Abstract
Hemophilia is a bleeding disorder with X-linked inheritance. Current prenatal diagnostic methods for hemophilia are invasive and pose a risk to the fetus. Cell-free fetal DNA analysis in maternal plasma provides a noninvasive mean of assessing fetal sex in such pregnancies. However, the disease status of male fetuses remains unknown if mutation-specific confirmatory analysis is not performed. Here we have developed a noninvasive test to diagnose whether the fetus has inherited a causative mutation for hemophilia from its mother. The strategy is based on a relative mutation dosage approach, which we have previously established for determining the mutational status of fetuses for autosomal disease mutations. In this study, the relative mutation dosage method is used to deduce whether a fetus has inherited a hemophilia mutation on chromosome X by detecting whether the concentration of the mutant or wild-type allele is overrepresented in the plasma of heterozygous women carrying male fetuses. We correctly detected fetal genotypes for hemophilia mutations in all of the 12 studied maternal plasma samples obtained from at-risk pregnancies from as early as the 11th week of gestation. This development would make the decision to undertake prenatal testing less traumatic and safer for at-risk families.
Introduction
Current prenatal diagnostic methods such as chorionic villus sampling and amniocentesis are invasive and present an approximately 1% risk of miscarriage.1-3 The discovery of cell-free fetal DNA in maternal plasma has offered new opportunities for noninvasive prenatal diagnosis.4,5 Several promising clinical applications have been developed on the basis of the detection of paternally inherited genetic traits in maternal plasma. For example, the noninvasive detection of fetal sex and RHD status are useful for the clinical management of sex-linked diseases and RhD incompatibility.6,7 For monogenic diseases such as achondroplasia and β-thalassemia, the detection of the presence or absence of paternally inherited mutations in maternal plasma would allow one to diagnose autosomal-dominant diseases or exclude autosomal-recessive diseases of the fetuses, respectively.8-10
Despite the rapid development of the field, it has remained difficult to detect fetal alleles that are inherited from mothers who are carriers for the mutations. The difficulty is caused by the coexistence of fetal and maternal DNA in maternal plasma, and the maternally inherited fetal allele is indistinguishable from the background maternal DNA.5 To overcome this problem, we reasoned that the contribution of cell-free fetal DNA to the maternal plasma DNA pool would lead to an increase in total DNA concentration for the allele inherited by the fetus. Hence, the fetal genotype could be deduced by detecting whether there is an overrepresentation of the amount of mutant or wild-type allele DNA in maternal plasma. However, the feasibility of this approach is constrained by the low fractional circulating fetal DNA concentration, which typically constitutes only 10%-20% of total DNA in maternal plasma.11 As a result, the small degree of allelic overrepresentation would be difficult to differentiate by conventional technologies.
Digital polymerase chain reaction (PCR)12 offers a highly sensitive and precise method for quantifying cell-free DNA in maternal plasma.11 We have developed digital PCR-based methods for measuring allelic imbalance in maternal plasma for the prenatal detection of trisomy 21 and autosomal-monogenic diseases. The strategies for analyzing allelic imbalances for the 2 disease classes are different. For chromosome dosage detection, an overrepresentation of chromosome 21 DNA sequences with reference to unaffected autosomal sequences would indicate a trisomy 21 fetus.13 For autosomal diseases such as β-thalassemia, we have developed the digital relative mutation dosage (RMD) approach to determine whether there is a dosage imbalance between mutant and wild-type alleles in the plasma of heterozygous pregnant women. An allelic balance would be expected for a heterozygous fetus, whereas an allelic imbalance would be observed for a homozygous fetus. The overrepresented allele would be the one inherited by the fetus.14
In this study, we further explored the application of the RMD approach in prenatal diagnosis of X-linked disorders by using hemophilia as a disease model. Because female fetuses are affected as carriers, except in rare scenarios such as skewed X-inactivation,15 we focused on at-risk women who are carriers for the mutations and are carrying male fetuses. The principal difference between the RMD analyses for autosomal diseases and X-linked diseases is that for the former there are 3 possible fetal genotypes (ie, homozygous normal, homozygous mutant, and heterozygous), whereas for the latter there are only 2 possible fetal genotypes. In the context of X-linked diseases, a male fetus possesses only one chromosome X, and thus it would be of either mutant or wild-type genotype. The lower complexity of RMD for X-linked disorders would likely translate to a greater diagnostic accuracy. In this study, we specifically adapted the digital RMD methodology for use in hemophilia, a recessive X-linked disorder.
Hemophilias A and B are caused by heterogeneous mutations in the genes on chromosome X that encode for the coagulation factor VIII (F8)16 and coagulation factor IX (F9),17 respectively. There is a 25% chance for a pregnant hemophilia carrier to have an affected male fetus in each pregnancy. Prenatal diagnosis is an important aspect of reproductive choices for women in families with hemophilia.18 In addition, it is also beneficial for appropriate obstetric management during labor and delivery because prolonged labor, invasive monitoring techniques, and instrumental deliveries should be avoided in affected fetuses to minimize potential fetal and neonatal hemorrhagic complications.18 Therefore, the development of a noninvasive prenatal diagnostic approach for hemophilia is beneficial to both obstetricians and hemophilia families.
Methods
Subjects
Seven women who were carriers of hemophilia (3 carriers of hemophilia A, 4 carriers of hemophilia B) and pregnant with male fetuses were recruited from the Royal Free Hospital, London, United Kingdom. We also recruited 20 pregnant women (noncarriers of hemophilia), each pregnant with a singleton healthy male fetus. Ten were recruited from the Royal Free Hospital, London, United Kingdom, and the other 10 were recruited from the Prince of Wales Hospital, Hong Kong. Clinical information of the cases is shown in Table 1.19 All women were recruited with informed consent in accordance with the Declaration of Helsinki. Ethical approvals were granted by the respective institutional boards of all participating institutions. Ten milliliters of peripheral blood samples were collected into ethylenediaminetetraacetic acid tubes from each pregnant woman. For 5 of the pregnant hemophilia carriers, peripheral blood samples were taken on 2 occasions during their pregnancies (Table 1). None of the pregnant hemophilia carriers in this study had invasive prenatal testing. Fetal sex and hemophilia status were confirmed after delivery. For the 10 unaffected pregnant women recruited in Hong Kong, placental tissues also were collected after deliveries.
Clinical information for the 7 pregnant women who are carriers of hemophilia mutations
| Sample (gestation, wk) . | Affected gene . | Mutation* . | Severity of hemophilia . | Fetal hemophilia status . |
|---|---|---|---|---|
| H9 (36) | F9 | c.874delC (p.Gln292Lysfs) | Severe | Affected |
| H12† (a, 18; b, 34) | F8 | c.6278A>G (p.Asp2093Gly) | Mild | Affected |
| H15† (a, 34; b, 38) | F9 | c.1144T>C (p.Cys382Arg) | Severe | Affected |
| H17 (28) | F8 | c.826G>A (p.Val276Met) | Mild | Unaffected |
| H25† (a, 23; b, 32) | F9 | c.802T>A (p.Cys268Ser) | Moderate/severe | Affected |
| H26† (a, 11; b, 23) | F8 | c.1171C>T (p.Arg391Cys) | Severe | Unaffected |
| H30† (a, 32; b, 40) | F9 | c.1069G>A (p.Gly357Arg) | Moderate/severe | Unaffected |
| Sample (gestation, wk) . | Affected gene . | Mutation* . | Severity of hemophilia . | Fetal hemophilia status . |
|---|---|---|---|---|
| H9 (36) | F9 | c.874delC (p.Gln292Lysfs) | Severe | Affected |
| H12† (a, 18; b, 34) | F8 | c.6278A>G (p.Asp2093Gly) | Mild | Affected |
| H15† (a, 34; b, 38) | F9 | c.1144T>C (p.Cys382Arg) | Severe | Affected |
| H17 (28) | F8 | c.826G>A (p.Val276Met) | Mild | Unaffected |
| H25† (a, 23; b, 32) | F9 | c.802T>A (p.Cys268Ser) | Moderate/severe | Affected |
| H26† (a, 11; b, 23) | F8 | c.1171C>T (p.Arg391Cys) | Severe | Unaffected |
| H30† (a, 32; b, 40) | F9 | c.1069G>A (p.Gly357Arg) | Moderate/severe | Unaffected |
All fetuses were male.
Mutation nomenclature is based on the guidelines of the Human Genome Variation Society.19 The reference sequences for hemophilia A and B mutations are F8 mRNA variant 1 (Genbank accession NM_000132.3) and F9 mRNA (NM_000133.3), respectively. Nucleotide position +1 corresponds to the A nucleotide of the ATG translation initiation codon. Amino acid changes deduced from the DNA mutations are shown in the brackets. The reference sequences correspond to coagulation factor VIII isoform a precursor (NP_000123.1) and coagulation factor IX preproprotein (NP_000124.1), respectively. The translation initiator methionine is numbered as position +1.
Peripheral blood samples were taken on 2 occasions (a and b) from the same women during their pregnancies.
Sample processing
We centrifuged the blood samples at 1600g for 10 minutes at 4°C. The plasma portion was recentrifuged at 16 000g for 10 minutes at 4°C. Maternal plasma and buffy coat samples were stored at −20°C until further processing. All samples collected in the United Kingdom were processed and stored frozen locally and were shipped on dry ice to Hong Kong. We extracted DNA from maternal plasma with the QIAamp DSP DNA Blood Mini Kit (QIAGEN) following the manufacturer's instructions. Buffy coat DNA was extracted by use of the Illustra DNA Extraction Kit (GE Healthcare) following the manufacturer's protocol.
Genotyping of rs6528633 single nucleotide polymorphism and hemophilia mutations
For the rs6528633 single nucleotide polymorphism (SNP) on chromosome X, the fetal and maternal genotypes were determined by the use of DNA obtained from the placental and maternal buffy coat samples, respectively. Genotyping was performed by the use of MassARRAY homogenous MassEXTEND assays (Sequenom) as previously described.20 Genomic DNA obtained from the peripheral blood samples of the pregnant hemophilia carriers was used for hemophilia mutation detection. PCRs were performed for all exons covering coding regions, intron/exon boundaries, promoter, and 3′ UTR. Cycle sequencing was performed by the use of Big Dye Terminators V1.1 (Applied Biosystems) and analyzed on an Applied Biosystems 3100 Avant Genetic Analyser.
Digital RMD reactions for maternal plasma analyses
The experimental workflow of digital RMD is illustrated in Figure 1. We measured the fractional fetal DNA concentrations in the maternal plasma samples by using the previously described digital ZFY/X assay, which quantified the homologous ZFY and ZFX gene loci located on chromosomes Y and X, respectively.11,14 For the rs6528633 SNP, a real-time PCR assay with 2 allele-specific TaqMan probes (Applied Biosystems) was designed to distinguish the 2 SNP alleles. For the mutations of the pregnant cases at risk for hemophilia, a real-time PCR assay for allelic discrimination was designed for each mutation. Each assay contained 2 allele-specific TaqMan probes for the mutant and the wild-type alleles. The primer and probe sequences are listed in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
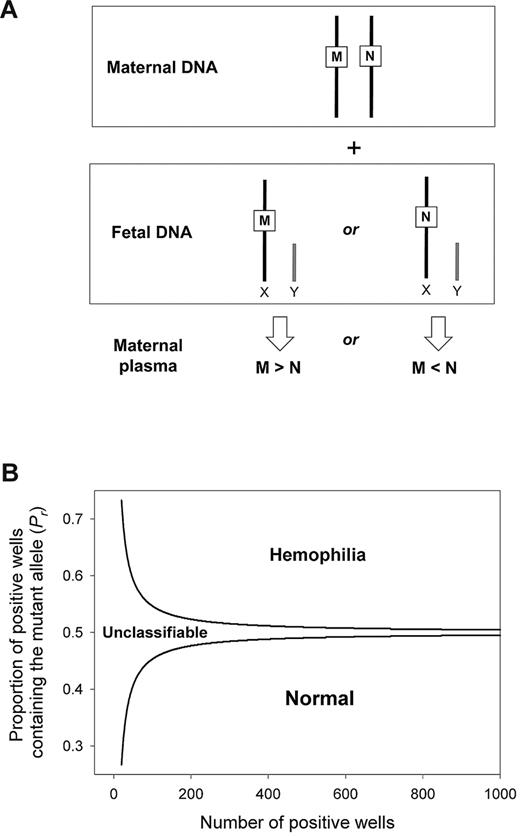
Illustration of the experimental steps for digital RMD analysis. For each maternal plasma DNA sample, both the mutant DNA proportion (Pr) and the fractional fetal DNA concentration are determined by digital PCRs. (A) Pr is determined by the use of the real-time PCR assay targeting the mutation carried by the mother, whereas fetal DNA proportion is determined by the use of the real-time PCR assay for the homologous ZFY and ZFX gene region. (B) Digital PCR is carried out on a microfluidics Digital Array (Fluidigm), which consists of 12 panels with each panel further partitioned into 765 reaction chambers. Each DNA sample is analyzed with 6 panels (ie, 765 × 6 = 4590 chambers). The PCR mixture is first manually added into the sample inlet of each panel. The mixture is next aliquoted into 765 chambers in each panel automatically by an Integrated Microfluidics Circuit Controller (Fluidigm). Each chamber contains a final reaction volume of 6 nL. The cell-free DNA concentration in maternal plasma is typically very low such that there is less than one template molecule per chamber on average. Hence, the distribution of template molecules to the chambers follows the Poisson distribution. (C) Real-time PCR is performed on the BioMark System (Fluidigm). Most of the chambers contain zero or one template DNA molecule, and the molecules are amplified and detected individually. (D) After PCR, the concentrations of mutant and wild-type DNA, as well as ZFY and ZFX DNA, are calculated. The number of chambers showing positive amplifications for the corresponding allele is counted. The concentration is then Poisson-corrected using the equation [ − ln((N − P)/N)]*N, where N is the total number of reaction chambers analyzed, P is the number of chambers positive for the allele, and ln is the natural logarithm. (E) Pr is calculated from the Poisson-corrected mutant and wild-type DNA concentrations. Fractional fetal DNA concentration is calculated from the Poisson-corrected ZFY and ZFX DNA concentrations. (F) SPRT is used to compare the experimental Pr with the expected Pr at the observed fractional fetal DNA concentration and average reference template concentration (mr). SPRT classification will be made if the mutant allele is over- or underrepresented when compared to the wild-type allele, which implies an affected or nonaffected fetus, respectively.
Illustration of the experimental steps for digital RMD analysis. For each maternal plasma DNA sample, both the mutant DNA proportion (Pr) and the fractional fetal DNA concentration are determined by digital PCRs. (A) Pr is determined by the use of the real-time PCR assay targeting the mutation carried by the mother, whereas fetal DNA proportion is determined by the use of the real-time PCR assay for the homologous ZFY and ZFX gene region. (B) Digital PCR is carried out on a microfluidics Digital Array (Fluidigm), which consists of 12 panels with each panel further partitioned into 765 reaction chambers. Each DNA sample is analyzed with 6 panels (ie, 765 × 6 = 4590 chambers). The PCR mixture is first manually added into the sample inlet of each panel. The mixture is next aliquoted into 765 chambers in each panel automatically by an Integrated Microfluidics Circuit Controller (Fluidigm). Each chamber contains a final reaction volume of 6 nL. The cell-free DNA concentration in maternal plasma is typically very low such that there is less than one template molecule per chamber on average. Hence, the distribution of template molecules to the chambers follows the Poisson distribution. (C) Real-time PCR is performed on the BioMark System (Fluidigm). Most of the chambers contain zero or one template DNA molecule, and the molecules are amplified and detected individually. (D) After PCR, the concentrations of mutant and wild-type DNA, as well as ZFY and ZFX DNA, are calculated. The number of chambers showing positive amplifications for the corresponding allele is counted. The concentration is then Poisson-corrected using the equation [ − ln((N − P)/N)]*N, where N is the total number of reaction chambers analyzed, P is the number of chambers positive for the allele, and ln is the natural logarithm. (E) Pr is calculated from the Poisson-corrected mutant and wild-type DNA concentrations. Fractional fetal DNA concentration is calculated from the Poisson-corrected ZFY and ZFX DNA concentrations. (F) SPRT is used to compare the experimental Pr with the expected Pr at the observed fractional fetal DNA concentration and average reference template concentration (mr). SPRT classification will be made if the mutant allele is over- or underrepresented when compared to the wild-type allele, which implies an affected or nonaffected fetus, respectively.
We performed digital PCR analyses on the BioMark System (Fluidigm) by using the 12.765 Digital Arrays (Fluidigm).11 Six of the 12 panels on the Digital Array were used for each DNA sample, which corresponded to 4590 individual PCRs. The reaction for one sample (6 panels) was set up with the 2X TaqMan Universal PCR Master Mix (Applied Biosystems) in a reaction volume of 52 μL. The reactions were set up according to the manufacturer's protocol; the primer and probe compositions are listed in supplemental Table 1. Each reaction mix contained 18.2 μL of the DNA sample. The reaction mixture was automatically loaded onto the Digital Array by the NanoFlex IFC Controller (Fluidigm). The reactions were performed on the BioMark System (Fluidigm). The reactions were initiated at 50°C for 2 minutes, followed by 95°C for 10 minutes, and 45 cycles of 95°C for 15 seconds, and assay-specific annealing temperatures (supplemental Table 1) for 1 minute. For a sample that remained unclassified by the RMD with data from one 4590-well digital PCR set, additional 4590-well digital PCR sets were performed until a genotype call could be made.
Digital PCR data interpretation
We have previously established a strategy for interpreting quantitative data from digital PCR analysis of circulating nucleic acids.13,14 For any mutation on chromosome X, there is always an allelic imbalance between the concentrations of the mutant and the wild-type alleles in the plasma of heterozygous women carrying male fetuses. The overrepresented allele is the one inherited by the fetus (Figure 2A). The calculation of the degree of allelic imbalance is illustrated in supplemental Table 2. We further calculated the ratio of the number of mutant alleles to the total number of mutant and wild-type alleles, denoted by Pr, present in the plasma samples. The Pr is dependent on the fractional fetal DNA concentration in the samples. The theoretically expected Pr for an affected fetus is 1/(2 − x) and is (1 − x)/(2 − x) for a nonaffected fetus, where x is the fractional fetal DNA concentration. We calculated the fractional fetal DNA concentration as previously described.11 In brief, we counted the number of wells that showed positive signals for the ZFY or the ZFX gene. Because there was less than one template molecule per reaction well, the actual number of template molecules distributed to each reaction well followed the Poisson distribution. Hence, the template concentrations were Poisson-corrected with the equation [− ln((N − P)/N)]*N, where N is the total number of digital PCR wells analyzed, P is the number of wells positive for the allele, and ln is the natural logarithm. The fractional fetal DNA concentration was then calculated using the equation [(2Y)/(X + Y)] × 100%, where Y is the Poisson-corrected concentration for the ZFY gene, and X is the Poisson-corrected concentration for the ZFX gene.
Fetal genotyping for rs6528633 in maternal plasma by digital RMD
| Sample . | Gestation, wks . | SNP genotype* . | Digital PCR result . | Fetal %‡ . | m§ . | SPRT classification¶ . | |||
|---|---|---|---|---|---|---|---|---|---|
| Mother . | Fetus . | Total wells . | A count† . | T count† . | |||||
| M5193P | 173/7 | AT | A | 4590 | 410 | 335 | 7.5 | 0.09 | A |
| M5280P | 174/7 | AT | A | 4590 | 204 | 144 | 10.7 | 0.05 | A |
| M5269P | 192/7 | AT | T | 4590 | 337 | 409 | 11.2 | 0.08 | T |
| M5297P | 193/7 | AT | A | 13 770‖ | 1482 | 1368 | 5.4 | 0.11 | A |
| M5244P | 226/7 | AT | T | 4590 | 139 | 163 | 8.5 | 0.03 | T |
| M5240P | 382/7 | AT | T | 4590 | 396 | 484 | 23.7 | 0.09 | T |
| M5241P | 382/7 | AT | T | 4590 | 706 | 920 | 13.9 | 0.17 | T |
| M4817P | 384/7 | AT | A | 4590 | 502 | 425 | 17.2 | 0.12 | A |
| M4847P | 393/7 | AT | A | 4590 | 379 | 312 | 14.9 | 0.09 | A |
| M4846P | 40 | AT | A | 4590 | 700 | 516 | 17.5 | 0.17 | A |
| Sample . | Gestation, wks . | SNP genotype* . | Digital PCR result . | Fetal %‡ . | m§ . | SPRT classification¶ . | |||
|---|---|---|---|---|---|---|---|---|---|
| Mother . | Fetus . | Total wells . | A count† . | T count† . | |||||
| M5193P | 173/7 | AT | A | 4590 | 410 | 335 | 7.5 | 0.09 | A |
| M5280P | 174/7 | AT | A | 4590 | 204 | 144 | 10.7 | 0.05 | A |
| M5269P | 192/7 | AT | T | 4590 | 337 | 409 | 11.2 | 0.08 | T |
| M5297P | 193/7 | AT | A | 13 770‖ | 1482 | 1368 | 5.4 | 0.11 | A |
| M5244P | 226/7 | AT | T | 4590 | 139 | 163 | 8.5 | 0.03 | T |
| M5240P | 382/7 | AT | T | 4590 | 396 | 484 | 23.7 | 0.09 | T |
| M5241P | 382/7 | AT | T | 4590 | 706 | 920 | 13.9 | 0.17 | T |
| M4817P | 384/7 | AT | A | 4590 | 502 | 425 | 17.2 | 0.12 | A |
| M4847P | 393/7 | AT | A | 4590 | 379 | 312 | 14.9 | 0.09 | A |
| M4846P | 40 | AT | A | 4590 | 700 | 516 | 17.5 | 0.17 | A |
PCR indicates polymerase chain reaction; RMD, relative mutation dosage; SNP, single-nucleotide polymorphism; and SPRT, sequential probability ratio test.
SNP genotypes were determined by mass spectrometry.
A count indicates number of wells positive for the A allele; and T count, number of wells positive for the T allele.
Fetal DNA proportions were determined by the digital ZFY/X assay.
Average reference template concentration per PCR well. The reference template referred to the allele with the lesser count in each sample.
Classification of fetal genotypes by SPRT.
M5297P was unclassifiable by SPRT with data from the first 4590-well digital PCR set. Two additional 4590-well digital PCR sets were performed after which classification could be made.
We used the sequential probability ratio test (SPRT) to determine whether the dosage imbalance between the mutant and the wild-type alleles in maternal plasma was statistically significant.13,14 SPRT simultaneously tested 2 alternative hypotheses in the case of RMD for chromosome X mutations. The 2 hypotheses were (1) the mutant allele was over-represented when compared to the wild-type allele, and (2) the mutant allele was underrepresented when compared to the wild-type allele. SPRT was performed by constructing a pair of curves, which defined the probabilistic boundaries for accepting or rejecting the hypotheses (Figure 2B).21,22 The SPRT curves delineated the required Pr (y-axis) for a given total number of positive reactions (x-axis) for classifying a fetal genotype. Hypothesis (1) or (2) was accepted if the experimental Pr was above the upper boundary or below the lower boundary, respectively. The equations for calculating the SPRT boundaries were adapted from El Karoui et al,23 with the level of statistical confidence adjusted to a threshold likelihood ratio of 8.21,22 The cutoff Pr values of the SPRT curves were sample-specific. The cutoff Pr values were dependent on the fractional fetal DNA concentration as described above, as well as the average reference template concentration per PCR well (mr).13,14 The reference template referred to the allele that showed the lesser positive amplification counts in the sample.
Results
Principle of digital RMD for X-linked polymorphisms
We used digital PCR to measure the concentration difference between the total amount (maternal- plus fetal-derived) of mutant and wild-type alleles in the plasma of heterozygous pregnant women carrying male fetuses. Because a male fetus possesses a single chromosome X, the relative concentration between the wild-type and the mutant allele is always in dosage imbalance (Figure 2A). An overrepresentation or underrepresentation of the mutant allele represents an affected or nonaffected fetus, respectively. We used SPRT to test for dosage imbalance. A pair of SPRT curves was constructed (Figure 2B). Samples with data points above the upper curve or below the lower curve were classified as affected or nonaffected, respectively. Samples with data points in between the 2 curves were not classified because of insufficient statistical power, and additional digital PCRs were performed.
Digital RMD for noninvasive detection of X-linked diseases in maternal plasma. (A) Digital RMD for X-linked diseases. The male fetus inherits either the mutant allele (M) or the wild-type allele (N) from his mother, leading to an overrepresentation of the M or the N allele, respectively, in maternal plasma. (B) SPRT for hemophilia detection. Samples with mutant allele proportion (Pr) above the upper boundary and below the lower boundary are classified as mutant and wild-type, respectively. Samples with Pr between 2 curves are unclassifiable and require additional digital PCR analysis.
Digital RMD for noninvasive detection of X-linked diseases in maternal plasma. (A) Digital RMD for X-linked diseases. The male fetus inherits either the mutant allele (M) or the wild-type allele (N) from his mother, leading to an overrepresentation of the M or the N allele, respectively, in maternal plasma. (B) SPRT for hemophilia detection. Samples with mutant allele proportion (Pr) above the upper boundary and below the lower boundary are classified as mutant and wild-type, respectively. Samples with Pr between 2 curves are unclassifiable and require additional digital PCR analysis.
Noninvasive determination of the fetal genotype for a SNP on chromosome X
We used a SNP, rs6528633 (A/T polymorphism), on chromosome X as a model to assess the practical feasibility of the RMD approach for determining the fetal genotype of a locus on chromosome X. The current RMD analysis is relevant to at-risk pregnant cases, ie, pregnant women who are heterozygous for mutations on chromosome X and are carrying male fetuses. Hence, we studied the plasma samples from 10 pregnant women who were heterozygous for the SNP on chromosome X and were carrying male fetuses. We developed an allele-discriminative digital real-time PCR assay to measure the concentrations of the A- and T-allele in each sample. We further measured the fractional fetal DNA concentrations with the ZFY/X assay. The digital RMD result is shown in Table 2. For all of the cases, the fetal SNP genotypes were concordant with the SPRT classification. The fractional fetal DNA concentrations (fetal % in Table 2) ranged from 5% to 24%. Hence, the result confirmed the feasibility of the digital RMD strategy.
Digital RMD for hemophilia mutation detection in DNA mixtures
We next applied the digital RMD approach for hemophilia mutation detection. We developed 7 duplex digital real-time PCR assays to detect 3 mutations in the F8 gene, 4 mutations in the F9 gene, and their corresponding wild-type counterparts. We evaluated the performance of the digital PCR assays by constructing artificial DNA mixtures that simulated the composition of maternal plasma samples with a minority male fetal DNA component among a majority maternal DNA background. We mixed 10% or 20% of placental DNA obtained from a nonaffected male fetus with blood cell DNA obtained from women heterozygous for the corresponding mutations. As shown in Table 3, the genotypes of the placental DNA, which mimicked the fetal DNA in maternal plasma, were correctly detected in all of the DNA mixtures by digital RMD analysis.
Validation of digital RMD assays with artificial DNA mixtures
| Mutation . | Fetal DNA proportion, %* . | Digital PCR result . | mr‡ . | SPRT classification§ . | ||
|---|---|---|---|---|---|---|
| Total wells . | Mutant count† . | Wild-type count† . | ||||
| F8 | 20 | 4590 | 1495 | 1721 | 0.47 | Wild-type |
| c.826G>A | 10 | 4590 | 2917 | 3006 | 1.06 | Wild-type |
| F8 | 20 | 4590 | 406 | 507 | 0.12 | Wild-type |
| c.1171C>T | 10 | 4590 | 435 | 483 | 0.11 | Wild-type |
| F8 | 20 | 4590 | 485 | 593 | 0.14 | Wild-type |
| c.6278A>G | 10 | 4590 | 857 | 892 | 0.22 | Wild-type |
| F9 | 20 | 4590 | 88 | 109 | 0.02 | Wild-type |
| c.802T>A | 10 | 4590 | 1640 | 1740 | 0.48 | Wild-type |
| F9 | 20 | 4590 | 375 | 463 | 0.11 | Wild-type |
| c.874delC | 10 | 4590 | 388 | 438 | 0.10 | Wild-type |
| F9 | 20 | 4590 | 308 | 365 | 0.05 | Wild-type |
| c.1144T>C | 10 | 4590 | 380 | 408 | 0.04 | Wild-type |
| F9 | 20 | 4590 | 673 | 835 | 0.20 | Wild-type |
| c.1069G>A | 10 | 4590 | 966 | 1046 | 0.26 | Wild-type |
| Mutation . | Fetal DNA proportion, %* . | Digital PCR result . | mr‡ . | SPRT classification§ . | ||
|---|---|---|---|---|---|---|
| Total wells . | Mutant count† . | Wild-type count† . | ||||
| F8 | 20 | 4590 | 1495 | 1721 | 0.47 | Wild-type |
| c.826G>A | 10 | 4590 | 2917 | 3006 | 1.06 | Wild-type |
| F8 | 20 | 4590 | 406 | 507 | 0.12 | Wild-type |
| c.1171C>T | 10 | 4590 | 435 | 483 | 0.11 | Wild-type |
| F8 | 20 | 4590 | 485 | 593 | 0.14 | Wild-type |
| c.6278A>G | 10 | 4590 | 857 | 892 | 0.22 | Wild-type |
| F9 | 20 | 4590 | 88 | 109 | 0.02 | Wild-type |
| c.802T>A | 10 | 4590 | 1640 | 1740 | 0.48 | Wild-type |
| F9 | 20 | 4590 | 375 | 463 | 0.11 | Wild-type |
| c.874delC | 10 | 4590 | 388 | 438 | 0.10 | Wild-type |
| F9 | 20 | 4590 | 308 | 365 | 0.05 | Wild-type |
| c.1144T>C | 10 | 4590 | 380 | 408 | 0.04 | Wild-type |
| F9 | 20 | 4590 | 673 | 835 | 0.20 | Wild-type |
| c.1069G>A | 10 | 4590 | 966 | 1046 | 0.26 | Wild-type |
The artificial mixtures were constructed to simulate the fetal and maternal DNA compositions in maternal plasma. Sequential probability ratio test (SPRT) classification of “fetal genotypes,” which was mimicked by the normal placental DNA in the artificial mixtures, was wild-type. SNP genotypes were determined by mass spectrometry.
PCR indicates polymerase chain reaction; and RMD, relative mutation dosage.
Fetal DNA was mixed in the specified proportions with maternal DNA. Fetal DNA was obtained from the placenta of a normal male fetus. Maternal DNA was obtained from the blood cells of pregnant women heterozygous for the corresponding mutations.
Mutant count indicates the number of wells positive for the mutant allele; and wild-type count, the number of wells positive for the wild-type allele.
Average reference template concentration per PCR well. The reference template referred to the allele with the lesser count in each sample.
SPRT classification of “fetal genotypes”, which was mimicked by the normal placental DNA in the artificial mixtures.
Detection of fetal hemophilia mutations in maternal plasma
We tested the digital RMD method for detecting fetal genotypes for the hemophilia mutations through maternal plasma DNA analysis. We performed digital PCR on 12 plasma samples obtained from 7 pregnant women heterozygous for the causative mutations (Table 1). All of the cases involved male fetuses. We also measured the fractional fetal DNA concentrations in the maternal plasma samples by the ZFY/X assay. The digital RMD results are shown in Table 4. The fetal genotypes were correctly classified in all studied cases by the SPRT algorithm (supplemental Figure 1). For 3 of the cases (H26a, H25a, and H12a), the fetal DNA proportions were less than 10%. Hence, the degree of quantitative difference between the amount of mutant and the wild-type alleles was too small to be classified with data from one 4590-well digital PCR set. Additional 4590-well digital PCR sets were therefore performed until classifications could be made.
Noninvasive detection of fetal hemophilia mutations in maternal plasma by digital RMD
| Plasma sample . | Fetal hemophilia status . | Digital PCR result . | Fetal %† . | mr‡ . | SPRT classification§ . | ||
|---|---|---|---|---|---|---|---|
| Total wells . | Mutant count* . | Wild-type count* . | |||||
| H9 | Affected | 4590 | 1022 | 801 | 14.8 | 0.19 | Mutant |
| H26a | Unaffected | 9180 | 1590 | 1710 | 3.8 | 0.21 | Wild-type |
| H26b | Unaffected | 4590 | 590 | 650 | 6.8 | 0.15 | Wild-type |
| H15a | Affected | 4590 | 573 | 435 | 10.5 | 0.10 | Mutant |
| H15b | Affected | 4590 | 2506 | 1956 | 10.7 | 0.56 | Mutant |
| H17 | Unaffected | 4590 | 329 | 342 | 14.0 | 0.08 | Wild-type |
| H30a | Unaffected | 4590 | 611 | 780 | 18.2 | 0.19 | Wild-type |
| H30b | Unaffected | 4590 | 1839 | 2017 | 11.4 | 0.58 | Wild-type |
| H25a | Affected | 9180 | 1160 | 1108 | 4.6 | 0.13 | Mutant |
| H25b | Affected | 4590 | 223 | 166 | 15.0 | 0.04 | Mutant |
| H12a | Affected | 9180 | 511 | 464 | 9.0 | 0.05 | Mutant |
| H12b | Affected | 4590 | 293 | 230 | 25.1 | 0.05 | Mutant |
| Plasma sample . | Fetal hemophilia status . | Digital PCR result . | Fetal %† . | mr‡ . | SPRT classification§ . | ||
|---|---|---|---|---|---|---|---|
| Total wells . | Mutant count* . | Wild-type count* . | |||||
| H9 | Affected | 4590 | 1022 | 801 | 14.8 | 0.19 | Mutant |
| H26a | Unaffected | 9180 | 1590 | 1710 | 3.8 | 0.21 | Wild-type |
| H26b | Unaffected | 4590 | 590 | 650 | 6.8 | 0.15 | Wild-type |
| H15a | Affected | 4590 | 573 | 435 | 10.5 | 0.10 | Mutant |
| H15b | Affected | 4590 | 2506 | 1956 | 10.7 | 0.56 | Mutant |
| H17 | Unaffected | 4590 | 329 | 342 | 14.0 | 0.08 | Wild-type |
| H30a | Unaffected | 4590 | 611 | 780 | 18.2 | 0.19 | Wild-type |
| H30b | Unaffected | 4590 | 1839 | 2017 | 11.4 | 0.58 | Wild-type |
| H25a | Affected | 9180 | 1160 | 1108 | 4.6 | 0.13 | Mutant |
| H25b | Affected | 4590 | 223 | 166 | 15.0 | 0.04 | Mutant |
| H12a | Affected | 9180 | 511 | 464 | 9.0 | 0.05 | Mutant |
| H12b | Affected | 4590 | 293 | 230 | 25.1 | 0.05 | Mutant |
PCR indicates polymerase chain reaction; RMD, relative mutation dosage; SPRT, sequential probability ratio test.
Mutant count, number of wells positive for the mutant allele. Wild-type count, number of wells positive for the wild-type allele.
Fetal DNA proportions were determined by the digital ZFY/X assay.
Average reference template concentration per PCR well. The reference template referred to the allele with the lesser count in each sample.
Classification of fetal genotypes by SPRT.
As controls, we also studied 5 maternal plasma samples obtained from normal pregnant women by using each of the mutation-specific assays. As shown in supplemental Table 3, no mutant alleles were detected in most of the cases. For 6 of the 35 studied maternal plasma cases, the positive wells containing the mutant alleles constituted less than 0.3% of the total number of positive wells in the experiments. These positive signals might have resulted from cross hybridizations of the fluorescent probes during PCR. Nonetheless, such low numbers of mutant-positive wells would not skew the allelic ratio between mutant and wild-type alleles to an extent that would alter the RMD classification by SPRT.
Discussion
Current prenatal diagnosis for hemophilia largely relies on invasive procedures such as chorionic villus sampling, which poses a finite risk to the fetuses.1,2,24 Consequently, many pregnant women from at-risk families do not consent to invasive testing because of the associated risks.25,26 Noninvasive fetal sex determination by the use of circulating fetal DNA has provided one safe alternative for managing such pregnancies.27 Through the detection of chromosome Y DNA sequences in maternal plasma, male fetuses could be identified with an accuracy of greater than 97% from the 7th week of gestation onwards.28,29 Unnecessary invasive testing could be avoided for female fetuses because they are either unaffected or are disease carriers.18,27
However, invasive diagnostic testing is still required for one-half of the pregnant cases involving male fetuses. In this study, we have developed a noninvasive prenatal diagnostic strategy to directly detect causative mutations carried by male fetuses in at-risk pregnancies. By using the digital RMD approach for genetic loci on chromosome X, we have accurately identified the mutant or the wild-type alleles inherited by the male fetuses in all of the 12 studied maternal plasma samples from 7 pregnant carriers of hemophilia (Table 4). The fetal genotypes could be detected as early as the 11th week of gestation (Table 1), demonstrating the potential for early diagnostic use of the method.
This noninvasive prenatal mutation detection method could be combined with the existing noninvasive fetal sex determination test to further minimize the number of at-risk pregnant cases that would require invasive diagnostic testing. The identification of affected fetuses could also facilitate subsequent obstetric management for pregnant women who would not otherwise consider invasive prenatal testing. A total of 3%–4% of infants with hemophilia experience a cranial bleed30 that occurs during labor and delivery. Prolonged labor and difficult instrumental deliveries are the main risk factors for this complication25,26 and should be avoided for delivery of affected fetuses.18 It is also recommended that affected fetuses are delivered in a tertiary unit with an affiliated hemophilia center to ensure availability of necessary expertise and resources for their management.18
Recently, prenatal diagnosis by third trimester amniocentesis has been suggested to help appropriate planning of the mode and place of delivery for parents who are unwilling to accept the risk of fetal loss associated with earlier prenatal testing.31 If a fetus is unaffected, labor and delivery can be managed without any restrictions in local maternity units. However, third trimester amniocentesis is also an invasive procedure and associated with potential risks and complications.32,33 Fetal DNA concentration is the greatest during the third trimester of pregnancy11 ; thus, digital RMD testing can offer an accurate noninvasive alternative to third trimester amniocentesis for this purpose.
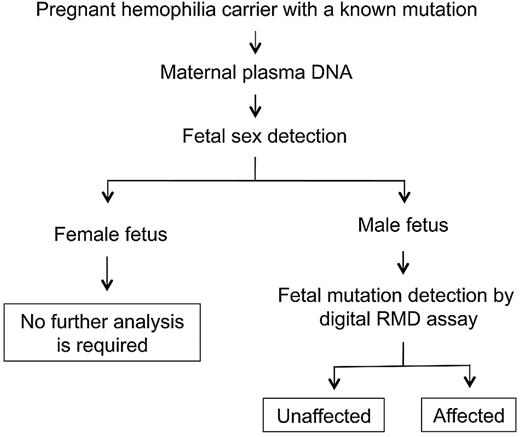
The workflow of noninvasive prenatal hemophilia assessment may be set up as illustrated in Figure 3. Maternal plasma sample is collected from a pregnant carrier receiving obstetric care. The sex of the fetus is then determined, eg, by plasma DNA analysis, and no further testing is required for a pregnancy involving a female fetus. For a pregnancy with a male fetus, digital RMD testing is offered to determine whether the fetus has inherited the maternal mutant allele. The digital RMD result could be confirmed, if necessary, by a second maternal plasma sample taken at a later stage of pregnancy when fetal DNA concentrations are greater,11 allowing for more robust RMD testing. The digital PCRs involved in the current RMD testing were performed on a microfluidics platform. Microfluidics automates the digital reaction setup by channeling nanoliter aliquots of PCR mixture into thousands of amplification chambers.11 The actual setup of a 4590-well digital PCR set involves only a few manual pipetting steps and could conveniently be performed in a routine clinical setting. In addition, the reagents consumed for digital PCRs are relatively inexpensive because only a nanoliter scale of reaction volumes are involved in such reaction chambers. Finally, the system developed in this report can also be applied for the noninvasive prenatal diagnosis of other sex-linked disorders.
Schematic illustration of the workflow for noninvasive prenatal diagnosis of hemophilia by circulating DNA analysis in maternal plasma.
Schematic illustration of the workflow for noninvasive prenatal diagnosis of hemophilia by circulating DNA analysis in maternal plasma.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Innovation and Technology Fund (ITS/378/09) of the Government of the Hong Kong Special Administrative Region and Women with Bleeding Disorders Research Fund, The Royal Free Hospital.
Authorship
Contribution: N.B.Y.T., R.A.K., C.C., R.W.K.C., and Y.M.D.L. designed the research; N.B.Y.T, R.A.K., K.C.A.C., C.C., G.M., E.G.T., T.Y.L., T.K.L., R.W.K.C., and Y.M.D.L. performed the research; N.B.Y.T. wrote the first draft of the manuscript; and all authors reviewed and approved the final version of the manuscript.
Conflict-of-interest disclosure: N.B.Y.T., K.C.A.C., R.W.K.C., and Y.M.D.L. have filed patent applications on the detection of fetal nucleic acids in maternal plasma for noninvasive prenatal diagnosis. Part of this patent portfolio has been licensed to Sequenom. Y.M.D.L. is a consultant and holds equities in Sequenom. The remaining authors declare no competing financial interests.
Correspondence: Y. M. Dennis Lo, Department of Chemical Pathology, The Chinese University of Hong Kong, Rm 38023, 1/F, Clinical Sciences Building, Prince of Wales Hospital, 30-32 Ngan Shing St, Shatin, Hong Kong SAR, China; e-mail: loym@cuhk.edu.hk.

![Figure 1. Illustration of the experimental steps for digital RMD analysis. For each maternal plasma DNA sample, both the mutant DNA proportion (Pr) and the fractional fetal DNA concentration are determined by digital PCRs. (A) Pr is determined by the use of the real-time PCR assay targeting the mutation carried by the mother, whereas fetal DNA proportion is determined by the use of the real-time PCR assay for the homologous ZFY and ZFX gene region. (B) Digital PCR is carried out on a microfluidics Digital Array (Fluidigm), which consists of 12 panels with each panel further partitioned into 765 reaction chambers. Each DNA sample is analyzed with 6 panels (ie, 765 × 6 = 4590 chambers). The PCR mixture is first manually added into the sample inlet of each panel. The mixture is next aliquoted into 765 chambers in each panel automatically by an Integrated Microfluidics Circuit Controller (Fluidigm). Each chamber contains a final reaction volume of 6 nL. The cell-free DNA concentration in maternal plasma is typically very low such that there is less than one template molecule per chamber on average. Hence, the distribution of template molecules to the chambers follows the Poisson distribution. (C) Real-time PCR is performed on the BioMark System (Fluidigm). Most of the chambers contain zero or one template DNA molecule, and the molecules are amplified and detected individually. (D) After PCR, the concentrations of mutant and wild-type DNA, as well as ZFY and ZFX DNA, are calculated. The number of chambers showing positive amplifications for the corresponding allele is counted. The concentration is then Poisson-corrected using the equation [ − ln((N − P)/N)]*N, where N is the total number of reaction chambers analyzed, P is the number of chambers positive for the allele, and ln is the natural logarithm. (E) Pr is calculated from the Poisson-corrected mutant and wild-type DNA concentrations. Fractional fetal DNA concentration is calculated from the Poisson-corrected ZFY and ZFX DNA concentrations. (F) SPRT is used to compare the experimental Pr with the expected Pr at the observed fractional fetal DNA concentration and average reference template concentration (mr). SPRT classification will be made if the mutant allele is over- or underrepresented when compared to the wild-type allele, which implies an affected or nonaffected fetus, respectively.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/13/10.1182_blood-2010-10-310789/4/m_zh89991168300001.jpeg?Expires=1769083343&Signature=Ma~xOFxnRstvrfG-0JkyKnehbo3Bv-yU04bFmtfUC2fN~57Qqk2wiEzOv5tT3LCB4Hvjj8FPjTqJnuPjm61bhuvk1zvGKFLBH9Ec1-7q2CrY7snF6Tevyc67Wo9ueqjsca72HkWl5GFKF5lrwH9VqRRaIQ~x~tjEwDoJh5WP6u8fWLrF5~GLsBzBZ3JkxVorQmDdeE5AXejQ7tRCeiR-t9iqmE25R44ec39LSCpP-5Tok5QkdeUxk~yWEluKmyhZqoFoO7T9CUO~wgL3Glh08vYmSIyu0-wwBnkc16pS-jDhrEBSVemxUi6FFn9Sk8wuBcq7-ivuTl0-38P6u-j~AQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal