Abstract
In allogeneic HSCT, NK-cell alloreactivity is determined by the presence in the donor of NK cells expressing inhibitory killer cell Ig-like receptors (KIRs) that recognize HLA class I allotypes present in the donor but lacking in the recipient. Dominant KIR ligands are the C1 and C2 epitopes of HLA-C. All HLA-C allotypes have either the C1 epitope, the ligand for KIR2DL2/L3, or the C2 epitope, the ligand for KIR2DL1/S1. Here, we show that, in alloreactive NK-cell responses, KIR2DS1 expression represents a remarkable advantage as it allows efficient killing of C2/C2 or C1/C2 myelomonocitic dendritic cells (DCs) and T-cell blasts. When DCs or T-cell blasts were derived from C2/C2, Bw4/Bw4 donors, the activating signals delivered by KIR2DS1 could override the inhibition generated by NKG2A or KIR2DL2/L3 expressed on the same NK-cell clone. Furthermore, substantial lysis of C2/C2, Bw4/Bw6 targets was mediated by KIR2DS1+ NK cells coexpressing KIR3DL1. Importantly, in the case of C1/C2 targets, KIR2DS1+ NK cells were inhibited by the coexpression of KIR2DL2/L3 but not of NKG2A. Thus, KIR2DS1 expression in HSC donors may substantially increase the size of the alloreactive NK-cell subset leading to an enhanced ability to limit GVHD and improve engrafment.
Introduction
HSCT has proven to be an effective treatment for hematologic malignancies, primarily high risk leukemias, including both adult myeloid leukemias (AML) and pediatric lymphoblastic leukemias (ALL).1 However, GVHD is a frequent complication of allogenic HSCT and has limited its overall effectiveness.2 GVHD is mediated by the activation and proliferation of donor's alloreactive T cells leading to tissue damage in the host, primarily in the gastrointestinal tract, liver, and skin. Previous studies in mice revealed that natural killer (NK) cells can suppress the development of GVHD while inducing an antileukemia response.3,4 These experiments revealed that the anti-GVHD effect of murine NK cells is secondary to the ablation of host APCs which are critical for inducing donor T-cell activation.5 In agreement with this concept, in vitro studies showed that human alloreactive, KIR-mismatched, NK-cell clones can kill both immature and mature monocyte-derived dendritic cells (DCs).6 In mice, prevention of GVHD was also attributed to NK cell–mediated killing of donor's T-cell blasts.7 On the other hand, it has also been shown that murine, alloreactive NK cells may prevent rejection of MHC-mismatched BM transplantations through ablation of recipient T lymphocytes.3
It is now well established that functional responses of NK cells are controlled by a set of NK-cell receptors that recognize HLA class I molecules. Some of these receptor/ligand interactions are conserved, such as the interaction between CD94/NKG2A and HLA-E,8,9 while those between polymorphic killer cell Ig-like receptors (KIRs) and polymorphic epitopes of HLA-A, -B, and -C (referred to as KIR ligands) are largely variable.10-17 In particular, KIR2DL1 recognizes HLA-C alleles characterized by Lys at position 80 (defined as C2 epitope) and KIR2DL2/L3 recognizes HLA-C alleles characterized by Asn at position 80 (C1 epitope), but it can also recognizes the C2 epitope although with low affinity. In addition, KIR3DL1 recognizes HLA-A and HLA-B alleles sharing the Bw4 public epitope.10-17 A direct consequence of the genetic variation of KIR and KIR ligands is that donor-derived alloreactive NK cells may display alloreactivity and mediate beneficial GVL reactions in HSCT. In this context, NK-cell alloreactivity can be predicted by the analysis of donor's KIR gene profile16,18 and by differences in KIR ligands between donor and recipient.4 In T cell-depleted haploidentical HSCT, donor-versus-recipient NK-cell alloreactivity has been shown to reflect mismatch between inhibitory receptors for self-HLA class I molecules expressed on a fraction of “licensed”19 donor NK cells (“alloreactive” NK cells) and the HLA class I ligands expressed on recipient cells. As a consequence, alloreactive NK cells can lyse leukemic cells as they sense the missing expression of self-HLA class I molecules.4 The presence and the size of alloreactive NK-cell subsets can be assessed by the combined use of appropriate anti-KIR–specific mAbs.20
Diverse KIR haplotypes can be simplified into 2 groups: group A haplotypes that have a fixed number of genes which encode inhibitory receptors with the exception of KIR2DS4 and group B haplotypes that have variable gene contents including additional activating receptor genes. All individuals can be categorized as having the following KIR genotypes: A/A, which is homozygous for group A KIR haplotypes, or B/x, which contains either 1 (A/B heterozygotes) or 2 (B/B homozygotes) group B haplotypes.18 Recent studies suggested that donor's group B haplotypes, compared with group A, yield significantly superior protection against leukemic relapses and improved disease-free survival (DFS) in patients undergoing T cell–depleted HSCT for AML.17,21-23 Moreover, in T cell–repleted HSCT both centromeric and telomeric B motifs contributed to relapse protection and improved survival.23
In this context, it has been recently shown that KIR2DS1 (C2-specific) is involved in the NK cell–mediated killing of EBV-transformed cell lines24-27 and can play a role also in alloreactive responses against leukemic blasts. Thus, in a cohort of pediatric patients with high-risk ALL, KIR2DS1 was found to play an important role in inducing NK-mediated lysis of C2/C2 leukemic blasts in KIR/KIR-ligand mismatched haplo-HSCT.20 Remarkably, triggering signals, generated by the interaction of KIR2DS1 with HLA-C expressed by leukemic blasts, could override not only the inhibitory signals generated on NKG2A engagement, but also those generated by KIR2DL2/L3 on HLA-C2 recognition.20
In the present study, we show that expression of KIR2DS1 confers a remarkable advantage in the ability of NK cells to kill allogeneic DCs and T-cell blasts. These data suggest that KIR2DS1+ NK cells may play an important role not only in the ablation of leukemic blasts, but also in preventing GVHD and graft rejection, respectively.
Methods
KIR gene profile and KIR ligand analyses
DNA of the tested samples was extracted using the QIAamp DNA Blood Mini kit (QIAGEN) according to the manufacturer's instruction. The KIR genes profile and KIR ligands detection were performed using Olerup SSP-PCR (sequence-specific polymorphism PCR) KIR genotyping and KIR HLA ligand kits (GenoVision), respectively. In particular, KIR genotyping kit allowed the detection of the presence/absence of all the KIR genes identified so far. Moreover, by the use of KIR HLA ligand kit, we typed HLA-C alleles on the basis of the dimorphism present at position 80 (analysis of C1 or C2 epitope), we detected HLA-B alleles characterized by Bw4 motif (dividing the Bw4+ alleles in 2 groups according to the aminoacidic residue present at position 80) as well as the presence of HLA-A alleles carrying the Bw4 public epitope. To detect the presence of HLA-B alleles characterized by the Bw6 epitope, RT-PCR analysis was performed. Total RNA was extracted from phytohemagglutinin (PHA) blasts using the RNeasy micro kit (QIAGEN) according to the manufacturer's instruction and the cDNA synthesis was performed on ∼ 1 μg of RNA using oligo-dT oligonucleotides. The PCR primer sequences used in this experiments were HLA-Bw6 up: CCT GCG GAA CCT GCG CG and HLA-B reverse: TCC GAT GAC CAC AAC TGC T.
KIR2DL3 allele identification
The cDNA obtained by retrotranscription of the RNA extracted from a polyclonal NK population derived by donor P61 were amplified using a set of primers KIR2DL2/L3/S2 up: CAT GTC GCT CAT GGT CGT C and E down: GTT CCG YGT ACA CGA TGA to detect the sequences coding for KIR2DL1/L2/L3. The amplification product was cloned into pcDNA3.1/V5/His TOPO vector using the Eukaryotic TOPO TA Cloning kit (Invitrogen) and sequenced. DNA sequencing was performed using d-Rhodamine Terminator Cycle Sequencing kit and a 3100 ABI automatic sequencer (PerkinElmer/Applied Biosystems).
Generation of NK-cell clones
Buffy coats from healthy donors were obtained from the Immunohematology and Transfusion Center at the S. Martino Hospital (Genova, Italy). Human PBMCs were isolated from a C1/C1 Bw4/Bw4 KIR2DS1+ healthy volunteer by Ficoll/Hypaque gradients. Highly purified NK cells (97%-99% purity) were isolated by depletion of non-NK cells, using the Miltenyi NK cell isolation kit (Miltenyi Biotec). NK cells were cultured on irradiated feeder cells in the presence of 100 U/mL rhIL-2 (Proleukin; Chiron Corporation) and 2 μg/mL PHA (Life Technologies) to obtain activated polyclonal NK-cell populations or, after limiting dilution, NK-cell clones as previously described.6
Phenotypic analysis of NK-cell clones
Cells were incubated with appropriate mAb followed by PE-conjugated isotype-specific goat anti–mouse secondary reagent (Southern Biotechnology Associates). The list of mAbs used in this study is available online as a data supplement (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Samples were analyzed by cytofluorimetric analysis on a FACScan with the CellQuest program (both from BD Biosciences).
Generation of iDCs, mDCs, and PHA blasts from PBMCs
PBMCs from HLA-typed healthy donors were seeded in 25-mm2 plastic flasks (Corning Life Sciences) at a density of 5 × 106 cells/mL in RPMI 1640 containing 10% FCS. After 45 minutes at 37°C, nonadherent cells were removed. The adherent fraction was cultured in the presence of rGM-CSF (50 ng/mL; PeproTech) and rIL4 (20 ng/mL; PeproTech). After 6 days of culture, cells characterized by the CD14−CD1a+CD83− phenotype corresponding to immature DCs (iDCs) were obtained. To generate CD14−CD1a+CD83+CD86+ mature DCs (mDCs), iDCs were stimulated for 24 hours with ultra-pure LPS from Salmonella minnesota (1 μg/mL final concentration; Alexis).
PHA blasts were obtained by culturing for 1-day PBMCs from HLA-typed healthy donors with 2 μg/mL PHA (Life Technologies). After this period of time, cells were cultured in medium supplemented with 100 U/mL rhIL-2 (Proleukin; Chiron Corporation).
Cytolytic activity of NK-cell clones
Cells used as targets in the various cytolytic assays were the following: 221 (LCL721.221, human EBV-transformed B-cell line); 221 transfected with HLA-Cw3, -Cw4, -Cw6, or -B51 alleles; P815 (FcγR+ murine mastocytoma); PHA blasts, iDCs, and mDCs derived from different HLA-typed donors. The NK-mediated cytotoxicity was assessed in a 4-hour 51Cr-release assay as previously described. For masking experiments, NK cells were preincubated with mAbs specific to the various NK receptors or with anti-HLA class I mAb (A6/136 or F(ab′)2 6A4) 10 minutes before addition of target cells; mAb concentration was 10 μg/mL. For redirected killing assays, P815 were used as target cells in the presence of mAbs of IgG isotype at a concentration of 0.5 μg/mL. The E:T ratios are indicated in the text. PASW Statistics 18 (SPSS Italia) was used to process and analyze data.
Results
Isolation and functional characterization of KIR2DS1+ NK-cell clones from a C1/C1 donor
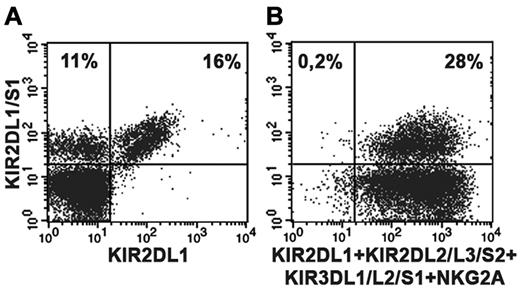
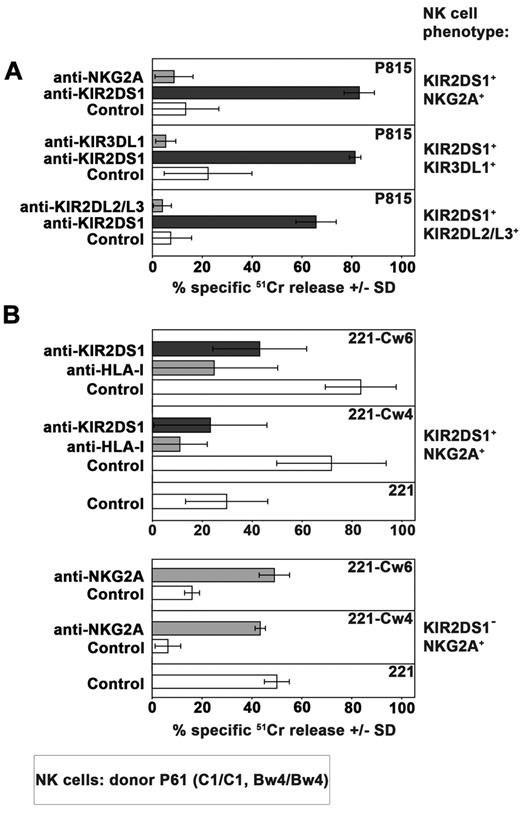
A series of healthy donors were screened on the basis of their KIR2DS1 expression by cytofluorimetric analysis. To distinguish KIR2DS1+ from KIR2DL1+ cells, purified peripheral blood NK cells were stained in double fluorescence analysis with 2 different mAbs: an anti-KIR2DL1–specific mAb (clone 143211) and a mAb specific for both KIR2DL1 and KIR2DS1 (clone 11PB6).20,28,29 Donors were further typed for KIR and KIR ligand to select suitable C1/C1 (or C1/C2) KIR2DS1+ individuals. Figure 1A shows the KIR2DS1+ NK-cell subset (143211− 11PB6+) of the C1/C1 donor (donor P61) used in the present study while in Tables 1 and 2 its KIR ligand and KIR typing are shown. Notably, further analysis indicated that this donor was KIR2DL3*001+ thus ruling out the possibility that 11PB6 mAb might react with KIR2DL3*005.30 As shown in Figure 1B, virtually all resting KIR2DS1+ NK cells isolated from peripheral blood of this donor coexpressed additional HLA-specific receptors (KIR and/or NKG2A). NK-cell clones derived from this donor were first screened on the basis of their surface phenotype. Selected KIR2DS1+ NK-cell clones were divided in different groups according to the coexpression of different HLA class I–specific inhibitory receptors including NKG2A, KIR3DL1, or KIR2DL2/L3 (Figure 2A). According to KIR and KIR-ligand genotype data, KIR3DL1 and KIR2DL2/L3 are licensing receptors while KIR2DL1 is not. Notably, from this donor no NK-cell clones expressing only KIR2DL1 and KIR2DS1 could be obtained. The inhibitory activity of these receptors was validated both in a redirected killing assay against P815 (Figure 2A) and in experiments of cytotoxicity against B-EBV cell lines transfected with HLA-Cw3 (C1-epitope), -Cw4 (C2-epitope), -Cw6 (C2-epitope), or -B51 (Bw4 public epitope) alleles (not shown). In line with previous reports,24,25 KIR2DS1+ NK-cell clones efficiently killed C2+ transfectants such as 221-Cw4 and -Cw6. Lysis could be inhibited by mAb-mediated blocking of KIR2DS1 (Figure 2B). Notably, an efficient disruption of KIR2DS1/HLA-C interaction was detected also by the use of the anti-HLA class I mAb 6A4 known to prevent HLA-C recognition by KIR2DL1+ cells31 (Figure 2B). It is also of note that the activating signals generated on engagement of KIR2DS1 by Cw4 or Cw6 could override the inhibitory signals generated by the NKG2A/HLA-E interaction in KIR2DS1+ NKG2A+ NK-cell clones. On the contrary, the HLA-E expression on HLA-Cw4 or -Cw6 transfectants was sufficient to induce strong inhibition of cytotoxicity by KIR2DS1−NKG2A+ NK-cell clones (Figure 2B).
Detection of the KIR2DS1+ NK-cell subset in a C1/C1 donor. Double fluorescence analysis was performed using purified peripheral blood NK cells derived from the C1/C1 Bw4/Bw4 donor P61. For staining, the following mAbs were used in combination: anti-KIR2DL1/S1-FITC (clone 11PB6) and anti-KIR2DL1-PE (clone143211). (A); anti-KIR2DL1/S1-FITC (clone 11PB6) and a mixture of anti-KIR2DL2/L3/S2 (Y249), anti-KIR3DL1/L2/S1 (AZ158), anti-NKG2A (Z199) followed by PE isotype-specific secondary reagents and anti-KIR2DL1-PE (clone 143211). (B). Numbers in upper quadrants indicate the percent of positive cells.
Detection of the KIR2DS1+ NK-cell subset in a C1/C1 donor. Double fluorescence analysis was performed using purified peripheral blood NK cells derived from the C1/C1 Bw4/Bw4 donor P61. For staining, the following mAbs were used in combination: anti-KIR2DL1/S1-FITC (clone 11PB6) and anti-KIR2DL1-PE (clone143211). (A); anti-KIR2DL1/S1-FITC (clone 11PB6) and a mixture of anti-KIR2DL2/L3/S2 (Y249), anti-KIR3DL1/L2/S1 (AZ158), anti-NKG2A (Z199) followed by PE isotype-specific secondary reagents and anti-KIR2DL1-PE (clone 143211). (B). Numbers in upper quadrants indicate the percent of positive cells.
KIR ligands carried by the NK-cell donor (P61) or by different mDCs and T-cell targets
| . | HLA class I epitope . | |||||
|---|---|---|---|---|---|---|
| C1* . | C2* . | HLA-Bw4 T80† . | HLA-Bw4 I80† . | HLA-Bw6† . | HLA-A Bw4+* . | |
| P61 | P | A | P | A | A | P |
| GF | A | P | P | A | P | A |
| SC | P | P | A | P | A | P |
| RB | A | P | A | P | A | A |
| CV | P | P | A | P | A | A |
| . | HLA class I epitope . | |||||
|---|---|---|---|---|---|---|
| C1* . | C2* . | HLA-Bw4 T80† . | HLA-Bw4 I80† . | HLA-Bw6† . | HLA-A Bw4+* . | |
| P61 | P | A | P | A | A | P |
| GF | A | P | P | A | P | A |
| SC | P | P | A | P | A | P |
| RB | A | P | A | P | A | A |
| CV | P | P | A | P | A | A |
DNA of the indicated samples have been tested by sequence-specific polymorphism PCR (SSP-PCR) approach to define KIR ligands. Results obtained using set of primers that allowed to detect the presence (P) or the absence (A) of HLA-C alleles coding for C1 (HLA-C N80) or C2 (HLA-C K80) epitope, of HLA-Bw4 alleles carrying a threonine (T) or a isoleucine (I) at position 80, and of HLA-A alleles characterized by the Bw4 public epitope.
RNA of the indicated samples have been tested by SSP-PCR approach to define KIR ligands. Results obtained using set of primers that selectively amplify HLA-B alleles sharing Bw6 epitope.
KIR genes profile analysis of P61 donor
| Inhibitory KIR . | Activating KIR . | Pseudogene . | |||
|---|---|---|---|---|---|
| KIR2DL1 | P | KIR2DS1 | P | KIR2DP1 | P |
| KIR2DL2 | † | KIR2DS2 | P | KIR3DP1 | P |
| KIR2DL3 | *001† | KIR2DS3 | P | ||
| KIR2DL4 | P | KIR2DS4 | P | ||
| KIR2DL5A | P | KIR2DS5 | A | ||
| KIR2DL5B | P | KIR3DS1 | P | ||
| KIR3DL1 | † | ||||
| KIR3DL2 | P | ||||
| KIR3DL3 | P | ||||
| Inhibitory KIR . | Activating KIR . | Pseudogene . | |||
|---|---|---|---|---|---|
| KIR2DL1 | P | KIR2DS1 | P | KIR2DP1 | P |
| KIR2DL2 | † | KIR2DS2 | P | KIR3DP1 | P |
| KIR2DL3 | *001† | KIR2DS3 | P | ||
| KIR2DL4 | P | KIR2DS4 | P | ||
| KIR2DL5A | P | KIR2DS5 | A | ||
| KIR2DL5B | P | KIR3DS1 | P | ||
| KIR3DL1 | † | ||||
| KIR3DL2 | P | ||||
| KIR3DL3 | P | ||||
Donor genotype was analyzed for the presence (P) or absence (A) of the indicated KIR genes using the sequence-specific polymorphism PCR approach.
001 indicates the KIR2DL3 allele typed.
The set of licensing KIR inhibitory receptors.
Functional analysis of HLA-specific receptors expressed on 3 different groups of KIR2DS1+ NK-cell clones. (A) 3 different groups of KIR2DS1+ NK cells clones (derived from the C1/C1 Bw4/Bw4 donor P61) characterized by the coexpression of NKG2A, KIR3DL1 or KIR2DL2/L3, were analyzed in a redirected killing assay against P815 cell line in the absence (white bars) or in the presence of 11PB6 mAb specific for KIR2DS1/L1 (black bars) or in the presence of mAb specific for the indicated HLA-specific inhibitory receptor (gray bars). The E:T ratio used was 4:1. Average of 3 independent experiments and SD (mean ± SD) are indicated. (B) Comparison between the cytolytic activity of KIR2DS1+NKG2A+ and KIR2DS1−NKG2A+ NK-cell clones derived from the same donor against untransfected, Cw4- or Cw6-transfected 221 cell line in the absence or in the presence of mAb to the indicated molecules. The E:T ratio used was 4:1. Average of 4 independent experiments and SD (mean ± SD) are indicated.
Functional analysis of HLA-specific receptors expressed on 3 different groups of KIR2DS1+ NK-cell clones. (A) 3 different groups of KIR2DS1+ NK cells clones (derived from the C1/C1 Bw4/Bw4 donor P61) characterized by the coexpression of NKG2A, KIR3DL1 or KIR2DL2/L3, were analyzed in a redirected killing assay against P815 cell line in the absence (white bars) or in the presence of 11PB6 mAb specific for KIR2DS1/L1 (black bars) or in the presence of mAb specific for the indicated HLA-specific inhibitory receptor (gray bars). The E:T ratio used was 4:1. Average of 3 independent experiments and SD (mean ± SD) are indicated. (B) Comparison between the cytolytic activity of KIR2DS1+NKG2A+ and KIR2DS1−NKG2A+ NK-cell clones derived from the same donor against untransfected, Cw4- or Cw6-transfected 221 cell line in the absence or in the presence of mAb to the indicated molecules. The E:T ratio used was 4:1. Average of 4 independent experiments and SD (mean ± SD) are indicated.
Killing of C2/C2 myeloid DCs and T-cell blasts by KIR2DS1+ NKG2A+ NK-cell clones from a C1/C1 donor
KIR2DS1+ NK-cell clones (derived from the C1/C1 Bw4/Bw4 donor P61) were further analyzed for their ability to kill allogeneic DCs and PHA blasts, derived from the C2/C2, Bw4/Bw4 donor RB (see Table 1). These experiments were performed either in the absence or in the presence of mAb specific for KIR2DS1, NKp30, and DNAM-1 to evaluate the involvement of these activating receptors in the mechanism of recognition/killing of the different target cell analyzed.
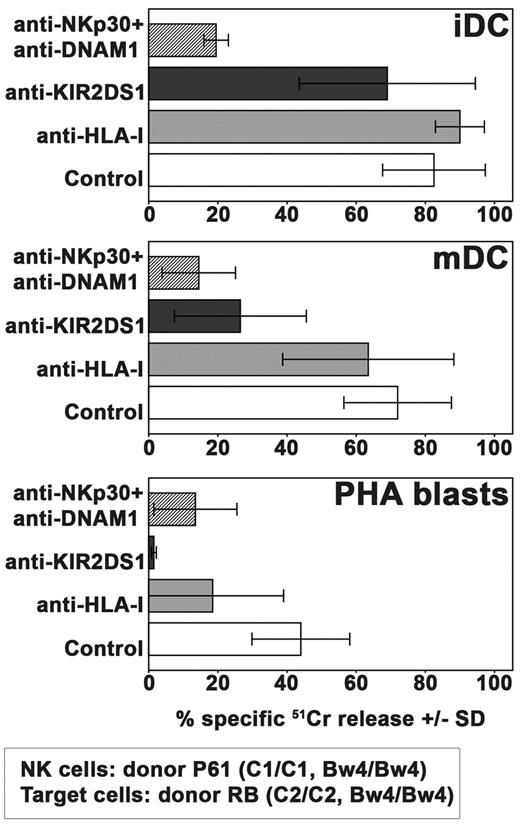
In Figure 3, we show that KIR2DS1+ NKG2A+ NK-cell clones were able to kill both iDCs and mDCs and, to a lower extent, PHA blasts. Lysis of iDCs was poorly affected by mAb-mediated masking of KIR2DS1 whereas, in line with previous reports, it was virtually abrogated by mAb-mediated masking of the non-MHC–specific activating receptors NKp30 and DNAM-1.32,33 On the contrary, killing of mDCs was inhibited by masking either KIR2DS1 or NKp30/DNAM-1. Similar to mDCs, PHA blasts were also killed on engagement of different receptors including KIR2DS1. In this case, however, the role of KIR2DS1 was even more evident because its blocking with the specific mAb could abrogate killing.
Role of KIR2DS1 in killing of allogeneic iDCs, mDCs, or PHA blasts by NKG2A+KIR2DS1+ NK-cell clones. The cytolytic activity of NKG2A+KIR2DS1+ NK-cell clones (derived from the C1/C1 Bw4/Bw4 donor P61) was analyzed against iDCs, mDCs, and PHA blasts derived from the C2/C2 Bw4/Bw4 donor RB either in the absence or in the presence of mAb to the indicated molecules. The E:T ratio used against iDCs and mDCs was 15:1 whereas that used against PHA blasts was 20:1. Average of 3 independent experiments and SD (mean ± SD) are indicated.
Role of KIR2DS1 in killing of allogeneic iDCs, mDCs, or PHA blasts by NKG2A+KIR2DS1+ NK-cell clones. The cytolytic activity of NKG2A+KIR2DS1+ NK-cell clones (derived from the C1/C1 Bw4/Bw4 donor P61) was analyzed against iDCs, mDCs, and PHA blasts derived from the C2/C2 Bw4/Bw4 donor RB either in the absence or in the presence of mAb to the indicated molecules. The E:T ratio used against iDCs and mDCs was 15:1 whereas that used against PHA blasts was 20:1. Average of 3 independent experiments and SD (mean ± SD) are indicated.
Effect of HLA-specific inhibitory receptors on KIR2DS1-dependent NK-cell activation against mDCs or T-cell blasts
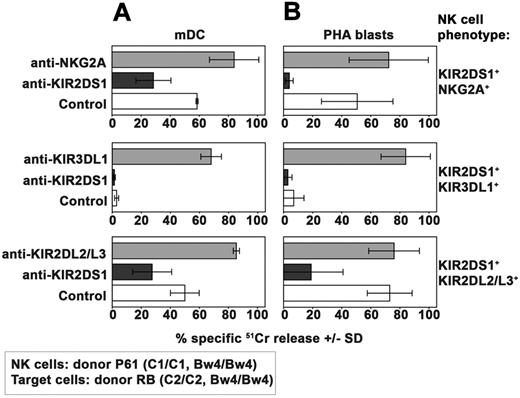
Because KIR2DS1 plays a major role in the process of killing of both mature DCs and PHA blasts, we further analyzed whether and how the NK-cell activation induced by KIR2DS1 could be controlled by different HLA-specific inhibitory receptors. To this end, NK-cell clones (from donor P61) expressing KIR2DS1 in combination with different inhibitory receptors were analyzed for their ability to kill mDCs (Figure 4A) and PHA blasts (Figure 4B) expressing the C2/C2 Bw4/Bw4 phenotype (derived from donor RB, see Table 1). In line with data shown in Figure 3, KIR2DS1+NKG2A+ NK-cell clones displayed cytotoxicity against mDCs (Figure 4A). Moreover, cytolytic activity was inhibited by anti-KIR2DS1 mAb and incremented by anti-NKG2A mAb.
Effect of HLA-specific inhibitory receptors on KIR2DS1-dependent NK-cell activation against allogeneic C2/C2 Bw4/Bw4 mDCs and T cells. Analysis of the cytolytic activity of different groups of KIR2DS1+ NK-cell clones (derived from donor P61) against C2/C2 Bw4/Bw4 mDCs (A) or PHA blasts (B) derived from donor RB either in the absence or in the presence of mAb to the indicated molecules. These KIR2DS1+ clones coexpressed either NKG2A or KIR3DL1 or KIR2DL2/L3. The E:T ratio used against mDCs was 15:1 whereas that used against PHA blasts was 20:1. Average of 3 independent experiments and SD (mean ± SD) are indicated.
Effect of HLA-specific inhibitory receptors on KIR2DS1-dependent NK-cell activation against allogeneic C2/C2 Bw4/Bw4 mDCs and T cells. Analysis of the cytolytic activity of different groups of KIR2DS1+ NK-cell clones (derived from donor P61) against C2/C2 Bw4/Bw4 mDCs (A) or PHA blasts (B) derived from donor RB either in the absence or in the presence of mAb to the indicated molecules. These KIR2DS1+ clones coexpressed either NKG2A or KIR3DL1 or KIR2DL2/L3. The E:T ratio used against mDCs was 15:1 whereas that used against PHA blasts was 20:1. Average of 3 independent experiments and SD (mean ± SD) are indicated.
On the contrary, NK-cell clones expressing KIR2DS1 in combination with KIR3DL1 were unable to kill C2/C2 Bw4/Bw4 mDCs (Figure 4A). That lack of killing was because of KIR3DL1-mediated recognition of Bw4 was shown by the complete restoration of lysis induced by the addition of anti-KIR3DL1 mAb.
KIR2DS1+KIR2DL2/L3+ NK-cell clones express an inhibitory (KIR2DL2/L3) receptor which displays low but significant capability of recognizing C2 ligands.34 These clones killed C2/C2 Bw4/Bw4 mDCs. Lysis was reduced by anti-KIR2DS1 mAb (Figure 4A), suggesting that KIR2DS1-mediated recognition of C2 on mDCs can override the inhibitory signals delivered by KIR2DL2/L3.
The same set of NK clones was then analyzed for cytotoxicity against PHA blasts derived from the same C2/C2 Bw4/Bw4 donor used to derive DCs (donor RB, see Table 1). Both KIR2DS1+NKG2A+ and KIR2DS1+KIR2DL2/L3+ NK clones displayed strong cytotoxicity against these target cells (Figure 4B). Again, lysis could be virtually abrogated by mAb-mediated masking of KIR2DS1. On the contrary, KIR2DS1+KIR3DL1+ NK-cell clones did not kill C2/C2 Bw4/Bw4 PHA blasts (Figure 4B). However mAb-mediated masking of KIR3DL1 fully restored their killing.
Taken together, these data show that KIR2DS1-mediated recognition of C2 on C2/C2 Bw4/Bw4 mDCs and PHA blasts triggers NK alloreactivity when KIR2DS1 is expressed in combination with NKG2A or KIR2DL2/L3. Thus, the expression of KIR2DS1 on donor NK cells may represent an added value not only in the amplification of the size of the alloreactive subset but also in mediating potent anti-GVHD effects and in preventing graft rejection. An exception is represented by NK cells coexpressing KIR3DL1, which (on engagement with Bw4) generates potent inhibition capable of dampening the triggering signal resulting from KIR2DS1/HLA-C2 interactions.
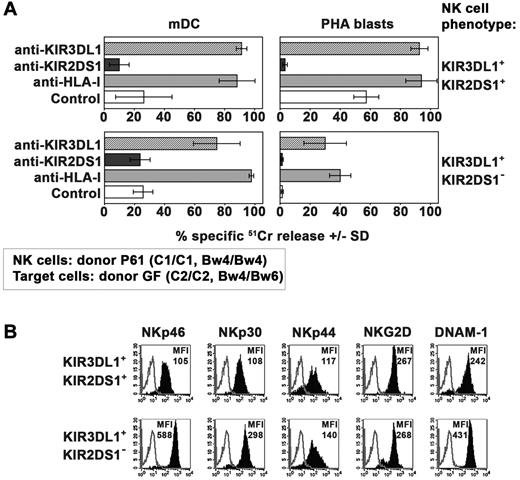
Killing of C2/C2, Bw4/Bw6 target cells by KIR3DL1+KIR2DS1+ NK-cell clones isolated from a C1/C1 donor
In these experiments, the same set of KIR3DL1+KIR2DS1+ NK-cell clones (derived from the donor P61) was analyzed for its cytolytic activity against mDCs and PHA blasts from the C2/C2 donor GF who was heterozygous for the expression of Bw4 (Table 1). Different from the results obtained with Bw4/Bw4 target cells (Figure 4) these NK-cell clones, although expressing KIR3DL1, killed C2/C2 Bw4/Bw6 mDCs and even more efficiently PHA blasts (Figure 5A). Because cytolytic activity could be further incremented in the presence of anti-KIR3DL1 mAb, this implies the occurrence of substantial inhibitory effects mediated by the interaction of KIR3DL1 with Bw4. However, this inhibition was only partial and insufficient to efficiently block the triggering effect mediated via KIR2DS1. Indeed also in these experiments inhibition of lysis was observed in the presence of anti-KIR2DS1 mAb. In this regard our experiments, in agreement with data described by Foley et al,35 indicate that KIR3DL1 efficiently recognize Bw4 alleles carrying a threonine at position 80 (T80) (expressed by donor GF, see Table 1) as demonstrated by the reconstitution of lysis obtained by the addition of anti-KIR3DL1 mAb (Figure 5).
Killing of allogeneic C2/C2, Bw4/Bw6 target cells by KIR3DL1+ KIR2DS1+ NK-cell clones. (A) Killing by KIR3DL1+ KIR2DS1+ or KIR3DL1+ KIR2DS1− NK-cell clones (derived from donor P61) against mDCs and PHA blasts (derived from the C2/C2 Bw4/Bw6 donor GF) was analyzed either in the absence or in the presence of mAb to the indicated molecules. The E:T ratio used against mDCs was 15:1 whereas that used against PHA blasts was 20:1. Average of 3 independent experiments and SD (mean ± SD) are indicated. (B) Representative KIR3DL1+KIR2DS1+ and KIR3DL1+KIR2DS1− NK-cell clones (derived from donor P61) were analyzed by cytofluorimetric analysis for the expression of the indicated molecules (black profiles). White profiles refer to cells incubated with the second reagent only. The MFI is indicated.
Killing of allogeneic C2/C2, Bw4/Bw6 target cells by KIR3DL1+ KIR2DS1+ NK-cell clones. (A) Killing by KIR3DL1+ KIR2DS1+ or KIR3DL1+ KIR2DS1− NK-cell clones (derived from donor P61) against mDCs and PHA blasts (derived from the C2/C2 Bw4/Bw6 donor GF) was analyzed either in the absence or in the presence of mAb to the indicated molecules. The E:T ratio used against mDCs was 15:1 whereas that used against PHA blasts was 20:1. Average of 3 independent experiments and SD (mean ± SD) are indicated. (B) Representative KIR3DL1+KIR2DS1+ and KIR3DL1+KIR2DS1− NK-cell clones (derived from donor P61) were analyzed by cytofluorimetric analysis for the expression of the indicated molecules (black profiles). White profiles refer to cells incubated with the second reagent only. The MFI is indicated.
We also compared KIR3DL1+ NK-cell clones coexpressing or not KIR2DS1. It is of note that only those coexpressing KIR2DS1 displayed a substantial cytotoxic activity against C2/C2, Bw4/Bw6 T-cell blasts (Figure 5A). This difference was not because of differential expression of other activating receptors such as NCR, NKG2D, and DNAM-1. Indeed, these receptors were in most instances expressed at higher density on KIR2DS1− NK-cell clones (Figure 5B). These data suggest a major role played by KIR2DS1 in rendering KIR3DL1+ NK cells alloreactive against these target cells.
The differential behavior of KIR3DL1/KIR2DS1+ NK clones against mDCs and T-cell blasts expressing either the Bw4/Bw4+ or the Bw4/Bw6+ phenotype suggests that the expression level of HLA-Bw4 on target cells may influence the magnitude of inhibitory signals generated by KIR3DL1. This effect, however, might be influenced also by the amino acid present at position 80 in the Bw4 public epitope (T80 vs I80)35,36 or by the type of KIR3DL1 allele.37
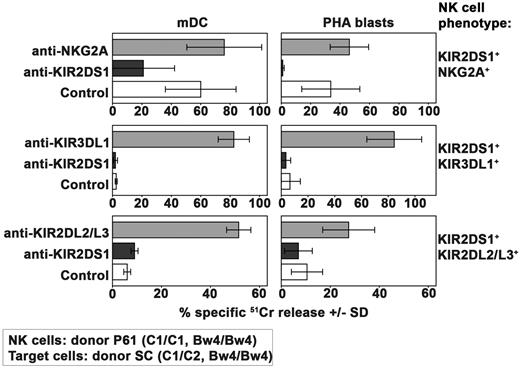
Alloreactivity mediated by KIR2DS1+ NK cells against C1/C2 target cells
Having established that the type and the number of inhibitory interactions may play a crucial role in the inhibition of KIR2DS1-dependent NK-cell activation, we further evaluated to what extent the number of activating interactions between this receptor and its C2 ligand may modify the behavior of the various NK-cell clones described in Figure 2A (all derived from donor P61). To this end, in this set of experiments, target cells expressing the C2/C2, Bw4/Bw4 phenotype (Figure 4) were substituted by C1/C2, Bw4/Bw4 targets (derived from donor SC, see Table 1).
As shown in Figure 6, KIR2DS1+NKG2A+ NK-cell clones killed both mDCs and PHA blasts from the C1/C2 donor. In addition, target cell lysis could be inhibited by mAb-mediated blocking of KIR2DS1. Because no substantial difference could be detected comparing these results with those obtained against C2/C2 cells (Figure 4), it is conceivable that the heterozygous expression of C2 on target cells may not represent a critical disadvantage in the case of KIR2DS1+NKG2A+ NK-cell clones.
Alloreactivity mediated by KIR2DS1+ NK cells against C1/C2 Bw4/Bw4 target cells. Analysis of the cytolytic activity of different groups of KIR2DS1+ NK-cell clones (derived from donor P61) against mDCs or PHA blasts derived from the C1/C2 Bw4/Bw4 donor SC either in the absence or in the presence of mAb to the indicated molecules. These KIR2DS1+ clones coexpressed either NKG2A or KIR3DL1 or KIR2DL2/L3. The E:T ratio used against mDCs was 15:1 whereas that used against PHA blasts was 20:1. Average of 3 independent experiments and SD (mean ± SD) are indicated.
Alloreactivity mediated by KIR2DS1+ NK cells against C1/C2 Bw4/Bw4 target cells. Analysis of the cytolytic activity of different groups of KIR2DS1+ NK-cell clones (derived from donor P61) against mDCs or PHA blasts derived from the C1/C2 Bw4/Bw4 donor SC either in the absence or in the presence of mAb to the indicated molecules. These KIR2DS1+ clones coexpressed either NKG2A or KIR3DL1 or KIR2DL2/L3. The E:T ratio used against mDCs was 15:1 whereas that used against PHA blasts was 20:1. Average of 3 independent experiments and SD (mean ± SD) are indicated.
As expected, the group of NK-cell clones expressing the KIR2DS1+KIR3DL1+ phenotype that was unable to kill C2/C2 Bw4/Bw4 target cells (Figure 4) did not kill mDCs and PHA blasts derived from a C1/C2 Bw4/Bw4 donor as well (Figure 6).
Regarding the group of KIR2DS1+KIR2DL2/L3+ NK-cell clones, they were unable to kill mDCs and PHA blasts derived from the C1/C2 donor (Figure 6) whereas they killed the C2/C2+ ones (Figure 4). This difference was because of inhibition mediated by KIR2DL2/L3 as revealed by the reconstitution of lysis in the presence of specific mAbs. Thus, in the presence of high-affinity interaction between inhibitory KIRs and their KIR ligands (C1), the activation mediated by KIR2DS1 cannot override the inhibitory signals.
In conclusion the only difference in the ability of the 3 different types of KIR2DS1+ NK-cell clones in killing C1/C2 or C2/C2 (Bw4/Bw4) target cells appears to be represented by the lack of alloreactivity of KIR2DS1+KIR2DL2/L3+ NK-cell clones against C1/C2 target cells.
Discussion
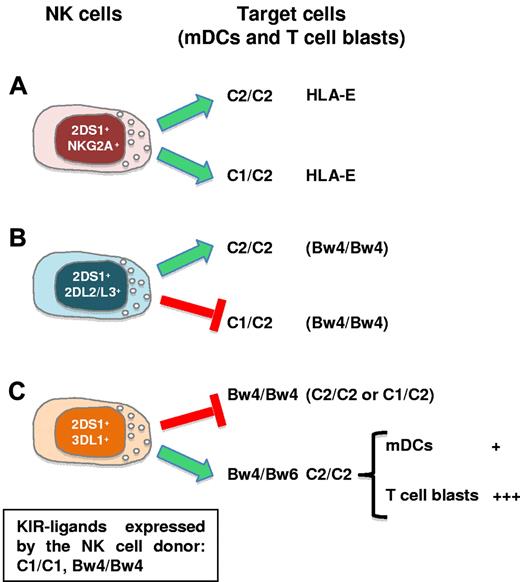
Our present study indicates that a fraction of KIR2DS1+ NK cells can mediate potent alloreactivity against mDCs and PHA-induced T-cell blasts. Accordingly, they may play a crucial role in the prevention and/or therapy of GVHD and graft rejection (host vs graft [HVG]), in KIR/KIR ligand mismatched haploidentical HSCT when recipients are carrying the C2 epitope of HLA-C alleles.
We show that both C2/C2 and C1/C2 mature myelomonocytic DCs and T-cell blasts are killed by KIR2DS1+ NK-cell clones derived from C1/C1 donors. However, this effect is tightly regulated by inhibitory receptors in ways that are diverse and dependent on gene dose. In particular, KIR2DS1+ clones coexpressing only NKG2A could kill both C2 homozygous and heterozygous cells (Figure 7A).
Killing of allogeneic mDCs and T-cell blasts by KIR2DS1+ NK cells. KIR2DS1 expression in HSC donors increases the size of the alloreactive NK-cell subset during allogeneic HSCT. KIR2DS1 triggering by its KIR ligand (C2) promotes the ability of NK cells to limit GVHD and improve engraftment. (A) KIR2DS1+ clones coexpressing NKG2A kill both C2 homozygous and heterozygous T-cell blasts and mDCs. (B) KIR2DS1+ clones coexpressing KIR2DL2/L3 selectively kill C2 homozygous cells. (C) KIR2DS1+ clones coexpressing KIR3DL1 kill C2+ cells only when derived from Bw4 heterozygous (Bw4/Bw6) donors. In this case, NK cells preferentially killed T-cell blasts (+++) compared with mDCs (+). For each type of NK-target interaction the relevant KIR ligands (and/or HLA-E) recognized by the HLA class I–specific receptors expressed by the NK clones are shown. The box indicates the KIR-ligands responsible for licensing in the NK-cell donor.
Killing of allogeneic mDCs and T-cell blasts by KIR2DS1+ NK cells. KIR2DS1 expression in HSC donors increases the size of the alloreactive NK-cell subset during allogeneic HSCT. KIR2DS1 triggering by its KIR ligand (C2) promotes the ability of NK cells to limit GVHD and improve engraftment. (A) KIR2DS1+ clones coexpressing NKG2A kill both C2 homozygous and heterozygous T-cell blasts and mDCs. (B) KIR2DS1+ clones coexpressing KIR2DL2/L3 selectively kill C2 homozygous cells. (C) KIR2DS1+ clones coexpressing KIR3DL1 kill C2+ cells only when derived from Bw4 heterozygous (Bw4/Bw6) donors. In this case, NK cells preferentially killed T-cell blasts (+++) compared with mDCs (+). For each type of NK-target interaction the relevant KIR ligands (and/or HLA-E) recognized by the HLA class I–specific receptors expressed by the NK clones are shown. The box indicates the KIR-ligands responsible for licensing in the NK-cell donor.
On the contrary, KIR2DS1+ clones coexpressing KIR2DL2/L3 selectively killed C2 homozygous cells (Figure 7B). Finally, KIR2DS1+ clones coexpressing KIR3DL1 killed C2+ cells only when derived from Bw4 heterozygous (Bw4/Bw6) donors (Figure 7C).
The observation that “nonalloreactive” NKG2A+ NK cells as well as certain NK cells expressing inhibitory KIRs can actually become “alloreactive” against DCs and T cells if they coexpress KIR2DS1 is particularly important in haplo-HSCT. Indeed, in given donor/recipient pairs, KIR2DS1 expression may considerably amplify the size of the alloreactive NK subset. In addition KIR2DS1+ NKG2A+ NK cells might play a relevant role even in the case of non-allo HSCT (patients expressing all the main KIR-ligands including C1, C2, and Bw4).
Alloreactive NK cells have been shown to play a crucial role in the successful therapy of high-risk acute leukemias in the haplo-HSCT setting.38-41 Indeed, alloreactive NK cells cannot only prevent leukemic relapses but also promote engraftment and reduce GVHD. On the other hand, alloreactive NK cells would spare normal cells because these cells lack ligands recognized by non-MHC–specific activating NK receptors.42,43 The best characterized alloreactive NK cells are those that express only inhibitory KIR that are not engaged by the HLA class I alleles present on allogeneic target cells.10,44 Therefore, alloreactive NK cells should also lack CD94/NKG2A because HLA-E is present on all HLA class I+ cells. In vitro human NKG2A− alloreactive NK cells expressing inhibitory KIRs kill both immature and mature monocyte-derived DCs while autologous KIR+ NK cells are unable to kill both cell types. On the other hand, most NKG2A+ NK cells kill iDCs while sparing mDCs.6
The possible contribution of activating KIR to DC lysis was not previously addressed. Our present data demonstrate that KIR2DS1 participate in the recognition and lysis of both iDCs and mDCs. However, while killing of iDCs is mainly dependent on signals delivered via NKp30 and DNAM1 (in line with previous reports32,33 ), lysis of mDCs is highly influenced by KIR2DS1 as revealed by the sharp inhibition of lysis by mAbs able to disrupt its interaction with HLA-C. This finding is likely to reflect the higher levels of HLA-class I expression on mDCs compared with iDCs.6 Thus, KIR2DS1+ NK cells may represent important alloeffectors to clear host mDCs, that is, the main source of APCs in priming donor's alloreactive T cells. Notably, in respect to “classic” alloreactive NK cells, KIR2DS1+ NK cells may have the advantage of killing mDC, in spite of the coexpression of inhibitory receptors engaged by HLA class I molecules at the mDC's cell surface.
As recently shown, alloreactive NK cells can acquire CCR7 after interaction with DC45 ; this capability would allow KIR2DS1+ NK cells to reach lymph nodes, that is, the site where donor's T cells can be activated by host mDCs and become responsible for GVH reactions.
In haplo-HSCT, the presence and persistence over time of alloreactive NK cells in the recipient is documented by different studies.3,20 Importantly, these cells display cytolytic activity even though they express inhibitory KIRs that are not engaged by HLA class I alleles of the patient. This finding is relevant, because during maturation NK cells require recognition of MHC class I molecules to acquire full effector function (“licensing” signal).19 Accordingly, in this case, the lack of this interaction should result in hyporesponsiveness rather than in the acquisition of cytolytic function. Both the generation and persistence of alloeffector cells in haplo-HSCT can be explained by the high doses (> 106 cells/kg) of HSCs infused into the patient.46 Thus, the hematopoietic microenvironment in which NK cells undergo maturation in the bone marrow is predominantly of donor type. Accordingly, donor NK cells develop from infused HSCs and acquire cytolytic activity against host hematopoietic target cells.
Several experimental evidences have previously shown that KIR2DS1 may induce NK alloreactivity. However in most studies KIR2DS1+ NK cells were analyzed against EBV cell lines transfected or not with HLA-C2 whereas no information is available regarding their reactivity against normal allogeneic cells expressing physiologic levels of HLA-C molecules such as DCs or T-cell blasts. Early studies from our laboratory showed that KIR2DS1 could induce potent NK cell–mediated cytotoxicity against HLA-Cw4+ EBV-transformed target cells.24 Additional reports showed that KIR2DS1 can recognize different alleles belonging to the C2 specificity expressed on EBV cell lines25-27,47 thus confirming and extending this concept. A recent study by Pende et al20 provided direct evidence that KIR2DS1 may play an important role also in the lysis of C2/C2+ blasts in pediatric leukemia. These data support the notion of a substantial clinical relevance of KIR2DS1 in mediating GVL effects.20 Based on these data it will be important to accurately verify the possible impact of KIR2DS1 in patients receiving allogeneic HSCT. In this context, a direct evidence for the clinical relevance of activating KIRs in HSCT was previously provided by Venstrom et al who showed that donor KIR3DS1 was associated with lower-grade II-IV acute GVHD in patients receiving unrelated allogeneic HSCT.48 Interestingly, Verheyden et al found that the presence of 2 activating KIRs, 2DS1 and 2DS2, in the donor genotype was significantly associated with a decreased leukemic relapse rate.49
A recent study by Fauriat et al,29 analyzed in detail the process of NK-cell education/licensing via KIR2DS1. They showed that in donors homozygous for C2 this type of education can tune down the responsiveness of NK cells to stimulation by target cells. This tuning was detectable in KIR2DS1+ NK cells lacking inhibitory KIRs and CD94/NKG2A, and in KIR2DS1+ NK cells coexpressing either CD94/NKG2A or KIR2DL3, but not in KIRD2S1+ NK cells coexpressing KIR2DL1.
In our present study, we used a set of phenotypically different KIR2DS1+ NK clones derived from a C1/C1 donor. In this case, KIR2DS1+ NK cells were educated (in a C2-negative environment) via NKG2A or additional inhibitory KIRs coexpressed at the cell surface together with KIR2DS1. Indeed, these clones were strongly cytolytic and activated by C2 cell transfectants as well as by C2 DCs or T-cell blasts. Interestingly, in line with the results of Fauriat et al,29 additional KIR2DS1+ NK clones derived from C1/C2 donors were characterized by a type of education similar to that of C1/C1 individuals as these cells were highly cytolytic and expressed a fully functional C2-specific activating receptor (not shown). In this context, the KIR2DS1+ NK clones analyzed by Pende et al after HSCT displayed strong KIR2DS1-dependent cytolytic activity against patient-derived C2/C2 leukemic blasts.20 These data further emphasize that NK-cell education after haplo-HSCT is mediated by a microenvironment of donor type; otherwise, according to Fauriat et al the function of KIR2DS1 would be down-modulated by the C2 homozygous environment.29
The potential importance of KIR2DS1 in haplo-HSCT was also underlined by experiments in which the various NK-cell clones analyzed were assessed for their ability to kill C2/C2 or C1/C2 T-cell blasts. In early studies, T-cell blasts were extensively used as cellular targets for the definition of different groups of alloreactive NK cells expressing inhibitory KIRs.44,50 On the other hand, their susceptibility to NK clones expressing activating KIRs was not further addressed. Our present data demonstrate that in most instances, their susceptibility to lysis is comparable with that of DCs. Remarkably, the contribution of KIR2DS1 to NK cell–mediated lysis of T-cell blasts appears even higher than that detected in mDC lysis (as shown by the blocking effect of specific mAbs on lysis).
Altogether, these data suggest that in KIR/KIR ligand mismatched haplo-HSCT a remarkable advantage exists in selecting KIR2DS1+ donors carrying either the C1 homozygous (recipient C1/C2) or the C1/C2 heterozygous (recipient C2/C2) phenotype. This advantage is related to the ability of KIR2DS1 to increase the fraction of alloreactive NK cells. These alloeffectors can mediate not only antileukemic but also anti-GVHD and anti-HVG activity, thanks to the potent cytotoxicity exerted against mDCs and T-cell blasts.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the equipe of Immunohematology and Transfusion Center, S. Martino Hospital (Genova, Italy) for helpfulness and efficient collaboration.
This work was supported by grants awarded by Associazione Italiana Ricerca sul Cancro: MFAG project n. 6383 (S.S.), IG project n. 4725 (L.M.), IG project n. 10 643 (A.M.), and Special Project 5 × 1000 n. 9962 (A.M., L.M.); Ministero dell'Istruzione, Università e Ricerca: MIUR-FIRB 2003 project RBLA039LSF-001/003 (L.M. and A.M.); Ministero della Salute: RF2006-Ricerca Oncologica-Project of Integrated Program 2006-08, agreements n. RO strategici 3/07 (L.M., A.M.) and Ricerca Finalizzata 2007 (M.F.). S.C. is the recipient of a fellowship awarded by FIRC.
Authorship
Contribution: S.S., S.C., and M.F. designed and performed research, interpreted data, and wrote the paper; E.R. performed research; L.M. revised the paper; and A.M. designed research, interpreted data, and wrote the paper.
Conflict-of-interest disclosure: A.M. is founder and shareholder of Innate-Pharma (Marseille, France). The remaining authors declare no competing financial interests.
Correspondence: Alessandro Moretta, MD, Dipartimento di Medicina Sperimentale, Sezione di Istologia, Via G.B. Marsano 10, 16132 Genova, Italy; e-mail: alemoret@unige.it.
References
Author notes
S.S. and S.C. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal