macrophages. Macrophages were infected by an R5-tropic HIV-1 isolate (Ba-L) and treated with NGF (10 ng/mL) or TNF-α (10 ng/mL) for 48 hours. Total RNA was then extracted using the QIAamp RNA kit, and qRT-PCR for the detection of MS (A) and FL (B) HIV-1 transcripts were performed as described previously.29 Data represent means ± SEM of 3 independent experiments. **P < .01.
NGF down-modulates the synthesis of A3G
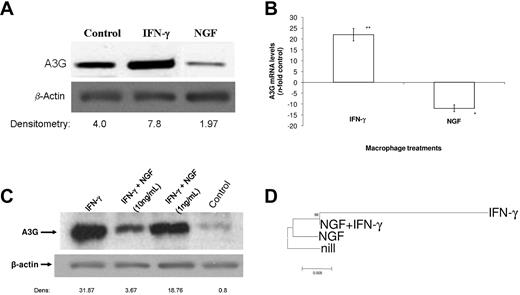
Because some cytokines, such as IFNs, exert their effect on HIV-1 replication through modulation of host restriction factors such as A3G, we evaluated whether NGF was endowed with the opposite effects compared with IFNs. We first measured A3G levels by immunoblotting, and found a reduction of approximately 50% of A3G basal protein content after cell treatment with NGF (Figure 6A). For comparison, IFN-γ, which induces A3G expression, doubled the cellular content of this protein (Figure 6A). The NGF-induced A3G reduction occurred not only at the protein level, but also transcriptionally (Figure 6B). NGF reduced the synthesis of A3G mRNA by 12-fold, whereas IFN-γ produced a 22-fold increase in A3G gene expression (Figure 6B).
NGF down-modulates A3G synthesis. Macrophages were treated with NGF (10 ng/mL unless otherwise indicated) alone or in combination with IFN-γ (10 ng/mL) for 48 hours. Cells were either lysed for immunoblotting (A,C) or RNA extraction and qRT-PCR were performed as described previously.30 (B). Alternatively, HIV-1–infected macrophages were treated with these molecules at the above-mentioned concentrations, and after 48 hours, cells were lysed, total DNA extracted, and the integrated HIV-1 LTR was sequenced. The phylogenetic relationship between the proviral DNA derived from these samples is presented in a tree rooted by the LTR sequence from the untreated HIV-1–infected cell (D). In panels A and C, representative immunoblots with their respective densitometries (A3G/β-actin) are displayed (n = 3). Data in panel B represent mean ± SEM of 3 independent experiments, **P < .01. In panel D, the bootstrap probability is indicated for each interior branch and the scale bar indicates the number of nucleotide changes per site.
NGF down-modulates A3G synthesis. Macrophages were treated with NGF (10 ng/mL unless otherwise indicated) alone or in combination with IFN-γ (10 ng/mL) for 48 hours. Cells were either lysed for immunoblotting (A,C) or RNA extraction and qRT-PCR were performed as described previously.30 (B). Alternatively, HIV-1–infected macrophages were treated with these molecules at the above-mentioned concentrations, and after 48 hours, cells were lysed, total DNA extracted, and the integrated HIV-1 LTR was sequenced. The phylogenetic relationship between the proviral DNA derived from these samples is presented in a tree rooted by the LTR sequence from the untreated HIV-1–infected cell (D). In panels A and C, representative immunoblots with their respective densitometries (A3G/β-actin) are displayed (n = 3). Data in panel B represent mean ± SEM of 3 independent experiments, **P < .01. In panel D, the bootstrap probability is indicated for each interior branch and the scale bar indicates the number of nucleotide changes per site.
Because HIV-1–infected macrophages may be targeted by a simultaneous stimulation by IFN-γ and NGF, we analyzed A3G synthesis when macrophages were treated in vitro by these modulators in combination. Macrophages were treated with IFN-γ at 10 ng/mL and with NGF at 10 or 1 ng/mL alone or together with IFN-γ, and examined the A3G protein levels. We found that at 10 and 1 ng/mL, NGF reduced IFN-γ-induced A3G synthesis by ∼ 90% and 40%, respectively (Figure 6C), suggesting an additional role of this neurotrophin in the maintenance of the HIV-1 reservoir.
To investigate the effects of NGF on A3G activity and further effects on HIV-1 replication, HIV-1–infected macrophages were treated with NGF, IFN-γ, or NGF + IFN-γ, and viruses produced under these experimental conditions were genotyped 48-72 hours after infection to measure G → A mutations. We chose to evaluate the proviral HIV-1 DNA to verify changes that would affect the virus life cycle directly. As seen in Figure 6D and supplemental Figure 3, IFN-γ induced a high frequency of G → A hypermutation, leading to a rapid viral evolution. Such a divergence did not occur in NGF-treated cells and was prevented because of NGF treatment in cells boosted with IFN-γ. Because the genome sequences of HIV-1 proviral DNA derived from NGF-treated cells and IFN-γ + NGF–treated cells clustered together (Figure 6D), this suggests that the HIV-1 provirus generated from cells under both treatments present the same nature of polymorphisms (G → A mutations); however, mutations in provirus from IFN-γ + NGF–treated cells occurred at a lower frequency compared with provirus from IFN-γ–treated cells. Therefore, in addition to enhancing HIV-1 replication through transcription activation, NGF also allows the host cell to produce a well-fitted virus, preventing the restriction imposed by IFN-γ/A3G.
Discussion
NGF was first described as a neurotrophic factor able to promote survival and neuritogenesis from sensory and sympathetic neurons.36 It is known today that the physiologic effects of NGF, which include cell survival/death, activation, and differentiation, occur in many biologic systems. This neurotrophin exerts many effects on the immune system, such as mast-cell dysregulation chemotaxis of granulocytes, differentiation and cytotoxic activity of monocytes and microglia, T-cell proliferation, release of cytokines, and B-cell differentiation22,23 ; therefore, the biology of pathogens that affect the functions of the cells of the immune system may be influenced by NGF. Because HIV-1 replicates in CD4+ cells, and considering that NGF is an autocrine factor secreted by HIV-1–infected macrophages necessary for cell survival and virus replication, we further investigated the mechanisms by which NGF affects HIV-1 replication. We show that NGF, as well as other neurotrophins, enhances HIV-1 replication in human primary macrophages. NGF-TrKA engagement triggered a calcium-dependent PKC activation, followed by NF-κB, ERK, and p38 kinase signaling. The activation of multiple pathways enhances HIV-1 transcription of both MS and FL mRNAs and down-regulates A3G synthesis.
Initially, we tested the effect of NGF, BDNF, and proNGF on HIV-1 production in human primary macrophages, and found that these neurotrophins reached maximum stimulation of HIV-1 replication at 10 ng/mL. Macrophages have the potential to become an HIV-1 reservoir, producing virus for long periods of time,1 and might contribute to HIV-1 evolution and genetic divergence in different compartments of the human body.37 Certainly, such maintenance of HIV-1 reservoirs is due to the physiologic effects of several molecules.38 Our results suggest that NGF, through its ability to sustain macrophage survival and HIV-1 replication in this cell type, is an important factor in the generation and maintenance of the HIV-1 reservoir.
Garaci et al26 and our results in the present study have shown that NGF stimulates virus production, whereas Samah et al34 found no increase in HIV-1 production after NGF treatment. There could be many reasons for these opposite results, such as macrophage maturation with M-CSF and GM-CSF, followed by treatment with and migration toward CXCL12, and, more importantly, exposure of a subfraction of macrophages to NGF before HIV-1 infection.
HIV-1–infected macrophages overexpress the NGF high-affinity receptor TrKA, whereas they down-regulate the NGF low-affinity receptor p75NTR.26 This is in agreement with the proposed action of NGF as a survival factor for HIV-1–infected macrophages, because p75NTR is a TNF superfamily receptor involved in the triggering of cell death events.14 In our study, TrKA signaling could have been pharmacologically and immunologically linked to our model of NGF-induced enhancement of HIV-1 replication, because we demonstrated the involvement of the classic NGF signaling (PKC, ERK, p38K, and NF-κB), triggered by TrKA,35 in HIV-1–infected human macrophages. This pathway may lead to both gene expression–dependent and –independent mechanisms to modulate cellular function.39 The gene expression–dependent mechanisms could regulate HIV-1 and A3G transcription because of the regulation of both cis and trans elements triggered by NGF.39 These elements are expected to be gene inducers and/or suppressors.40 In the model we used to study cell signaling, we treated HIV-1–infected macrophages 13-14 days after infection with the pharmacologic inhibitors 10 minutes before NGF addition. Therefore, by the time NGF was added to the cell cultures, the pathways involved in its signaling would already be inhibited. To avoid cytotoxicities caused by pharmacologic inhibitors, once they were added, experiments were carried out for only 48 hours.
To better understand the earliest step of the HIV-1–replicative cycle that would be amplified by NGF, we measured HIV-1 adsorption/penetration, cDNA synthesis, integrated provirus, and transcription after NGF treatments within short time frames. We did not find any effect of NGF addition on viral adsorption, penetration, and integration. In agreement with the findings that NGF signaling polarizes to the nucleus, we found a strong activation of HIV-1 transcription of both MS and FL viral RNAs. The NGF enhancement of HIV-1 transcription was more powerful than the TNF-α stimulation, a classic HIV-1 LTR activator.9 It has already been proposed that NGF could activate HIV-1 LTR41,42 ; however, these previous works were performed using glial and neuronal cell lines transfected with HIV-LTR-CAT vectors. Therefore, our study included a more physiologic model of induction of HIV-1 transcription on NGF stimulation. The NGF-induced enhancement of MS and FL HIV-1 transcripts might suggest a role of this neurotrophin in de novo cellular infection and transcription activation of old/recently integrated genomes.
Because some studies have shown that NGF induces HIV-1 transcription in neuronal and glial transfected cell lines41,42 and that this neurotrophin is overexpressed in patients with AIDS-associated dementia,43 a possible connection between NGF and HIV-1 neuropathogenesis may be a matter of relevant speculation. Therefore, many investigators have used NGF and BDNF to rescue neuronal cells from apoptosis induced by HIV-1–related proteins.44-46 These investigators claim that NGF and BDNF would be beneficial factors for patients suffering from HIV-1–associated dementia.44-47 However, these studies did not take into account the effects of NGF on HIV-1 replication. We propose that it would be essential to review these studies and to conduct others to determine the equilibrium between NGF-induced HIV-1 replication and neurotrophin-mediated neuronal rescue from HIV-1–induced apoptosis to better understand the impact of NGF and other neurotrophins as protective or deleterious molecules for HIV-1–infected persons.
The cellular antiretroviral factor A3G restricts HIV-1 replication by packing within the virions and causing a G → A hypermutation in the viral genome, thus impairing consecutive rounds of viral infection within its host cell.48 Because A3G is packaged within HIV-1 virions during late events of virus replication, overlapping with NGF effects on HIV-1 transcription and viral production, we analyzed the ability of this neurotrophin to modulate the synthesis of this restriction factor. In fact, NGF diminished the basal levels of A3G content in macrophages. Moreover, NGF counterbalanced the IFN-γ-induced A3G synthesis. This result could mean that NGF is not only an essential factor for the survival of HIV-1–infected macrophages26 able to stimulate HIV-1 transcription, but is also a mediator that allows this HIV-1 reservoir to be resistant to innate antiviral defenses. Consolidating this finding and this interpretation, we found that the NGF-mediated reduction in A3G synthesis also diminished the introduction of G → A hypermutations in HIV-1 genome by INF-γ–mediated A3G synthesis (supplemental Figure 3), and that NGF prevented the virus evolution secondary to IFN-γ–induced hypermutations, suggesting that this neurotrophin could favor the production of well-fitted viruses. A potential explanation for the effects of NGF on APOBEC activity may be the ability of NGF to activate the transcriptional factor NF-IL6 (also known as C/EBPβ),24 which, once activated, can promote HIV-1 replication by reducing A3G function.49,50 Based on these findings, one might expect that NGF would allow the growth of ΔVif-HIV-1. However, we did not observe ΔVif-HIV-1 growth in an NGF-treated HEK-293T cell line transfected with A3G (supplemental Figure 5). In addition, NGF did not down-regulate significantly the synthesis of A3F (supplemental Figure 4). Both conditions associated presume that ΔVif-HIV-1 would not replicate in primary cells treated with NGF. Nonetheless, we observed under the same experimental condition that NGF increased the infectivity of a wild-type virus in the presence of A3G (supplemental Figure 5). The apparent paradox in the observation that wild-type virus infectivity is enhanced in NGF-treated macrophages, even in unaffected A3F expression, could be explained by the diminished APOBEC pressure on the generation of virus carrying G → A mutations (Figure 6 and supplemental Figure 3). Taking into account a scenario that NGF reduces significantly the levels of A3G but does not affect A3F, an additional explanation would be a synergy or additive effect between NGF signaling and Vif effects on APOBEC degradation (supplemental Figure 5). Considering that NGF weakens the restriction imposed by A3G, as judged by the phylogenetic analysis, we could say that this activity is a more relevant role of the effects of NGF toward HIV-1 replication.
In conclusion, we show here that a molecule with a broad range of physiologic and physiopathologic actions in many biologic systems12-14 is endowed with additional functional aspects. Considering that macrophage survival is highly influenced by NGF, its participation in the persistence of a chronic human viral infection may be a hallmark of the degree of adaptation/dependence established between a parasite and its host. Therefore, we propose that NGF, in addition to being secreted by HIV-1–infected human macrophages, contributes to the maintenance of virus replication and in the perpetuation of the HIV-1 reservoir. This phenomenon probably occurs because such a neurotrophin influences virus transcription and production and affects both basal levels and stimulated production of cellular A3G synthesis. Our data may stimulate additional studies to determine a possible role of NGF as a factor able to influence HIV-1 genetic diversity in the different compartments of the human body where HIV-1 replication occurs.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the Hemotherapy Service of the Hospital Clementino Fraga Filho (Federal University of Rio de Janeiro, Brazil) for providing buffy coats, and the AIDS Research and Reference Reagent Program (Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD) for providing sCD4 and the HIV-1 isolate Ba-L.
This study was supported by grants from PAPES/Fiocruz, CNPq, and Faperj.
Authorship
Contribution: T.M.L.S., E.G.A. and D.C.B.-H. designed research; T.M.L.S., D.Q.R., J.R.T., V.B.-S., R.S.A. and C.P.B.P. performed research; T.M.L.S., D.Q.R., V.B.-S., R.S.A., C.P.B.P., M.G.M., C.F.L.F., E.G.A. and D.C.B.-H. analyzed and interpreted data; and T.M.L.S. and D.C.B.-H. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Thiago Moreno L. Souza and Dumith Chequer Bou-Habib, Fundação Oswald Cruz, Av Brasil L365, 21045-900, Rio de Janeiro, RJ-Brazil; e-mail: tmoreno@ioc.fiocruz.br or Dumith@ioc.fiocruz.br.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal