Abstract
T cells originate from early T lineage precursors that have entered the thymus and differentiate through well-defined steps. Mice deficient for the BTB/POZ domain of zinc finger protein-1 (Miz-1) almost entirely lack early T lineage precursors and have a CD4−CD8− to CD4+CD8+ block causing a strong reduction in thymic cellularity. Miz-1ΔPOZ pro-T cells cannot differentiate in vitro and are unable to relay signals from the interleukin-7R (IL-7R). Both STAT5 phosphorylation and Bcl-2 up-regulation are perturbed. The high expression levels of SOCS1 found in Miz-1ΔPOZ cells probably cause these alterations. Moreover, Miz-1 can bind to the SOCS1 promoter, suggesting that Miz-1 deficiency causes a deregulation of SOCS1. Transgenic overexpression of Bcl-2 or inhibition of SOCS1 restored pro-T cell numbers and their ability to differentiate, supporting the hypothesis that Miz-1 is required for the regulation of the IL-7/IL-7R/STAT5/Bcl-2 signaling pathway by monitoring the expression levels of SOCS1.
Introduction
Hematopoietic precursors differentiate into mature blood cell lineages through a series of well-coordinated steps. T cells are generated in the thymus, which is continuously replenished with lymphoid progenitors from the bone marrow via the bloodstream.1 Early lymphoid progenitors (ELPs) enter the thymus and become early T lineage precursors (ETPs), defined as Lin−/low, CD117high, and CD25−.2 The capacity of ELPs to migrate to the thymus has been attributed to their expression of CCR9.3,4 In addition to CCR9+ ELPs, other progenitors, such as CLPs, may home to the thymus and generate T cells. Recently, Ly6D has been used to identify the branch point of CLPs that gives rise to the first stages of B-cell development, B cell-biased lymphoid progenitor (BLP), and all-lymphoid progenitor (ALP), which contribute to the T-cell development.5
The subsequent development of ETPs starts with CD4−CD8− double-negative 1 (DN1) cells. DN1s are subdivided into DN1a-e according to the expression of CD24 and CD117, DN1a/b corresponding to the ETP subset.6 DN1s give rise to DN2a-b cells, which differentiate into DN3s, subdivided into DN3a-b based on their size and CD27 expression.7 DN3a cells that have productively rearranged the T-cell receptor β-gene (TCR-β) become activated by TCR-dependent signals (β-selection), differentiate into DN3b, and become DN4 pre-T cells. The newly developed DN4s become CD4+CD8+ double-positive (DP) cells and undergo positive/negative selection before reaching the periphery as mature CD4+ or CD8+ T cells.8
Pro-T-cell differentiation steps depend on the expression of Notch ligands, mainly δ-like ligand 1 (DL1) and DL4 on thymic stroma,9 and on cytokines, such as interleukin-7 (IL-7).10 Notch signaling assures lineage commitment, survival, and development of ETPs into further DN subsets.11 The IL-7/IL-7R pathway drives proliferation, survival, and progression of pro-T cells,12 and also induces the rearrangement and transcription of the TCR-γ locus.13 The IL-7R signaling activates Janus kinase 1/3 (Jak1/3), which phosphorylate signal transducer and activator of transcription 5 (STAT5). Phosphorylated STAT5 then activates the transcription of IL-7–dependent target genes.14
A key player in IL-7R cascade is the maintenance of cell survival by promoting a favorable balance of B-cell lymphoma-2 (Bcl-2) family members.15 The expression of the antiapoptotic protein Bcl-2 is up-regulated after IL-7 stimulation. Some studies have shown that the up-regulation of Bcl-2 can be STAT5-dependent.16-18 Other studies have shown that STAT5-mediated activation of AKT protein regulates the glucose metabolism of the cell and maintains prosurvival and growth functions.19 Suppressor of cytokine signaling 1 (SOCS1) is known to inhibit phosphorylation of STAT proteins by directly binding to the Jak proteins and therefore inhibiting all further downstream signaling events to ensure a return to steady-state homeostasis after cytokine responses.20
Miz-1 (Zbtb17) is a transcription factor of 87 kDa that is composed of 13 zinc finger domains at its carboxy-terminal end and of a BTB/POZ domain at its N-terminus.21 It has originally been identified as an interacting partner of the c-Myc proto-oncogene.21 The BTB/POZ domain of Miz-1 is essential for its trans-activating functions, but not for its capacity to bind to the DNA.22 Miz-1 can activate or repress the transcription of its target genes depending on its interacting partner. For example, Miz-1 acts as a transcriptional trans-activator by binding to core elements of RNA Pol II–dependent target gene promoters and by recruiting coactivators, such as the histone deacetylase p300/CBP.22-24 Miz-1 can also be a transcriptional trans-repressor, for example, by recruiting c-Myc to an E-box-independent site around the initiator of its target gene promoters. Genes that encode the negative cell cycle regulators CDKN2b22,25 and CDKN1a26,27 have been validated as direct Miz-1 targets that are repressed by Miz-1/c-Myc complex.
Because Miz-1 deletion is lethal,28 we have used conditional Miz-1–deficient mice, in which the exons coding for the BTB/POZ domain29 are deleted via Cre recombinase.30 This deletion generates a truncated form of Miz-1 that lacks the BTB/POZ domain and thus eliminates specifically its activity as a transcriptional regulator in all hematopoietic cells.22,31 In this report, we describe findings that identify Miz-1 as a new regulator of early T-cell differentiation, at stages where the IL-7/IL-7R interaction assures survival and lineage commitment. Our data suggest that Miz-1 regulates the expression of SOCS1 and thus controls the activation of STAT5 phosphorylation in response to IL-7 to gauge the level of Bcl-2 expression required for the survival and development of ETP/DN1 and DN2 cells.
Methods
Mice
All mice used in this study are described in the supplemental Data (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Mice have been bred on C57BL/6 background for at least 10 generations and were maintained in Specific-Pathogen-Free Plus+. The Institutional Review Board approved all animal protocols, and experimental procedures were performed in compliance with the Institut de recherches cliniques de Montréal guidelines.
Antibodies and FACS analysis
Antibodies were from BD Biosciences, except when indicated. DN1-DN4, ETP, ELP, ALP, and BLP were analyzed using lineage marker-negative cells (Lin−) selected by staining with biotinylated antibodies described in the supplemental Data, followed by streptavidin-peridinin-chlorophyll protein-Cy5.5 or phycoerythrin-Cy5. Cells were analyzed with a FACSCalibur, FACSScan, or LSR (BD Biosciences). Cell sorting was performed using a MoFlo cell sorter (Cytomation).
Intracellular staining and cell activation
After 1 hour of incubation at 37°C to shut off endogenous signaling, cells were activated with or without 10 ng/mL IL-7 (PeproTech 217-17) for 18 hours to detect Bcl-2 or 15 minutes for pSTAT5. To verify Bcl-2, cells were fixed with Cyto Fixation/Permeabilization kit (BD 554714). For pSTAT5, cells were fixed with formaldehyde (BD cytofix 554655) and permeabilized with methanol (BD phosflow Perm III 558050). For gene expression profiling, DN1-DN2 cells were sorted, activated, and lysed in TRIZOL Reagent (Invitrogen). RNA was extracted as described in the supplemental Data.
OP9DL1 cocultures
OP9 stromal cells expressing DL132 were plated at 2.5 × 104 cells/well and cocultured with sorted DN1-4 and ETPs/ELPs. The cells were incubated in Opti-α-modified Eagle medium supplemented with 1 to 5 ng/mL IL-7 and 5 ng/mL Fms-like tyrosine kinase 3 ligand (PeproTech) and charcoal-stripped fetal bovine serum.
ChIP assay
Assays were performed using ChIP-IT Express (Active Motif) on purified primary CD4−CD8− DN cells (preparation purity > 90% by Auto-MACS), which rested at 37°C 1 hour in phosphate-buffered saline, or on SCID.adh murine thymic lymphoma (clone P6D4).33 Variation in the cell lysis was as follow: first lysis with 5mM piperazine-N, N-bis[2-ethanesulfonic acid], pH 8, 85mM KCl, 0.5% NP-40, 1mM phenylmethylsulfonyl fluoride, protease inhibitors cocktail (complete Mini; Roche Diagnostics), and second with 50mM Tris-HCl, 10mM ethylenediaminetetraacetic acid, and 1% sodium dodecyl sulfate, protease inhibitor cocktail, and 1mM phenylmethylsulfonyl fluoride. After sonication (Branson Digital Sonifier), immunoprecipitation was performed using salmon sperm DNA/Protein G-agarose (Upstate Biotechnology) and 10 μg of rabbit anti–Miz-1 (H190; Santa Cruz Biotechnology) or rabbit control IgG antibodies (Abcam).
SOCS1 knockdown
Sorted DN1 or Lin−Sca1+c-Kit+ (LSK) were cultured in Opti-α–modified Eagle medium supplemented with 1 ng/mL IL-7, 5 ng/mL Fms-like tyrosine kinase 3 ligand, and 10 ng/mL stem cell factor for 1 hour. Cells were then incubated with Endo-Porter reagent (Gene Tools) for delivering fluorescein isothiocyanate-morpholino oligo against SOCS1 mRNA or a control morpholino oligo in vitro. After 4 hours, the cells were transferred on OP9DL1 stroma layer and cultured for 6 to 20 days.
Statistical analysis
Two-tailed Student t tests were used to calculate P values. A P value < .05 was considered statistically significant.
Results
Miz-1ΔPOZ mice have severe defects in T-cell development
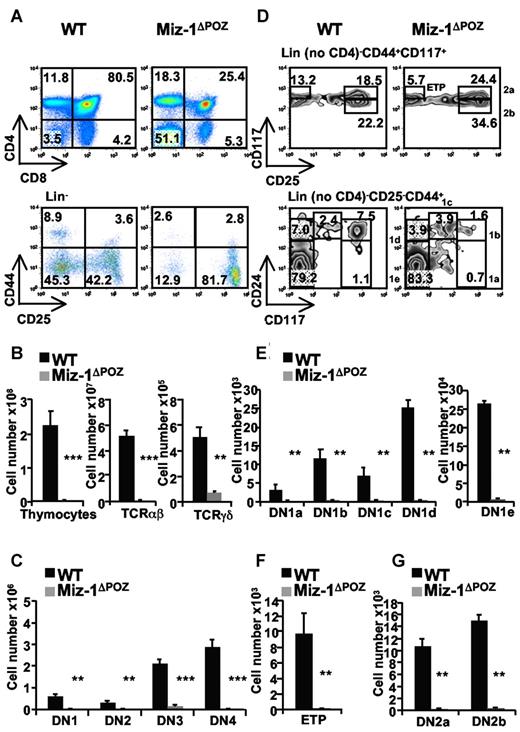
To investigate the role of Miz-1 during T-cell development, we used Vav-Cre Miz-1flox/flox mice (described in the supplemental Data, and hereafter named Miz-1ΔPOZ). We observed that Miz-1ΔPOZ mice have a block at the transition from DN to DP cells (Figure 1A), which results in a 100-fold reduced thymic cellularity compared with a wild-type (WT) littermate (Figure 1B). The deletion of the POZ domain of Miz-1 also caused a 1000-fold reduction of αβ-T cells compared with a 7-fold reduction of γδ-T cells (Figure 1B), and a significant reduction of all DN subpopulations compared with controls (Figure 1C).
The deletion of the POZ domain of Miz-1 disrupts T-cell development at the transition from DN to DP and at the ETP/DN1 stage. Flow cytometric (FACS) analysis (A,D) and total cell numbers (B,C,E-G) of thymic populations of wild-type (WT) and Miz-1ΔPOZ mice. (A) CD4 and CD8 surface staining (top panel) and lineage-negative (Lin−) cells (bottom panel), described in supplemental Methods are analyzed for the surface expression of CD44 and CD25 to assess DN1 (Lin−CD44+CD25−), DN2 (Lin−CD44+CD25+), DN3 (Lin−CD44−CD25+), and DN4 (Lin−CD44−CD25−). Numbers in quadrants indicate the percentage of cells. (B-C) Absolute numbers of thymocytes, TCR-αβ+, TCR-γδ+, and DN1 to DN4 cells are calculated relative to the live cells gated according to the FSC/SSC profile and expressed as absolute cell count. (D) FACS analysis of ETP (Lin− no CD4, CD25− CD44+CD117+), DN1a-e (Lin− no CD4, CD25−, CD44+, and CD24/CD117medium/high) and DN2a-b (Lin− no CD4, CD44+CD117+CD25medium/high) populations. (E-G) Percentages of positive cells in panel D are calculated relative to the total live cells and expressed as absolute cell count. Average counts of at least 8 mice and error bars representing the SD are shown. **P ≤ .01. ***P ≤ .001.
The deletion of the POZ domain of Miz-1 disrupts T-cell development at the transition from DN to DP and at the ETP/DN1 stage. Flow cytometric (FACS) analysis (A,D) and total cell numbers (B,C,E-G) of thymic populations of wild-type (WT) and Miz-1ΔPOZ mice. (A) CD4 and CD8 surface staining (top panel) and lineage-negative (Lin−) cells (bottom panel), described in supplemental Methods are analyzed for the surface expression of CD44 and CD25 to assess DN1 (Lin−CD44+CD25−), DN2 (Lin−CD44+CD25+), DN3 (Lin−CD44−CD25+), and DN4 (Lin−CD44−CD25−). Numbers in quadrants indicate the percentage of cells. (B-C) Absolute numbers of thymocytes, TCR-αβ+, TCR-γδ+, and DN1 to DN4 cells are calculated relative to the live cells gated according to the FSC/SSC profile and expressed as absolute cell count. (D) FACS analysis of ETP (Lin− no CD4, CD25− CD44+CD117+), DN1a-e (Lin− no CD4, CD25−, CD44+, and CD24/CD117medium/high) and DN2a-b (Lin− no CD4, CD44+CD117+CD25medium/high) populations. (E-G) Percentages of positive cells in panel D are calculated relative to the total live cells and expressed as absolute cell count. Average counts of at least 8 mice and error bars representing the SD are shown. **P ≤ .01. ***P ≤ .001.
One of the most striking phenotypes of Miz-1ΔPOZ mice was the reduction of the DN1 population. DN1a-e subsets were reduced by 70- to 130-fold; and the ETP subset (DN1a/b), which is the most affected, showed a reduction of 230-fold compared with WT (Figure 1D-F). Similarly, DN2a and DN2b cells were reduced by 100- and 40-fold (Figure 1G), respectively, whereas DN3a and DN3b cell numbers were only reduced by 3- and 10-fold, respectively, in Miz-1ΔPOZ mice (not shown), suggesting that Miz-1 has an important function in ETP/DN1 and DN2 cells.
Lack of early T-cell precursors in the thymus of Miz-1 mutant mice
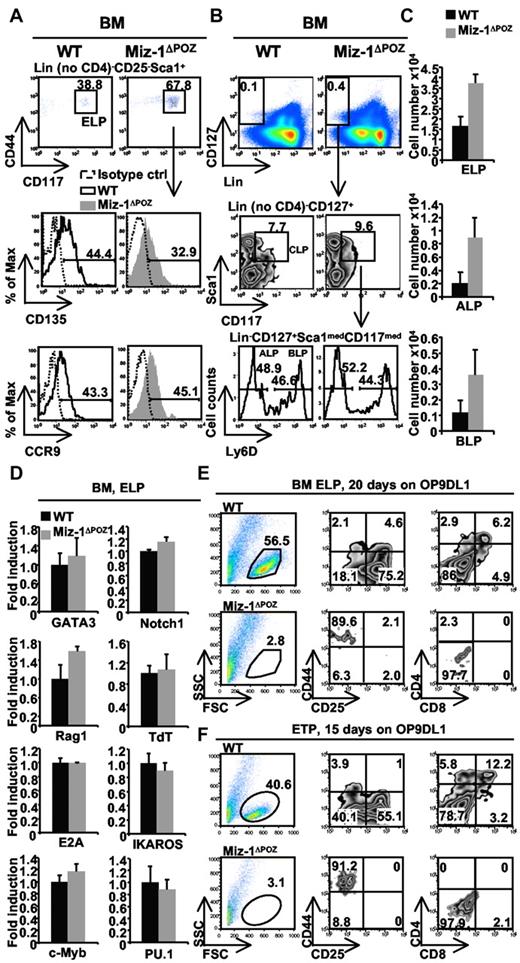
The few thymic ETPs that are present in Miz-1ΔPOZ mice were phenotypically normal according to the expression of CD117, CD44, CCR9, and CD135 (supplemental Figure 1A). Compared with ETPs, the frequencies of bone marrow ELPs or the ALP and BLP subsets of CLPs were not altered, and even present at higher frequencies compared with WT (Figure 2A-C). Miz-1ΔPOZ ELPs showed normal expression levels of CD117 and CCR9, with a small reduction in CD135 expression (Figure 2A), and ALPs and BLPs showed normal expression of Ly6D (Figure 2B). Moreover, Miz-1ΔPOZ ELPs sorted from the bone marrow expressed T-lineage specific genes, such as GATA3, Notch1, Rag1, TdT, and E2A, Ikaros, c-Myb, and PU.1 at WT levels (Figure 2D), suggesting that Miz-1 deficiency does not alter the expression program related to T-lineage specification.
Loss of Miz-1 POZ domain affects frequencies of bone marrow ELPs, ALPs, and BLPs and in vitro differentiation of ETPs and ELPs. (A) ELPs were gated on Lin− no CD4, CD25− Sca1+CD44+CD117+ (top panel) and further analyzed for CD135 and CCR9 expression (bottom panel). The plots are composed of an overlay of the CD135 or CCR9 staining in black (WT) or gray (Miz-1ΔPOZ) with the matching isotype control antibodies staining (dotted black; n = 4). (B) Bone marrow Lin−CD127+Sca1medCD117med CLPs were examined for the expression of Ly6D dividing the progenitor population into Ly6D− ALPs and Ly6D+ BLPs. (C) Percentages of positive cells in panels A and B are calculated relative to the total live cells and expressed as absolute cell count (n = 4 for ELPs and n = 2 for ALPs and BLPs). (D) Quantitative real-time PCR analysis of target genes involved in ELP development. RNA was extracted from 5000 sorted bone marrow ELPs from WT and Miz-1ΔPOZ mice. All values are presented as fold induction relative to values obtained with the respective wild-type control. Average of triplicate values and SD are shown (n = 3). FACS analysis indicating the development of sorted Lin− no CD4, CD25− Sca1+CD44+CD117+ ELPs (E) and ETPs (F) from the bone marrow and thymus of WT or Miz-1ΔPOZ mice. Fifty sorted ETPs or ELPs were cocultured on OP9DL1 stroma cells for 15 or 20 days in T-cell media. The developmental progression of the cells is evaluated by flow cytometry using T-cell markers CD44, CD25, CD4, and CD8 (n = 4).
Loss of Miz-1 POZ domain affects frequencies of bone marrow ELPs, ALPs, and BLPs and in vitro differentiation of ETPs and ELPs. (A) ELPs were gated on Lin− no CD4, CD25− Sca1+CD44+CD117+ (top panel) and further analyzed for CD135 and CCR9 expression (bottom panel). The plots are composed of an overlay of the CD135 or CCR9 staining in black (WT) or gray (Miz-1ΔPOZ) with the matching isotype control antibodies staining (dotted black; n = 4). (B) Bone marrow Lin−CD127+Sca1medCD117med CLPs were examined for the expression of Ly6D dividing the progenitor population into Ly6D− ALPs and Ly6D+ BLPs. (C) Percentages of positive cells in panels A and B are calculated relative to the total live cells and expressed as absolute cell count (n = 4 for ELPs and n = 2 for ALPs and BLPs). (D) Quantitative real-time PCR analysis of target genes involved in ELP development. RNA was extracted from 5000 sorted bone marrow ELPs from WT and Miz-1ΔPOZ mice. All values are presented as fold induction relative to values obtained with the respective wild-type control. Average of triplicate values and SD are shown (n = 3). FACS analysis indicating the development of sorted Lin− no CD4, CD25− Sca1+CD44+CD117+ ELPs (E) and ETPs (F) from the bone marrow and thymus of WT or Miz-1ΔPOZ mice. Fifty sorted ETPs or ELPs were cocultured on OP9DL1 stroma cells for 15 or 20 days in T-cell media. The developmental progression of the cells is evaluated by flow cytometry using T-cell markers CD44, CD25, CD4, and CD8 (n = 4).
Similar to ETPs, ELPs were reduced in the blood of Miz-1ΔPOZ mice compared with WT controls (supplemental Figure 1B). To evaluate whether a homing problem was responsible for the observed lack of thymic ETPs, LSK progenitor cells from WT mice were sorted and transplanted into Miz-1ΔPOZ irradiated mice. These cells successfully reconstituted the thymus of Miz-1ΔPOZ mice. Sorted Miz-1ΔPOZ LSKs transplanted into WT mice showed the same phenotype as Miz-1ΔPOZ mice, generating a hypocellular thymus (not shown). These data indicate that the effect is cell intrinsic and not caused by a defect in the thymic stroma. Furthermore, the absence of early T-cell differentiation in the thymus does not seem to be related to aberrant Notch1 signaling in Miz-1ΔPOZ mice. Indeed, the intracellular expression of Notch1 and Notch1 target genes, such as Notch1 itself and Hes1, were not reduced in Miz-1ΔPOZ cells (supplemental Figure 1C-D). In addition, no aberrant T-cell development in the bone marrow was noticeable, and no B-cell development was detected in the thymus of Miz-1ΔPOZ mice (not shown). Moreover, the expression of chemokine receptors CXCR4, CCR7, and CCR9 on DN subsets from Miz-1ΔPOZ mice was intact or even higher compared with WT cells (supplemental Figure 2). Therefore, the observed T-cell development defects are most probably cell autonomous.
Nevertheless, sorted bone marrow or blood (not shown) ELPs and thymic ETPs from Miz-1ΔPOZ mice were unable to differentiate into mature T-cell stages in the presence of IL-7 (Figure 2E-F). It is thus probable that Miz-1 affects cytokine-dependent survival or proliferation signals needed for the intrathymic differentiation of ETP/DN1 cells.
ETP and DN1 from Miz-1ΔPOZ mice do not differentiate in vitro because of increased apoptosis
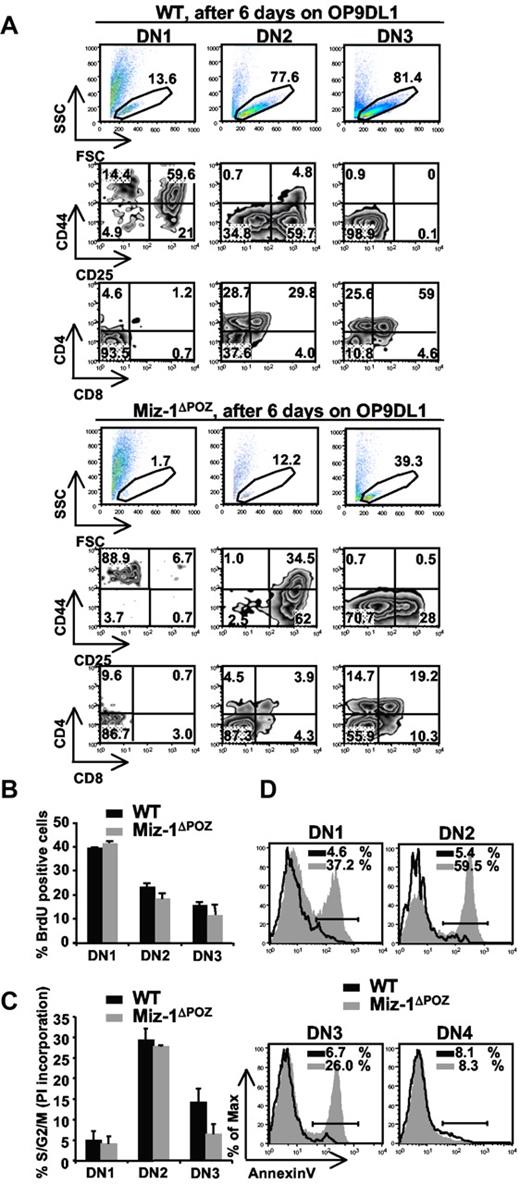
The developmental defect of ETPs and ELPs in vitro was also seen when sorted thymic DN1 cells from Miz-1ΔPOZ mice were cultured on OP9DL1. Sorted DN2 or DN3 cells from Miz-1ΔPOZ thymus survived better in vitro compared with DN1 cells but were still less efficient at generating DP cells compared with WT (Figure 3A). The development of γδ-T cells from Miz-1ΔPOZ sorted DN2 and DN3 was comparable with WT levels on OP9DL1 (not shown). This suggests that Miz-1 is important for ELP and ETP/DN1 survival and/or differentiation, but this requirement seems to decrease as cells reach the DN2 or DN3 stage. Indeed, the deletion of Miz-1 after the DN3 transition does not influence T-cell differentiation. The overall thymic cellularity (not shown) and development are normal in Lck-cre Miz-1ΔPOZ mice (supplemental Figure 3A-B).
DN1s from Miz-1ΔPOZ mice lack in vitro differentiation and/or survival signals on OP9DL1 cocultures. (A) Comparative differentiation kinetics of 500 cells sorted from DN1, DN2, and DN3 subsets after 6 days of culture on stromal OP9DL1 cells. Gated live cells (top panel) were further gated on CD4−CD8−TCR-γδ− and analyzed for CD44 and CD25 surface expression to assess DN stages of differentiation (middle panel). CD4 and CD8 surface expression shows developmental progression of more mature T cell (bottom panel). The numbers in dot plots are percentages of cells (n = 6). (B) Cell cycle analysis after in vivo BrdU labeling. Cells were stained for surface markers and anti-BrdU and gated on DN1, DN2, and DN3. Data show percentage of BrdU-positive cells (n = 2). (C) Cell cycle analysis using propidium iodide staining was performed on permeabilized, sorted DN1, DN2, and DN3 cells. Data show percentage of cells in S/G2/M phase and are representative of 4 independent experiments. (D) Single-cell suspensions of thymocytes were stained with antibodies against lineage markers, CD44 and CD25 followed by annexin V staining. Percentages of annexin V-positive cells are indicated (n = 4).
DN1s from Miz-1ΔPOZ mice lack in vitro differentiation and/or survival signals on OP9DL1 cocultures. (A) Comparative differentiation kinetics of 500 cells sorted from DN1, DN2, and DN3 subsets after 6 days of culture on stromal OP9DL1 cells. Gated live cells (top panel) were further gated on CD4−CD8−TCR-γδ− and analyzed for CD44 and CD25 surface expression to assess DN stages of differentiation (middle panel). CD4 and CD8 surface expression shows developmental progression of more mature T cell (bottom panel). The numbers in dot plots are percentages of cells (n = 6). (B) Cell cycle analysis after in vivo BrdU labeling. Cells were stained for surface markers and anti-BrdU and gated on DN1, DN2, and DN3. Data show percentage of BrdU-positive cells (n = 2). (C) Cell cycle analysis using propidium iodide staining was performed on permeabilized, sorted DN1, DN2, and DN3 cells. Data show percentage of cells in S/G2/M phase and are representative of 4 independent experiments. (D) Single-cell suspensions of thymocytes were stained with antibodies against lineage markers, CD44 and CD25 followed by annexin V staining. Percentages of annexin V-positive cells are indicated (n = 4).
To explain the decrease in DN differentiation, we sorted cells from Miz-1ΔPOZ mice and verified their expression of T-cell genes. Rag-1, Rag-2, and TdT and genes encoding for HEB, Idb2, E2A, and Egr1-3 were at WT levels, whereas Idb1, Tgfb1, Notch1, and IL-7R at levels slightly higher than WT (supplemental Figure 3C). This confirms again that Miz-1 does not regulate the expression of genes specifying the T-cell lineage. The negative cell cycle regulators CDKN2b and CDKN1a are direct Miz-1 target genes. Whereas CDKN2b could not be detected in thymocytes, the expression of CDKN1a was elevated in sorted DN1 and DN2 subsets (supplemental Figure 3D), where T-cell differentiation in Miz-1ΔPOZ mice is affected the most. Because CDKN1a regulates cell cycle, we next examined whether the thymic atrophy in Miz-1ΔPOZ mice and the absence of ETP/DN1 differentiation in vitro were the result of a reduction in cell division, cell proliferation, or an increase in apoptosis. In vivo bromodeoxyuridine (BrdU) labeling or propidium iodide staining did not show significant defects in cell cycle progression of Miz-1ΔPOZ pro-T cells (Figure 3B-C). Moreover, crossing Vav-cre Miz-1ΔPOZ mice with CDKN1a-deficient mice did not restore the DN to DP transition, confirming that the block seen in Miz-1ΔPOZ mice cannot be explained by a cell cycle defect (supplemental Figure 3E). By contrast, annexin V staining revealed increased apoptosis in DN1 (37%), DN2 (60%), and DN3 (26%) subpopulations of Miz-1ΔPOZ thymocytes compared with WT controls, but not in Miz-1ΔPOZ DN4 cells (8%; Figure 3D). These results indicated that Miz-1ΔPOZ pro-T cells exhibit an excessive cell death, particularly at the critical cytokine-dependent step (DN1 and DN2) of pro-T-cell differentiation.
Miz-1ΔPOZ pro-T cells lack an IL-7–dependent survival signal because of a deregulated Bcl-2 expression
IL-7 signaling assures survival and proliferation in DN1-3 subsets mainly by controlling the increased expression of antiapoptotic Bcl-2 and Mcl-1, and by redistributing the cell-death proteins Bax and Bad.15 In Miz-1ΔPOZ thymocytes, both CD127 (IL-7Rα) and CD132 (common γ-chain) are expressed at WT levels on all DN subsets (supplemental Figure 4). However, compared with the respective WT, mRNA level of Bax was elevated in particular in the DN2 subsets of Miz-1ΔPOZ mice, and Bcl-2 expression was either at the WT level (DN1) or increased (DN2; Figure 4A). Bcl-xL, Mcl-1, Bad, and Pim-1 expression levels were similar between WT and Miz-1ΔPOZ cells (Figure 4A). Miz-1 expression was also at comparable levels in DN1 and DN2 cells isolated from WT thymus, with a decrease in its expression in DN4 pre-T cells (supplemental Figure 5A). Miz-1 was also expressed in DP cells (supplemental Figure 5B), with a noticeable reduction in mature splenic CD3+ cells, consistent with a predominant role of Miz-1 in early pro-T-cell development. Because Miz-1 has been described to interact with c-Myc, we have evaluated c-Myc expression, which was at WT levels in all DN subsets isolated from Miz-1ΔPOZ mice (supplemental Figure 5C). Moreover, it has been shown that a mutated c-Myc allele, in which the valine residue (V) at position 394 is substituted by an aspartic acid (D), can no longer interact with Miz-1.24 We have generated homozygous c-MycV394D knockin mice (described in supplemental Data). These mice did not show any defects in thymic development or in the overall cellularity of the thymus, indicating that the phenotype we observe in Miz-1ΔPOZ mice is probably c-Myc-independent (supplemental Figure 5D).
Miz-1ΔPOZ DN thymocytes show aberrant IL-7-related gene expression profile. (A) RNA was extracted from 5 to 10 × 103 sorted DN1 and DN2 cells. All values are normalized to the expression of GAPDH gene and are presented as fold induction relative to values obtained with the respective wild-type control (set as 1-fold). Average of triplicate values ± SD are shown (n = 5). (B-C) Quantitative real-time PCR analyses of indicated genes in sorted DN1 and DN2 cells nonactivated (NA) or activated with IL-7 for 4 hours. Data are presented as fold induction relative to values obtained with the respective NA sample for wild-type control (left panel) and for Miz-1ΔPOZ (right panel). Average of triplicate values ± SD are shown (n = 3).
Miz-1ΔPOZ DN thymocytes show aberrant IL-7-related gene expression profile. (A) RNA was extracted from 5 to 10 × 103 sorted DN1 and DN2 cells. All values are normalized to the expression of GAPDH gene and are presented as fold induction relative to values obtained with the respective wild-type control (set as 1-fold). Average of triplicate values ± SD are shown (n = 5). (B-C) Quantitative real-time PCR analyses of indicated genes in sorted DN1 and DN2 cells nonactivated (NA) or activated with IL-7 for 4 hours. Data are presented as fold induction relative to values obtained with the respective NA sample for wild-type control (left panel) and for Miz-1ΔPOZ (right panel). Average of triplicate values ± SD are shown (n = 3).
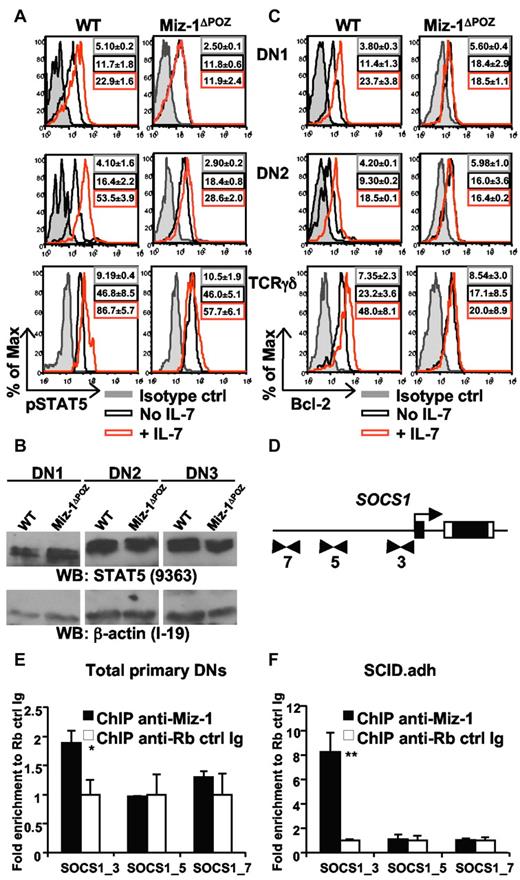
Expression levels of the Jak inhibitor SOCS1 and, to a smaller extent, SOCS3 were elevated in DN, but not in DP subsets of Miz-1ΔPOZ mice compared with controls (Figure 4A; supplemental Figure 5E). The already increased expression of SOCS1 was still further inducible by IL-7 in DN1-2 from Miz-1ΔPOZ mice, albeit at a much lower extent than in WT cells (Figure 4B-C), contributing to SOCS1 overexpression in the absence of a functional Miz-1. The induction of Bcl-2 expression by IL-7 was almost completely abrogated in all Miz-1ΔPOZ pro-T cell subsets compared with WT cells (Figure 4B-C). In contrast, Miz-1 mRNA expression was not induced by IL-7, making it doubtful that Miz-1 itself is an IL-7 effector gene (supplemental Figure 5F). These data suggest that the part of IL-7R signaling that is involved in protecting cells from apoptosis and promoting survival in early T-cell subsets is defective in Miz-1ΔPOZ cells. The elevated expression levels of SOCS1 could be the responsible factor that prevents IL-7 from initiating signal transduction in Miz-1ΔPOZ DNs.
Miz-1 controls the IL-7/IL-7R signaling pathway by regulating SOCS1
IL-7 stimulation did not activate STAT5 phosphorylation in Miz-1ΔPOZ DN1 cells and was also less efficient in DN2, DN3 (not shown), and TCR-γδ cells compared with the respective WT control cells, despite similar STAT5A/B protein expression levels (Figure 5A-B). Similarly, even if DN2 cells expressed higher Bcl-2 levels than DN1s, IL-7-mediated induction of Bcl-2 expression was completely blocked in all Miz-1ΔPOZ DN subsets at the protein level (Figure 5C). Given that Bcl-2 is a target of STAT5 and thus a downstream effector of IL-7R signaling, and that SOCS1 binds to the Jak proteins and thereby blocks IL-7R signaling, we reasoned that one mechanism that could explain the lack of signaling observed in Miz-1ΔPOZ thymocytes is the regulation of SOCS1 by Miz-1 itself.
Miz-1 is required for proper IL-7/IL-7R signaling and binds to the SOCS1 promoter. (A) Intracellular pSTAT5 detection in WT and Miz-1ΔPOZ thymocytes after ex vivo stimulation with IL-7. Histograms show isotype control (ctrl) antibodies staining in gray, and pSTAT5 antibody stainings in unstimulated (No IL-7) and stimulated with IL-7 (+ IL-7) cells. Mean fluorescence intensities ± SD are indicated; n = 4 for DN1 and DN2 and n = 3 for CD4−CD8−TCR-γδ+. (B) Total STAT5 proteins in DN1, DN2, and DN3 cells. Cells were sorted and whole protein extracts were evaluated by Western blot for STAT5 (top blot) and β-actin loading control (bottom blot; n = 2). (C) Bcl-2 detection in WT and Miz-1ΔPOZ thymocytes after ex vivo stimulation with IL-7. Mean fluorescence intensities ± SD are indicated. (D) ChIP analysis to identify Miz-1 binding to potential sites within SOCS1 promoter. Cells were rested at 37°C in phosphate-buffered saline for 1 hour, and ChIP was performed on primary DN cells (E) or on SCID.adh murine thymic lymphoma cells (F). Quantitative real-time PCR was performed using primers flanking the initiator region (SOCS1_3) or upstream (SOCS1_5 and _7) of SOCS1 promoter (indicated as arrows in panel D and described in supplemental Table 3). Data are fold enrichment of specific anti–Miz-1 ChIP over rabbit control Ig ChIP (set as 1-fold) from triplicates ± SD (n = 4). *P ≤ .05. **P ≤ .01.
Miz-1 is required for proper IL-7/IL-7R signaling and binds to the SOCS1 promoter. (A) Intracellular pSTAT5 detection in WT and Miz-1ΔPOZ thymocytes after ex vivo stimulation with IL-7. Histograms show isotype control (ctrl) antibodies staining in gray, and pSTAT5 antibody stainings in unstimulated (No IL-7) and stimulated with IL-7 (+ IL-7) cells. Mean fluorescence intensities ± SD are indicated; n = 4 for DN1 and DN2 and n = 3 for CD4−CD8−TCR-γδ+. (B) Total STAT5 proteins in DN1, DN2, and DN3 cells. Cells were sorted and whole protein extracts were evaluated by Western blot for STAT5 (top blot) and β-actin loading control (bottom blot; n = 2). (C) Bcl-2 detection in WT and Miz-1ΔPOZ thymocytes after ex vivo stimulation with IL-7. Mean fluorescence intensities ± SD are indicated. (D) ChIP analysis to identify Miz-1 binding to potential sites within SOCS1 promoter. Cells were rested at 37°C in phosphate-buffered saline for 1 hour, and ChIP was performed on primary DN cells (E) or on SCID.adh murine thymic lymphoma cells (F). Quantitative real-time PCR was performed using primers flanking the initiator region (SOCS1_3) or upstream (SOCS1_5 and _7) of SOCS1 promoter (indicated as arrows in panel D and described in supplemental Table 3). Data are fold enrichment of specific anti–Miz-1 ChIP over rabbit control Ig ChIP (set as 1-fold) from triplicates ± SD (n = 4). *P ≤ .05. **P ≤ .01.
To determine whether Miz-1 can bind to the SOCS1 promoter, we performed chromatin immunoprecipitation (ChIP) on purified primary DN cells from WT thymi. Quantitative polymerase chain reaction (PCR) analysis, using primers located around the initiator start site of SOCS1 (Figure 5D), revealed a significant 1.8-fold enrichment using anti–Miz-1 antibodies compared with control IgG (Figure 5E). DN1 and DN2 cells, where Miz-1 is actually promoting survival, represent a small percentage of the total DN purified, probably causing the little enrichment obtained. We therefore confirmed this result in SCID.adh murine thymic lymphoma clone P6D4 (Figure 5F; P ≤ .01), which highly expresses endogenous Miz-1 (supplemental Figure 6A-B) and validated the specific binding around the initiator using control primers upstream of SOCS1 where Miz-1 did not bind (Figure 5E-F). As an additional control, we did not detect any enrichment with an anti–Miz-1 ChIP using primers designed to detect binding sites near or upstream of the initiator of SOCS3 promoter (supplemental Figure 6C). These data indicate that Miz-1 is specifically binding to the initiator site of SOCS1 but not SOCS3 promoter in primary DN and P6D4 cells. To further validate the regulation of SOCS1 by Miz-1, we transduced P6D4 cells with a retroviral vector expressing Miz-1–IRES-GFP or a control empty vector (MIGR1-GFP; supplemental Figure 6D). Protein band intensities were quantified and, when GFP+ sorted cells were stimulated with IL-7, P6D4 cells overexpressing 5 times more Miz-1 were less efficient in up-regulating SOCS1 compared with control cells (supplemental Figure 6E). This experiment provided additional evidence that increased levels of Miz-1 repress SOCS1 expression.
Inhibition of SOCS1 or overexpression of Bcl-2 can restore the differentiation block of Miz-1ΔPOZ pro-T cells in vitro
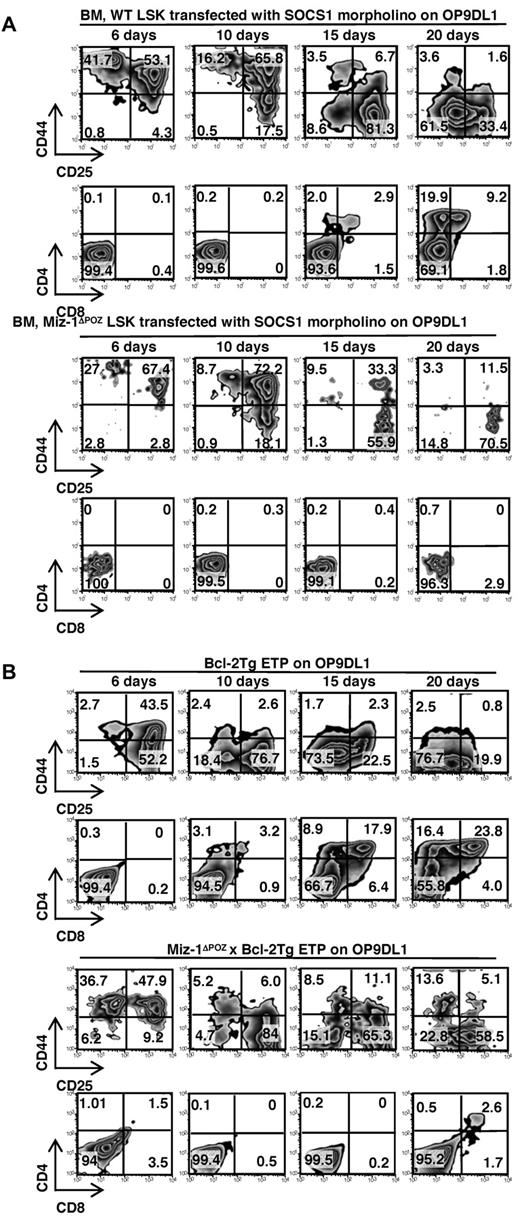
To further test our hypothesis, we treated sorted Miz-1ΔPOZ LSK or DN1 cells with either a fluorescein isothiocyanate-labeled morpholino-oligonucleotide against SOCS1 mRNA or a control morpholino and cocultured them on OP9DL1. After 6 to 20 days, Miz-1ΔPOZ LSK or DN1 cells treated with the morpholino against SOCS1 mRNA developed into DN3 cells, whereas the control morpholino-treated cells did not differentiate (Figure 6A; supplemental Figure 7A-B). The efficiency of the transfection of fluorescein isothiocyanate-labeled morpholinos was monitored by flow cytometry, which was diluted out as cells proliferated (supplemental Figure 7C). Miz-1ΔPOZ LSK or DN1 went through similar cycles of cell division after 19 days on OP9DL1 as WT cells, although their mean fluorescence intensities were slightly higher. The reason for that higher fluorescence may be attributable to fewer cells in culture, which are apoptotic. The cells did not survive past the DN3 stage and did not generate DP cells compared with WT (Figure 6A). This suggested that high SOCS1 levels might indeed be responsible for the observed block of differentiation or lack of survival seen in Miz-1 ΔPOZ ETP/DN1 cells by blocking IL-7R signaling.
Early T-cell development is restored in Miz-1ΔPOZ cells by SOCS1 knockdown or Bcl-2 overexpression in vitro. (A) A total of 1000 sorted LSK cells from WT or Miz-1ΔPOZ were incubated with morpholino against SOCS1 mRNA. Comparative in vitro differentiation kinetics of the cells was monitored after 6 to 20 days of coculture on OP9DL1. (B) A total of 500 sorted ETP cells from Bcl-2Tg or Miz-1ΔPOZ x Bcl-2Tg were analyzed by FACS after 6 to 20 days of culture. Gated live cells were further gated on CD4−CD8−TCR-γδ− and analyzed for CD44 and CD25 surface expression to assess DN stages of differentiation. CD4 and CD8 surface expression shows development progression of more mature T cells. Data are representative of 4 independent experiments for panel A and 5 experiments for panel B.
Early T-cell development is restored in Miz-1ΔPOZ cells by SOCS1 knockdown or Bcl-2 overexpression in vitro. (A) A total of 1000 sorted LSK cells from WT or Miz-1ΔPOZ were incubated with morpholino against SOCS1 mRNA. Comparative in vitro differentiation kinetics of the cells was monitored after 6 to 20 days of coculture on OP9DL1. (B) A total of 500 sorted ETP cells from Bcl-2Tg or Miz-1ΔPOZ x Bcl-2Tg were analyzed by FACS after 6 to 20 days of culture. Gated live cells were further gated on CD4−CD8−TCR-γδ− and analyzed for CD44 and CD25 surface expression to assess DN stages of differentiation. CD4 and CD8 surface expression shows development progression of more mature T cells. Data are representative of 4 independent experiments for panel A and 5 experiments for panel B.
It has previously been shown that overexpression of Bcl-2 restores defective T-cell development in mice lacking IL-7 or IL-7R.34,35 Hence, to further investigate whether Miz-1 controls the part of the IL7-R signaling that assures survival of early T-cell subsets through the induction of Bcl-2, we crossed Miz-1ΔPOZ mice with H2K-Bcl-2 transgenic mice, which express high constitutive Bcl-2 levels throughout hematopoiesis.36 In contrast to the ETPs from Miz-1ΔPOZ mice, ETPs from Miz-1ΔPOZ x Bcl-2Tg animals now survived and expanded in vitro on OP9DL1 stroma cells, differentiated into DN3-DN4 cells, and gave rise to a small number of DP cells (Figure 6B). Of note, ETPs from Miz-1ΔPOZ x Bcl-2Tg (Figure 6B), like sorted DN2 and DN3 cells from Miz-1ΔPOZ thymus (Figure 3A), survived better in vitro but were less efficient at generating DP cells compared with WT or Bcl-2Tg cells on OP9DL1. This indicates that a second differentiation block at the DN3/DN4 transition exists in Miz-1ΔPOZ thymus (Figure 1A-C) that can only partially be rescued by Bcl-2 overexpression.
Overexpression of Bcl-2 restores T-cell differentiation in Miz-1ΔPOZ mice
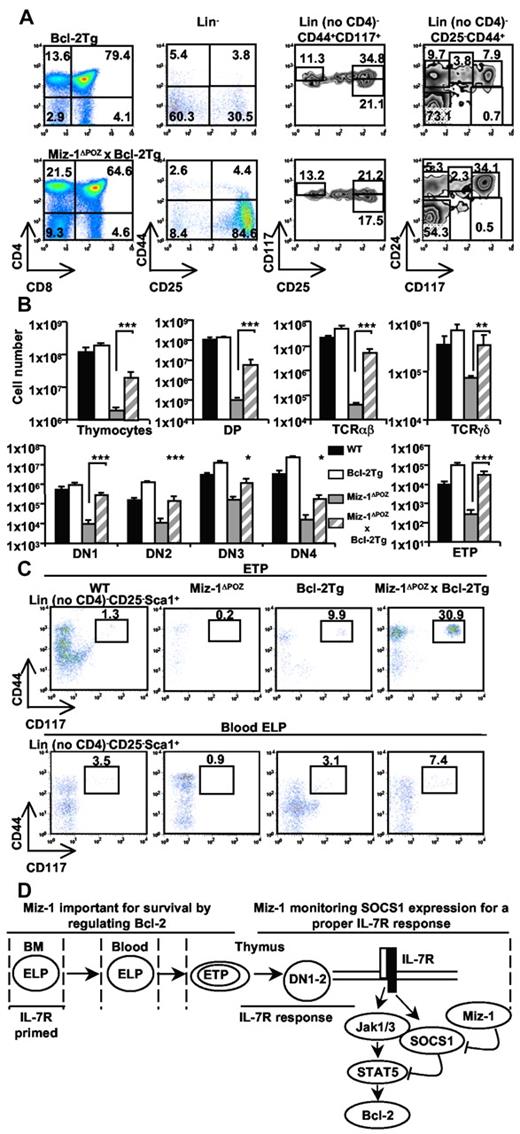
The introduction of the H2K-Bcl-2 transgene into Miz-1ΔPOZ mice reduced the DN to DP block observed in Miz-1ΔPOZ thymus, as demonstrated by CD4/CD8 fluorescence-activated cell sorter (FACS) analysis (Figure 7A), and restored the numbers of both αβ-T cells and TCR-γδ cells (Figure 7B). As αβ-T cells, DP cells were also significantly increased in Miz-1ΔPOZx Bcl-2Tg, but their cell numbers remained 20-fold lower than WT or Bcl2Tg mice, consistent with the few DP generated in vitro when Miz-1ΔPOZ x Bcl-2Tg ETPs were cocultured on OP9DL1. The second block observed at the DN3/DN4 transition in Miz-1ΔPOZ thymus was not rescued by Bcl-2 overexpression in vivo, as demonstrated by CD44/CD25 FACS analysis (Figure 7A-B). Nonetheless, the ETP and blood ELP subsets, which were almost undetectable in Miz-1–deficient mice, were completely restored in Miz-1ΔPOZ x Bcl-2Tg animals, reaching more than WT levels (Figure 7A-C). The number of cells in DN1 and DN2 subsets, as well as their subpopulations, was also completely restored (supplemental Figure 7D). Therefore, increased levels of Bcl-2 are sufficient to rescue Miz-1ΔPOZ early ETP/DN1/DN2 defects, allowing their survival and differentiation both in vitro and in vivo until the DN3 stage.
The early T-cell differentiation block in Miz-1ΔPOZ mice is overcome by Bcl-2 overexpression in vivo. FACS analysis (A,C) and total cell numbers (B) of thymic and blood lymphoid populations from Bcl-2Tg or Miz-1ΔPOZ x Bcl-2Tg mice (n = 7). (A) CD4 and CD8 surface staining is shown (left panel). CD4−CD8− DN cells gated on lineage-negative cells were further analyzed for the surface expression of CD44 and CD25 to assess the 4 DN populations (middle left panel). Within the DN populations, DN2s (Lin− no CD4, CD44+CD117+CD25medium/high), ETPs (Lin− no CD4, CD25− CD44+CD117+; middle right panel), and DN1s (Lin− no CD4, CD25−, CD44+, and CD24/CD117medium/high; right panel) were also characterized. (B) Numbers in rectangular gates or quadrants indicate the percentage of cells. Total cell numbers of the thymocyte subsets and of gated TCR-αβ+ and TCR-γδ+ cells are shown. (C) FACS analysis of Lin− no CD4, CD25− Sca1+CD44+CD117+ thymic ETPs (top panel) and peripheral blood ELPs (bottom panel; n = 5). (D) Schematic representation of Miz-1 implication throughout T-lineage progenitor and pro-T-cell development. *P ≤ .05. **P ≤ .01. ***P ≤ .001.
The early T-cell differentiation block in Miz-1ΔPOZ mice is overcome by Bcl-2 overexpression in vivo. FACS analysis (A,C) and total cell numbers (B) of thymic and blood lymphoid populations from Bcl-2Tg or Miz-1ΔPOZ x Bcl-2Tg mice (n = 7). (A) CD4 and CD8 surface staining is shown (left panel). CD4−CD8− DN cells gated on lineage-negative cells were further analyzed for the surface expression of CD44 and CD25 to assess the 4 DN populations (middle left panel). Within the DN populations, DN2s (Lin− no CD4, CD44+CD117+CD25medium/high), ETPs (Lin− no CD4, CD25− CD44+CD117+; middle right panel), and DN1s (Lin− no CD4, CD25−, CD44+, and CD24/CD117medium/high; right panel) were also characterized. (B) Numbers in rectangular gates or quadrants indicate the percentage of cells. Total cell numbers of the thymocyte subsets and of gated TCR-αβ+ and TCR-γδ+ cells are shown. (C) FACS analysis of Lin− no CD4, CD25− Sca1+CD44+CD117+ thymic ETPs (top panel) and peripheral blood ELPs (bottom panel; n = 5). (D) Schematic representation of Miz-1 implication throughout T-lineage progenitor and pro-T-cell development. *P ≤ .05. **P ≤ .01. ***P ≤ .001.
Discussion
ELPs initiate pro-T-cell differentiation in the thymus in response to the appropriate signaling pathways, such as Notch or IL-7. ELPs from the bone marrow are similar to thymic ETPs, exhibiting comparable gene expression patterns.37 Both Miz-1ΔPOZ ELPs and ETPs express normal critical markers and T-cell genes, suggesting that Miz-1 deficiency does not alter the expression program related to their T-lineage specification. Nevertheless, Miz-1ΔPOZ ELPs/ETPs and DN1s failed to differentiate into more mature pre-T cells. In this study, we present evidence that Miz-1 controls an IL-7–dependent survival step in pro-T cells, particularly at the ETP/DN1/DN2 stages, by regulating Bcl-2 induction through the control of SOCS1.
It is still not clear whether ETPs, contained in the Lin−CD117+IL-7Rα−/low fraction of DN1s, depend on IL-7R signaling for survival, but early T-cell expansion is severely reduced in IL-7−/− and IL-7R−/− mice,38,39 and more recent data showed that ETPs or their progenitors have encountered IL-7/IL-7R priming throughout their development.40 This history of IL-7R signaling makes it plausible to hypothesize that progenitor cells require IL-7 and benefit from IL-7 accessibility in the bone marrow.41 Miz-1 may be needed at or before thymic settling of T-cell progenitors to regulate Bcl-2 and help the cells benefit from IL-7 availability to survive (Figure 7D). This hypothesis would explain the lack of survival of blood ELPs and thymic ETPs in Miz-1ΔPOZ mice that can be rescued by Bcl-2 overexpression.
One outcome of IL-7R signaling is the maintenance of cell survival by promoting a positive balance of Bcl-2-family members.15 Our findings indicate that the balance between proapoptotic and antiapoptotic factors downstream of the IL-7R signaling is altered in Miz-1ΔPOZ pro-T cells. Miz-1–deficient DN2s express elevated levels of Bcl-2 mRNA but show higher levels of apoptosis. This may be the result of a selection of Miz-1ΔPOZ DN2 cells that express enough antiapoptotic Bcl-2, possibly through an IL-7-independent signal, to escape the lack of survival signals caused by Miz-1 deficiency at the ETP/DN1 stage. It is also possible that a residual and very inefficient IL-7–dependent signal could be responsible for the high Bcl-2 expression despite elevated SOCS1 expression levels because DN2 cells from Miz-1ΔPOZ still had a low level of STAT-5 phosphorylation in response to IL-7 stimulation.
The Bax/Bcl-2 ratio in Miz-1ΔPOZ DN1 and DN2 cells seems to be in favor of apoptosis rather than survival. Because Bax is a target gene of the Gfi1 transcriptional repressor and Miz-1 can recruit Gfi1 to target gene promoters, it is possible that up-regulation of Bax in Miz-1ΔPOZ cells is the result of a disruption of the Miz-1–Gfi1 complex,42,43 and future work would be required to clarify such regulation. Collectively, our data led us to conclude that Miz-1 deficiency probably interrupts the IL-7/IL-7R/STAT5/Bcl-2 axis, which assures cell survival.
Consistent with these observations, IL-7R deficiency causes a block in early T-cell differentiation that can be reversed by overexpressing Bcl-234,35 or by deleting the Bax gene.44 We could observe that transgenic overexpression of Bcl-2 restored most of the early αβ-T-cell deficiencies in Miz-1ΔPOZ mice. Previous reports proposed that Bcl-2 could be an effector gene of Miz-1.45,46 Our data show that Miz-1 deficiency inhibits Bcl-2 up-regulation on IL-7 treatment. However, a role of Miz-1 in Bcl-2 regulation in T cells is probably indirect because we did not detect binding of Miz-1 to the Bcl-2 promoter in primary sorted DN thymocytes (data not shown). It is however possible that such a direct regulation happens in T-cell progenitors that settle in the thymus because ELP survival can be rescued by Bcl-2 overexpression in Miz-1ΔPOZ mice.
In the IL-7–dependent DN1 and DN2 subsets, it is more likely that Miz-1 regulates SOCS1, which inhibits IL-7R signaling and is highly expressed in Miz-1ΔPOZ DNs. ChIP experiments suggest that Miz-1 binds to the SOCS1 promoter at the initiator site. Although additional experimental evidence would be required to definitively prove that Miz-1 directly represses SOCS1 transcription, our data show that Miz-1 overexpression inhibited SOCS1 up-regulation in response to IL-7. Moreover, Miz-1ΔPOZ LSKs and DN1 cells treated with a morpholino oligo against SOCS1 mRNA regain their ability to differentiate in vitro. Finally, high SOCS1 levels in Miz-1ΔPOZ cells correlated with an interruption of IL-7 signaling. We could also show that Miz-1 is highly expressed in WT DN thymocytes, where SOCS1 levels are normally low. Although DP thymocytes express the highest SOCS1 levels, we did not observe an inverse correlation between SOCS1 and Miz-1 expression in this subset, which is consistent with data from Lck-cre Miz-1ΔPOZ mice that argue against a role of Miz-1 in DP cells.
Because SOCS1 overexpression was only observed in Miz-1ΔPOZ DN not DP, and Miz-1 is not involved in T-cell development beyond the DN stage, our data support a model in which Miz-1 monitors SOCS1 expression levels to ensure a proper IL-7 response in ETP/DN1/DN2 cells (Figure 7D). It would have been conceivable that Miz-1 expression would be down-regulated in response to IL-7 to derepress SOCS1 transcription. However, Miz-1 expression level is not regulated after IL-7 stimulation, suggesting that Miz-1 itself is doubtful a direct IL-7–dependent effector gene.
Whether Miz-1 regulates IL-7R signaling in a c-Myc–dependent manner in T cells is an intriguing question because Miz-1 was originally identified as a c-Myc binding protein.21 It has been reported that a pre-T cell–specific deletion of c-Myc leads to a block at the transition between DN to DP cells.47,48 However, c-Myc–deficient mice do not show a strong reduction in their thymic cellularity.47 Moreover, the reported DN3 and DN4 cell frequencies in c-Myc–deficient mice are at WT levels, and the DN4 cells have high levels of cytoplasmic TCR-β chain, contrary to what we observed in Miz-1ΔPOZ mice (I.S., T.M., unpublished data, June 2010).
Moreover, the c-MycV394D knockin mice do not phenocopy Miz-1ΔPOZ mice and have a normal T-cell development. Although these data suggest a c-Myc-independent function of Miz-1, it could be argued that, in the absence of a functional Miz-1, there is an increase in c-Myc levels that do not form a complex with Miz-1, which would explain the developmental defects seen in Miz-1ΔPOZ mice. This also seems doubtful because: (1) constitutive c-Myc expression in T cells leads to T-cell lymphomas rather than to T-cell depletion49 ; and (2) higher c-Myc activity would lead to a higher proliferation of pre-T cells,50 which is not the case in Miz-1ΔPOZ mice.
In conclusion, we show that the BTB/POZ domain transcription factor Miz-1 regulates the part of IL-7R signaling that is involved in protecting cells from apoptosis and promoting differentiation of early pro-T cell subsets. Miz-1 deficiency causes a deregulation in SOCS1 expression levels, an interruption of Jak/STAT5 signaling, an unbalanced ratio of Bcl-2 to Bax, and a high rate of apoptosis. Our data indicate that Miz-1 is required for the regulation of the IL-7/IL-7R/STAT5/Bcl-2 signaling pathway in ETP/DN1/DN2 cells by monitoring the expression levels of SOCS1 in a c-Myc-independent manner. Our study not only establishes Miz-1 as a new factor in early T-lymphoid differentiation but also implies that these functions are linked to the BTB/POZ domain of Miz-1.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Ellen V. Rothenberg and Juan-Carlos Zuniga-Pflucker for providing SCID.adh and OP9DL1 cells, respectively; Alfred Singer, Ellen V. Rothenberg, Avinash Bhandoola, and Martin Pelletier for critical reading of the manuscript; Mathieu Lapointe and Rachel Bastien for technical assistance; and Eric Massicotte, Martine Dupuis, and Julie Lord for flow sorting.
This work was supported by the Université de Montréal (PhD scholarships; I.S), the Institut de recherches cliniques de Montréal (Michel F. Bélanger and Gérard Limoges fellowships; I.S.), the Deutsche Forschungsgemeinschaft (grant 435/10-4, 10-5), the Institut de recherches cliniques de Montréal, the Canadian Foundation for Innovation, the Canadian Institutes of Health Research (operating grant 84526), and a Canada Research Chair (Tier1; T.M.).
Authorship
Contribution: I.S., C.K., and T.M. designed the research and analyzed the results; I.S., C.K., and L.V. performed experiments; C.K. generated the mice; I.S. and T.M. wrote the paper; and T.M. provided funding.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tarik Möröy, Institut de recherches cliniques de Montréal, 110 Pine West Ave, Montréal, QC, H2W 1R7, Canada; e-mail: Tarik.Moroy@ircm.qc.ca.
References
Author notes
I.S. and C.K. contributed equally to this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal