Abstract
MicroRNAs (miRNAs) are small, noncoding RNAs that regulate gene expression by sequence-specific targeting of multiple mRNAs. Although lineage-, maturation-, and disease-specific miRNA expression has been described, miRNA-dependent phenotypes and miRNA-regulated signaling in hematopoietic cells are largely unknown. Combining functional genomics, biochemical analysis, and unbiased and hypothesis-driven miRNA target prediction, we show that lentivirally over-expressed miR-125b blocks G-CSF–induced granulocytic differentiation and enables G-CSF–dependent proliferation of murine 32D cells. In primary lineage-negative cells, miR-125b over-expression enhances colony-formation in vitro and promotes myelopoiesis in mouse bone marrow chimeras. We identified Stat3 and confirmed Bak1 as miR-125b target genes with approximately 30% and 50% reduction in protein expression, respectively. However, gene-specific RNAi reveals that this reduction, alone and in combination, is not sufficient to block G-CSF–dependent differentiation. STAT3 protein expression, DNA-binding, and transcriptional activity but not induction of tyrosine-phosphorylation and nuclear translocation are reduced upon enforced miR-125b expression, indicating miR-125b–mediated reduction of one or more STAT3 cofactors. Indeed, we identified c-Jun and Jund as potential miR-125b targets and demonstrated reduced protein expression in 32D/miR-125b cells. Interestingly, gene-specific silencing of JUND but not c-JUN partially mimics the miR-125b over-expression phenotype. These data demonstrate coordinated regulation of several signaling pathways by miR-125b linked to distinct phenotypes in myeloid cells.
Introduction
MicroRNAs (miRNAs) represent an emerging class of noncoding single-stranded RNAs of approximately 22 nucleotides1,2 that play an important role in posttranscriptional regulation of gene expression. miRNAs are processed from primary pri-miRNA transcripts to pre-miRNAs and mature miRNAs in a multistep process. Mature miRNAs are incorporated into and subsequently recruit a multi–protein effector complex RISC (RNA-induced silencing complex) to complementary miRNA-binding sites located preferentially within the 3′UTR of target mRNAs. Sequence-specific binding of RISC results in reduced mRNA translation and/or degradation through RNA interference (RNAi).3 The interaction between a miRNA and its target mRNAs usually requires complementarity only within so-called seed sequence (miRNA nucleotides 2-8). Hence, a single miRNA has the potential to regulate hundreds of proteins4-6 but resulting target protein repression is typically relatively mild.5,6 Thereby, the ratio of regulatory RNAs to target mRNAs may modulate the silencing activity with a negative correlation between target abundance and target down-regulation.7 To identify miRNA target genes, several prediction programs based on the hybridization energy of complementary miRNA/mRNAs sequences have been described.8-12 However, the use of these programs is error-prone and identification of miRNA targets still remains a fundamental task in miRNA-biology.
Lineage- and maturation-specific miRNA expression in hematopoietic cells was initially described by Chen and colleagues.13 In addition, aberrant miRNA expression has been reported in many diseases including solid and hematologic malignancies.14-17 Notably, miR-125b was reported to be up-regulated because of the chromosomal translocation t(2;11)(p21;q23) in myelodysplastic syndrome/acute myeloid leukemia (MDS/AML)18 and megakaryoblastic leukemia related to Down syndrome (DS-AMKL).19 In hematopoietic cells, however, miRNA-dependent phenotypes, underlying molecular mechanisms, and miRNA-regulated target genes are still largely unknown.
In the present work we analyzed miRNA-dependent phenotypes in a cell-culture model of G-CSF–induced granulocytic differentiation by combining bioinformatic target prediction,8-12 functional genetics, and biochemical analysis of potential candidate gene products. Initially, we used murine IL-3 dependent 32D cells to screen for miRNAs differentially expressed upon G-CSF treatment. Subsequent functional studies of gain- and loss-of-function phenotypes for selected miRNAs revealed distinct phenotypes, including a block of granulocytic differentiation and induction of G-CSF–dependent proliferation, only upon enforced expression of miR-125b. In addition, enforced miR-125b expression in bone marrow (BM) lineage-negative cells modulates colony formation in vitro, and supports myeloid cell development in mouse BM chimeras.
Several miRNA target prediction programs identified Bak1 and Stat3 as putative targets of miR-125b, and we confirmed Bak120,21 and, for the first time, Stat3 as direct targets of miR-125b. To study the functional contribution of each target gene to miR-125b–mediated phenotypes, we used gene-specific RNAi to reduce protein expression to similar levels as observed in the presence of enforced miR-125b expression. Depending on target down-regulation, gene-specific RNAi can mimic some but not all phenotypes observed upon miR-125b over-expression. Subsequent biochemical analyses of STAT3 activation and function suggested that miR-125b may also target a cofactor required for STAT3 DNA-binding and transcriptional activity. c-JUN is known to bind STAT322,23 and we identified miR-125b binding sites within the CDS and UTR regions of c-Jun and Jund. Furthermore, we demonstrated reduced protein expression of both genes in 32D cells upon enforced miR-125b expression. Interestingly, silencing of JUND but not c-JUN partially mimics the miR-125b over-expression phenotype in 32D cells. These data demonstrate that aberrant miRNA expression can induce robust disease-specific phenotypes in hematopoietic cells by targeting multiple signaling pathways in a coordinated manner. In addition, the combination of functional genetics, biochemical analysis and modified miRNA target prediction may help to identify new miRNA-regulated target genes. In this context, target down-regulation by miRNAs and gene-specific RNAi has to be adjusted to correctly define the contribution of individual gene products to miRNA-mediated phenotypes.
Methods
Cells, cell culture, and cytokines
Parental and transgenic 32D cells were maintained in RPMI 1640 (Gibco) with 10% FCS, 10% WEHI-3 conditioned media as a source of IL-3,24 and 1% penicillin/streptomycin (PS). 293T and NIH3T3 cells were cultured in DMEM (Gibco) with 10% FCS and 1% PS. Primary murine progenitor Lin− c-Kit+/Sca-1+ cells were isolated from bone marrow of 8- to 12-week-old C57BL/6 mice (Charles River Laboratories) and purified using a mouse lineage cell depletion panel (Miltenyi Biotec) according to the manufacturer's instructions. Subsequently, Lin− cells were cultured in serum-free expansion media (SFEM; Stemspan) supplemented with mSCF (100 ng/mL; CellSystems), mIL-3 (50 ng/mL; CellSystems) and mIL-6 (50 ng/mL; PromoKine) for 24h at 37°C in 5% CO2 before exposure to lentiviral particles.
G-CSF–induced granulocytic differentiation and growth factor deprivation
For granulocytic differentiation, 32D cells were washed 3 times with PBS (Gibco), resuspended in cytokine-free media and stimulated with hG-CSF (Lenograstim) at a final concentration of 50 ng/mL. For growth factor withdrawal, 32D cells were washed 3 times with PBS and maintained in the cytokine-free media. The number of viable cells was determined by trypan blue exclusion and flow cytometric analysis of APC-conjugated annexin-V (BD Pharmingen) and propidium iodide (PI) staining using BD FACSCanto II (BD Bioscience) and FlowJo Version 8.0 software. Cell morphology was evaluated by May-Grunwald-Giemsa staining.
DNA constructs, oligonucleotides, and site-directed mutagenesis
Chemically synthesized, self-complementary DNA oligonucleotides (BioSpring) encompassing the sequence of murine pre-miR-125b-1 and pre-miR-125b-2 (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) were cloned as previously described.25 S-miR30-IEW vector served as a control. All miRNA-lentiviral vectors encode eGFP (enhanced green fluorescent protein) as a reporter gene.
Anti-STAT3 shRNA (5′–CCAATTGTGATGCCTCCTT–3′),26 anti-BAK1 shRNA (5′–TGCCTACGAACTCTTCACCTT–3′),27 anti-JUN shRNAs (5′-GATGGAAACGACCTTCTA-3′ and 5′-CAGTAACCCTAAGATCCTA-3′ referred to as shRNA#1 and shRNA#2, respectively)28,29 lentiviral transgene plasmids pdcH1-shRNA-SR and pdcH1-Gl4-SR (shRNA-control) were cloned as previously described.30 All shRNA-lentiviral constructs encode RFP (red fluorescent protein) as a reporter gene.
Chemically synthesized, self-complementary DNA oligonucleotides (BioSpring) encompassing the sequence of murine Stat3 3′UTRs (supplemental Table 1) with overhang sequences from 5‘XbaI- and 3‘XbaI- restriction sites were hybridized and directionally cloned in the XbaI site of pGL3-Promoter vector (Promega), downstream of the Firefly luciferase open reading frame, thus generating reporter vectors wt-1 and wt-2 corresponding to pGL3-STAT3-3′UTR(1) and pGL3-STAT3-3′UTR(2), respectively. To generate reporter vectors with deletion of putative binding sites for miR-125b (referred as mut-1 and mut-2), site-directed mutagenesis (Stratagene) and overlapping primers listed in supplemental Table 2 (bottom panel) were used.
Lentiviral transduction
Lentiviral particles were produced and titrated, and lentiviral transductions were performed as previously described.25 Subsequently, shRNA-control and shRNA-STAT3 cells with low and high RFP fluorescence were sorted using BD FACSAria II (BD Biosciences). Murine primary Lin− cells were transduced twice in SFEM medium supplemented with mSCF (100 ng/mL), mIL-3 (50 ng/mL), mIL-6 (50 ng/mL) and 1x protamine sulfate. Forty-eight-hours posttransduction cells were either used for transplantation experiments or sorted (BD FACSAriaII) GFP+ and GFP− fractions were subjected to clonogenic assay.
Mice
B6.SJL-Ptprca Pepcb/BoyJ mice (C57BL/6 mice carrying the CD45.1 allele) were purchased from The Jackson Laboratory and crossed to C57BL/6 mice (Charles River Laboratories). (C57BL/6J x B6.SJL-Ptprca Pepcb/BoyJ)F1 mice (CD45.1/CD45.2 heterozygous) were bred at the animal facility of Hannover Medical School. Animals were maintained under specific-pathogen-free conditions. All animal experiments were conducted in accordance with local and institutional guidelines (Nds. Landesamt f. Verbraucherschutz und Lebensmittelsicherheit).
Bone marrow chimeras
After lentiviral transduction, 2 × 105 Lin− BM cells from C57BL/6 mice (CD45.2) were transferred intravenously into (C57BL/6 x B6.SJL-Ptprca Pepcb/BoyJ)F1 mice (CD45.1/CD45.2) that were lethally irradiated (8 Gy). Bone marrow, spleen, and thymus of recipient mice were isolated 10 weeks after transfer and analyzed by flow cytometry.
Antibodies and flow cytometry
Monoclonal antibodies specific for CD11c (N418), GR1 (RB6-8C5), erythroid cell marker (Ter-119), CD19 (1D3), CD11b (M1/70), NK1.1 (PK136), TCRβ (H57-597), CD45.1 (A20), CD117 (2B8, ACK2), Sca-1 (E13-161.7), CD135 (A2F10), CD11c (N418), were used purified or as biotin, PacificBlue, eFluor450, fluorescein isothiocyanate (FITC), Alexa488, phycoerythrin (PE), peridinin chlorophyll protein-Cy5.5 (PerCP-Cy5.5), PE-Cy7, allophycocyanin (APC), APC-Cy7, or APC-Alexa750 conjugates. Antibodies were purified from hybridoma supernatants or purchased from eBioscience, BD Biosciences or Biolegend. PE-Cy7 conjugated streptavidin (BD) was used to reveal staining with biotinylated mAb. Flow cytometric analysis was performed on a BD LSRII (BD) and data were analyzed with FACSDiva software Version 6.1.2 (BD). Lin− cells were isolated from total BM by staining cell suspensions with a lineage-specific antibody cocktail (anti-TCRβ, anti-CD19, anti-CD11b, anti–Gr-1, Ter-119, and DX5) followed by incubation with anti–rat-IgG-conjugated magnetic beads (Dynal; Invitrogen) and magnetic bead depletion of mature lineages.
Microscopy
Microscopy of cytospins and clonogenic assays was performed with an Olympus BX51 (Olympus) and a Nikon Eclipse TE300 (Nikon) microscopes, respectively. Digital images of cytospins and clonogenic assays were taken with an Olympus XC50 (Olympus) and a Basler A113C (Basler) cameras, archived, and processed with software packages CellF̂1244 (Olympus) and LuciaG 4.60 (Nikon), respectively.
Clonogenic assays
Lin− cells (1 × 105) per plate were cultured in methylcellulose (MethoCult M3234; StemCell Technologies) supplemented with 15% FBS (StemCell Technologies), 1% PS (Gibco) and 10 ng/mL mG-CSF (CellSystems). Lin−/GFP+ cells isolated from mouse chimeras were polled and 1 × 103 cells were plated in methylcellulose (MethoCult M3434; StemCell Technologies) supplemented with 15% FBS (StemCell Technologies), 1% PS (Gibco). Colonies were evaluated microscopically 10 days after plating by standard criteria.
Quantitative real time-PCR
RNA was extracted from cells using Trizol reagent (Invitrogen). For miR-qRT-PCR, total RNA was reverse-transcribed and subjected to Taqman miRNA Assay (Applied Biosystems) following the manufacturer's protocol using miRNA-specific primers and probes (sno-202 served as an internal control). For qRT-PCR of mRNAs, cDNA synthesis was performed with 1 μg of total RNA digested with DNaseI and subjected to SYBR green (Applied Biosystems) and TaqMan-based (Applied Biosystems) gene expression profiling following the manufacturer's protocol. Sequences of the primers and probes are listed in supplemental Table 2 (top panel), except for Hprt (Mm01545399_m1) and c-Jun (Mm00495062_s1*) primer/probe assays purchased from Applied Biosystems. Differential gene expression between conditions is expressed as ΔΔCT that corresponds to the log2 fold difference in mRNA or miRNA levels between the conditions compared. Real-time PCR was performed using an ABI7500 cycler (Applied Biosystems).
Luciferase reporter assay
NIH3T3 cells stably expressing miR-125b were cotransfected with pGL3-Promoter (Promega) empty vector or pGL3-STAT3-3′UTR (wt-1) or pGL3-STAT3-3′UTR (wt-2) reporter vectors together with pRL-SV40 using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. The luciferase activities were measured 36 hours after transfection using Dual-Luciferase Reporter Assay System (Promega) and a Mithras LB 940 (Berthold Technologies) luminometer. The same protocol was applied for mutated variants of pGL3-STAT3-3′UTR (mut-1 and mut-2) with deleted miR-125b-seed sequence.
Cell lysis and Western blotting
Cell extracts and Western blotting were performed as previously described.31 Primary antibodies (all rabbit) were obtained from Cell Signaling Technology: STAT3-pTyr705 (9145), STAT3 (4904), β-ACTIN (4970), BAK1 (3814), PUMA (4976); Santa Cruz Biotechnology: c-JUN (sc-45), JUND (sc-74); and eBioscience: BMF (14-6031). Horseradish peroxidase-conjugated anti–rabbit secondary antibody was purchased from Roche. To remove bound antibodies, membranes were incubated at room temperature for 1h in stripping buffer containing dH2O/10% glycine/1% HCl. Densitometric analysis of x-ray films was performed using VersaDoc 3000 Imaging system (Bio-Rad) and 1-D analysis software Quantity One Version 4.6.5 (Bio-Rad). The intensity ratio of the protein of interest band to the β-ACTIN band (loading control) was calculated to measure changes in protein levels. Value in the group of interest indicates level of the protein expression compared with the control group (protein expression in control group was set as 1).
Nuclear extracts and detection of STAT3 DNA-binding
32D cells were treated with G-CSF and nuclear extracts were prepared using Nuclear Extract Kit (Active Motif) following the manufacturer's protocol. STAT3 DNA-binding was measured and quantified using an ELISA-based transcription factor assay kit (STAT3 family) according to the manufacturer's instructions (Active Motif). The absorbance was measured using a Tecan Sunrise microplate reader.
Statistical analysis
Data are presented as mean values ± SEM. P values were obtained using the Student 2-sided t test. P values ≤ .05 were considered statistically significant.
Results
Several miRNAs are differentially expressed in 32D cells treated with G-CSF but only over-expression of miR-125b blocks granulocytic differentiation
To identify miRNAs involved in granulocytic differentiation, we used 32D progenitor cells that differentiate into mature granulocytes upon G-CSF treatment.32 In preliminary experiments, a miRNA-specific quantitative real-time PCR (miR-qRT-PCR) was used to quantify miRNA expression at different time points of G-CSF treatment (data not shown). Among differentially expressed miRNAs, miR-34a, 34b, 34c, 125b, 155, 181b, 223, 291a, and 370 were selected for further functional analysis. miRNAs and/or miRNA-complementary sequences, so called antagomirs, were expressed after lentiviral transduction,25,33 and morphology of transduced 32D cells cultured in the presence of IL-3 or G-CSF was analyzed (data not shown). We found that only over-expression of miR-125b affected granulocytic differentiation. Thus, this miRNA was selected for further analysis.
Over-expression of miR-125b blocks granulocytic differentiation, enables G-CSF–dependent cell proliferation, and delays cytokine withdrawal–induced cell death of 32D cells
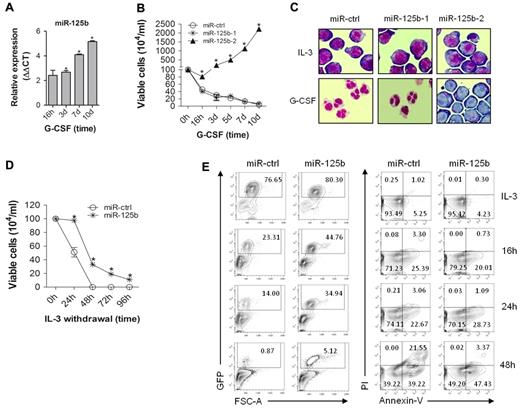
miR-125b is expressed at very low level in 32D cells cultured in the presence of IL-3 but its expression gradually increases up to 32-fold following G-CSF treatment (Figure 1A). Given that miR-125b can be processed from 2 different precursors located on separate chromosomes, 32D cells were transduced with lentiviral vectors encoding pre-miR-125b-1 (murine chromosome 9) and pre-miR-125b-2 (murine chromosome 16) embedded within identical heterologous miR-30 sequences (supplemental Figure 1A). miR-125b expression was significantly higher in 32D/miR-125b-2 compared with 32D/miR-125b-1 cells, 4 and 14 days after lentiviral transduction, despite of similar GFP reporter gene expression (supplemental Figure 1B-C). In addition, 32D/miR-125b-2 but not 32D/miR-125b-1 cells survive and proliferate in the presence of G-CSF (Figure 1B). Morphologic analysis revealed a complete block of granulocytic differentiation in the presence of G-CSF only in 32D/miR-125b-2 but not in 32D/miR-125b-1 or 32D/miR-ctrl cells (Figure 1C). Therefore, we focused only on 32D/miR-125b-2 cells (hereafter referred to as 32D/miR-125b cells) in the subsequent studies. In addition, over-expression of miR-125b confers a survival advantage upon growth factor depletion as analyzed by trypan blue (Figure 1D) and monitoring of GFP+ cells stained for annexin-V and propidium iodide (PI; Figure 1E).
Enforced expression of miR-125b blocks granulocytic differentiation, enables G-CSF–dependent cell proliferation, and delays cytokine withdrawal–induced cell death. (A) miR-qRT-PCR of mature miR-125b level at the indicated time points; sno-202 served as an endogenous control. (B) Proliferation kinetics of the 32D cells expressing miR-ctrl (circle), miR-125b-1 (star), and miR-125b-2 (triangle) in G-CSF-supplemented suspension cultures. (C) Morphology of 32D derivates (May-Grunwald-Giemsa staining) on day 7 of IL-3 (top panel) and G-CSF (bottom panel) supplemented cell cultures. A 60× magnification of a representative field is shown. (D) Viability of 32D/miR-ctrl and 32D/miR-125b cells after IL-3 deprivation analyzed by trypan blue dye exclusion, and (E) monitoring of GFP+ population (left panel), and propidium iodide (PI) and annexin-V stained GFP+ fraction (right panel). Mean of at least 3 (panels A,B,D) and representative experiments are shown, respectively. * P < .05 compared with control at corresponding time point of treatment.
Enforced expression of miR-125b blocks granulocytic differentiation, enables G-CSF–dependent cell proliferation, and delays cytokine withdrawal–induced cell death. (A) miR-qRT-PCR of mature miR-125b level at the indicated time points; sno-202 served as an endogenous control. (B) Proliferation kinetics of the 32D cells expressing miR-ctrl (circle), miR-125b-1 (star), and miR-125b-2 (triangle) in G-CSF-supplemented suspension cultures. (C) Morphology of 32D derivates (May-Grunwald-Giemsa staining) on day 7 of IL-3 (top panel) and G-CSF (bottom panel) supplemented cell cultures. A 60× magnification of a representative field is shown. (D) Viability of 32D/miR-ctrl and 32D/miR-125b cells after IL-3 deprivation analyzed by trypan blue dye exclusion, and (E) monitoring of GFP+ population (left panel), and propidium iodide (PI) and annexin-V stained GFP+ fraction (right panel). Mean of at least 3 (panels A,B,D) and representative experiments are shown, respectively. * P < .05 compared with control at corresponding time point of treatment.
Enforced miR-125b expression modulates granulocytic differentiation of Lin− bone marrow cells in vitro and promotes development of myeloid cells in vivo
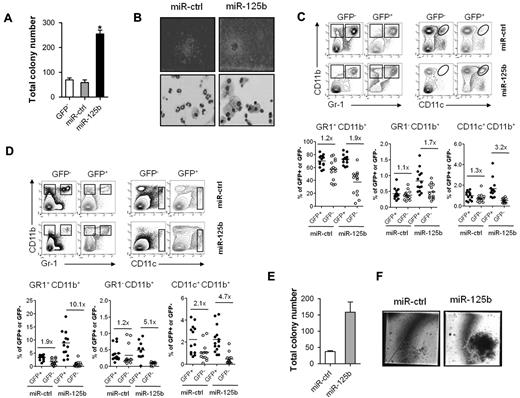
To study the effects of miR-125b over-expression in primary cells, purified Lin− c-Kit+/Sca-1+ cells were lentivirally transduced and subsequently sorted into GFP+ and GFP− fractions (supplemental Figure 2A). In G-CSF-supplemented clonogenic assays, Lin−/miR-125b cells generate more and greater colonies than control cells (Figure 2A and B top panel). In addition, colonies in control cultures consist mostly of mature neutrophils, whereas in miR-125b cultures colonies almost exclusively contain macrophages (Figure 2B bottom panel).
Enforced expression of miR-125b modulates granulocytic differentiation of murine Lin− cells in vitro and promotes myeloid cell development in vivo. (A) Colony formation by 1 × 105 Lin−/GFP−, Lin−/miR-ctrl and Lin−/miR-125b cells in methylcellulose cultures supplemented with G-CSF. (B) Microscopic images of colonies (top panel) and morphology of cells isolated from corresponding clonogenic assays (bottom panel); 4× and 60× magnifications of a representative field are shown, respectively. Analysis of (C) BM and (D) spleen cells of 10-week BM chimeras. The top panels show representative density plots of donor-derived cells stained for GR-1, CD11b, and CD11c and electronically gated on GFP− (left) and GFP+ (right) cells. In the bottom panels, lines indicate mean percentage of GFP− and GFP+ cells according to gates indicated in the corresponding top panels. Each dot represents an individual mouse. Data are pooled from 2 independent experiments. (E) Colony formation by Lin− cells isolated from BM chimeras described in panels C and D. 1 × 103 Lin−/miR-ctrl or Lin−/miR-125b cells (1 × 103) were cultured in methylcellulose containing SCF, IL-3, IL-6, and Epo. (F) Microscopic images of colonies from the experiment described in panel E; a 60× magnification of the representative field is shown. Mean of 4 (panel A), 2 (panels C,D,E) and representative experiments are shown, respectively. *P < .05 compared with Lin−/miR-ctrl cells.
Enforced expression of miR-125b modulates granulocytic differentiation of murine Lin− cells in vitro and promotes myeloid cell development in vivo. (A) Colony formation by 1 × 105 Lin−/GFP−, Lin−/miR-ctrl and Lin−/miR-125b cells in methylcellulose cultures supplemented with G-CSF. (B) Microscopic images of colonies (top panel) and morphology of cells isolated from corresponding clonogenic assays (bottom panel); 4× and 60× magnifications of a representative field are shown, respectively. Analysis of (C) BM and (D) spleen cells of 10-week BM chimeras. The top panels show representative density plots of donor-derived cells stained for GR-1, CD11b, and CD11c and electronically gated on GFP− (left) and GFP+ (right) cells. In the bottom panels, lines indicate mean percentage of GFP− and GFP+ cells according to gates indicated in the corresponding top panels. Each dot represents an individual mouse. Data are pooled from 2 independent experiments. (E) Colony formation by Lin− cells isolated from BM chimeras described in panels C and D. 1 × 103 Lin−/miR-ctrl or Lin−/miR-125b cells (1 × 103) were cultured in methylcellulose containing SCF, IL-3, IL-6, and Epo. (F) Microscopic images of colonies from the experiment described in panel E; a 60× magnification of the representative field is shown. Mean of 4 (panel A), 2 (panels C,D,E) and representative experiments are shown, respectively. *P < .05 compared with Lin−/miR-ctrl cells.
To assess the function of miR-125b over-expression in vivo, we performed competitive repopulation experiments to generate chimeric mice over-expressing either miR-ctrl or miR-125b in hematopoietic cells. Equal numbers of lentivirally transduced and nontransduced Lin− cells were transplanted into lethally irradiated recipients (supplemental Figure 2B). Ten weeks after transplantation, monocytes/macrophages (GR1−CD11b+), granulocytes (GR1+CD11b+), and dendritic cells (CD11c+CD11b+) but not lymphoid cells over-expressing miR-125b exhibited a competitive advantage over nontransduced cells as demonstrated by the ratios of control transduced versus nontransduced cells (Figure 2C-D, and supplemental Figure 2C). Moreover, this effect was more pronounced in spleen than in bone marrow. In addition, Lin−/miR-125b cells isolated from bone marrow 10 weeks after transplantation generated more and larger colonies compared with Lin−/miR-ctrl cells (Figure 2E-F, and supplemental Figure 2D). The enhanced engraftment and colony formation may suggest that miR-125b over-expression confers a survival and/or proliferative advantage to myeloid cells.
Over-expression of miR-125b reduces transcription of granulocytic genes
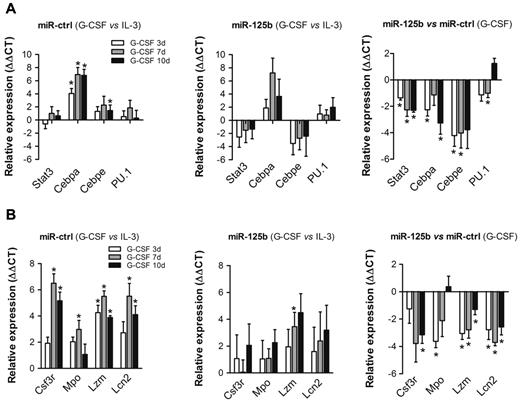
We next quantified mRNA expression of granulocyte-related transcription factors in 32D cells upon over-expression of miR-125b. G-CSF treatment significantly induces mRNA expression of Cebpa and Cebpe in 32D/miR-ctrl cells, whereas Stat3 and PU.1 levels are only slightly influenced compared with cultures supplemented with IL-3 (Figure 3A left panel). In contrast, transcription of Cebpa is slightly, and transcription of Stat3 and Cebpe is markedly reduced in 32D/miR-125b cells on G-CSF treatment compared with controls (Figure 3A middle and right panels). This may indicate that Cebpa is not sufficiently expressed or a cofactor is missing to induce Cebpe transcription. Beside transcription factors, G-CSF–mediated expression of granulocyte-colony stimulating factor receptor (Csf3r) and components of early and late granules, such as myeloperoxidase (Mpo), lysozyme-2 (Lzm) and lipocalin-2 (Lcn2) is induced by G-CSF in 32D/miR-ctrl (Figure 3B left panel). On the contrary, transcription of the aforementioned genes is reduced in 32D/miR-125b cells compared with controls (Figure 3B middle and right panels).
Over-expression of miR-125b changes the transcriptional profile of genes related to granulocytic differentiation. Transcriptional profiling of (A) myeloid transcription factors and (B) primary and secondary granule components in 32D/miR-ctrl cells (left panels compare G-CSF vs IL-3 conditions), 32D/miR-125b cells (middle panels compare G-CSF vs IL-3 conditions), and the fold difference between 32D/miR-ctrl and 32D/miR-125b cells in the presence of G-CSF (right panels compare 32D/miR-125b vs 32D/miR-ctrl) at the indicated time intervals. Hprt served as an endogenous control. Mean of 4 experiments is shown. *P < .05 compared with corresponding control at indicated time point of treatment.
Over-expression of miR-125b changes the transcriptional profile of genes related to granulocytic differentiation. Transcriptional profiling of (A) myeloid transcription factors and (B) primary and secondary granule components in 32D/miR-ctrl cells (left panels compare G-CSF vs IL-3 conditions), 32D/miR-125b cells (middle panels compare G-CSF vs IL-3 conditions), and the fold difference between 32D/miR-ctrl and 32D/miR-125b cells in the presence of G-CSF (right panels compare 32D/miR-125b vs 32D/miR-ctrl) at the indicated time intervals. Hprt served as an endogenous control. Mean of 4 experiments is shown. *P < .05 compared with corresponding control at indicated time point of treatment.
BAK1 and STAT3 are direct targets of miR-125b
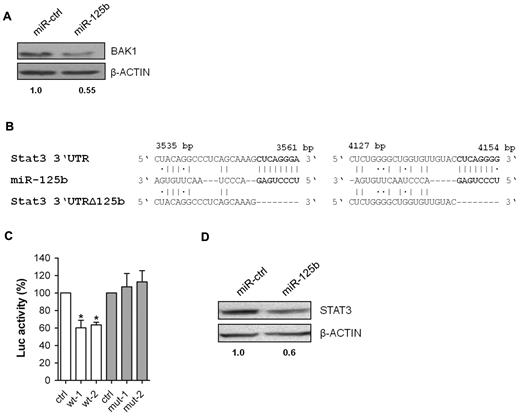
We used the prediction programs DIANA microT v 0.3,8 miRDB,9,10 Target Scan12 and RNA2211 to identify potential miR-125b target genes. Among approximately 500 predicted genes, we initially focused on genes potentially related to the observed phenotypes (supplemental Table 3). Members of the Bcl2 family, such as Bak1, Bmf and PUMA are predicted and/or validated targets for miR-125b.20,21,34 However, we failed to detect repression of BMF and PUMA in 32D/miR-125b cells (data not shown). In contrast, BAK1 protein expression was reduced by approximately 50% in 32D/miR-125b compared with 32D/miR-ctrl cells (Figure 4A).
BAK1 and STAT3 are direct targets of miR-125b. (A) Western blotting of BAK1 in 32D/miR-ctrl and 32D/miR-125b cells. (B) Two predicted miR-125b target sites within the 3′UTR of Stat3 (NM_011486.4), their deleted derivates, and the miRNA/mRNA pairing regions. (C) Luciferase activity resulting from transfection of NIH3T3/miR-125b cells with reporter plasmids containing wild-type (wt-1 and wt-2) or mutated (mut-1 and mut-2) 3′UTR fragments of Stat3. Firefly luciferase luminescence, normalized to Renilla luciferase, is presented, (n = 5). (D) Western blotting of STAT3 in 32D/miR-ctrl and 32D/miR-125b cells. A representative experiment out of 4 is shown (except for panel C). *P < .05 compared with control.
BAK1 and STAT3 are direct targets of miR-125b. (A) Western blotting of BAK1 in 32D/miR-ctrl and 32D/miR-125b cells. (B) Two predicted miR-125b target sites within the 3′UTR of Stat3 (NM_011486.4), their deleted derivates, and the miRNA/mRNA pairing regions. (C) Luciferase activity resulting from transfection of NIH3T3/miR-125b cells with reporter plasmids containing wild-type (wt-1 and wt-2) or mutated (mut-1 and mut-2) 3′UTR fragments of Stat3. Firefly luciferase luminescence, normalized to Renilla luciferase, is presented, (n = 5). (D) Western blotting of STAT3 in 32D/miR-ctrl and 32D/miR-125b cells. A representative experiment out of 4 is shown (except for panel C). *P < .05 compared with control.
In addition, we identified 2 potential miR-125b-binding sites within the 3′UTR of Stat3 (Figure 4B), a transcription factor strongly involved in granulocytic differentiation.35,36 To demonstrate direct regulation by miR-125b, luciferase reporter containing wild-type Stat3 3′UTR sequences or their mutant-derivates with deletion of the putative miR-125b-binding sites were transfected into NIH3T3 cells stably over-expressing miR-125b (NIH3T3/miR-125b; Figure 4B and supplemental Figure 3A). As shown in Figure 4C, miR-125b represses luciferase activity by approximately 40% depending on the presence of miR-125b binding in the Stat3 3′UTR. Finally, Western blotting revealed an approximately 30%-40% reduction of STAT3 protein expression in 32D/miR-125b compared with 32D/miR-ctrl cells (Figure 4D).
Repression of BAK1 and STAT3 may mimic miR-125b–induced phenotypes in a dose-dependent manner
To characterize the functional role of BAK1 and STAT3 in miR-125b–induced phenotypes, gene-specific short hairpin RNAs (shRNAs) were lentivirally over-expressed in 32D cells (supplemental Figure 3B). As depicted in Figure 5A (top panel), anti-BAK1 specific shRNA reduced BAK1 expression a little stronger as achieved by over-expression of miR-125b. 32D/shRNA-BAK1 cells display a delay in cell death upon G-CSF treatment compared with 32D/shRNA-ctrl cells but G-CSF–induced differentiation is not detectably blocked (Figure 5A bottom panel). Furthermore, 32D/shRNA-STAT3 and 32D/shRNA-ctrl cells were sorted according to different levels of reporter gene expression. As shown in Figure 5B (top panel), anti-STAT3 shRNA reduced STAT3 protein expression by approximately 35% and 86%, compared with 32D/shRNA-ctrl cells with an equivalent RFP expression, respectively. Interestingly, cell survival, proliferation and granulocytic differentiation in the presence of G-CSF clearly depend on the intracellular amount of STAT3 protein (Figures 5B bottom panel, and supplemental Figure 3C). Strong reduction of STAT3 expression enables G-CSF–dependent cell proliferation with a delay of some days and induces a complete block of differentiation. In contrast, a mild reduction of STAT3 protein expression, as induced by enforced miR-125b expression, results only in a slight delay of cell death but not in G-CSF–dependent proliferation or block of granulocytic differentiation.
Reduction of BAK1 and STAT3 expression may mimic miR-125b–induced phenotypes in a dose-dependent manner. (A) Western blotting (top panel) of BAK1 in 32D cells transduced with shRNA-ctrl and specific anti-BAK1 shRNA (shRNA-BAK1) lentiviruses and proliferation kinetics (bottom panel) of 32D cells expressing shRNA-ctrl (open circle) and shRNA-BAK1 (filled circle) during G-CSF treatment. (B) Western blotting (top panel) of STAT3 in 32D cells transduced with shRNA-ctrl and specific anti-STAT3 shRNA (shRNA-STAT3) lentiviruses with comparable RFP expression and proliferation kinetics (bottom panel) of 32D cells expressing shRNA-ctrl-low RFP+ (open circle), shRNA-STAT3-low RFP+ (filled circle), shRNA-ctrl-high RFP+ (open triangle), and shRNA-STAT3-high RFP+ (filled triangle) during G-CSF treatment. (C) Western blotting (top panel) of STAT3 and BAK1 in 32D cells transduced with shRNA-ctrl, shRNA-ctrl/BAK1 and shRNA-STAT3/BAK1 lentiviral vectors. Proliferation kinetics (bottom panel) of 32D/shRNA-ctrl (open circle) and 32D/shRNA-STAT3/BAK1 (filled circle) cells during G-CSF-treatment. All 32D/shRNA-STAT3 derivates were sorted for low RFP expression before lentiviral transduction with shRNA-BAK1. Mean of 4 experiments and representative Western blots are shown, respectively. *P < .05 compared with control.
Reduction of BAK1 and STAT3 expression may mimic miR-125b–induced phenotypes in a dose-dependent manner. (A) Western blotting (top panel) of BAK1 in 32D cells transduced with shRNA-ctrl and specific anti-BAK1 shRNA (shRNA-BAK1) lentiviruses and proliferation kinetics (bottom panel) of 32D cells expressing shRNA-ctrl (open circle) and shRNA-BAK1 (filled circle) during G-CSF treatment. (B) Western blotting (top panel) of STAT3 in 32D cells transduced with shRNA-ctrl and specific anti-STAT3 shRNA (shRNA-STAT3) lentiviruses with comparable RFP expression and proliferation kinetics (bottom panel) of 32D cells expressing shRNA-ctrl-low RFP+ (open circle), shRNA-STAT3-low RFP+ (filled circle), shRNA-ctrl-high RFP+ (open triangle), and shRNA-STAT3-high RFP+ (filled triangle) during G-CSF treatment. (C) Western blotting (top panel) of STAT3 and BAK1 in 32D cells transduced with shRNA-ctrl, shRNA-ctrl/BAK1 and shRNA-STAT3/BAK1 lentiviral vectors. Proliferation kinetics (bottom panel) of 32D/shRNA-ctrl (open circle) and 32D/shRNA-STAT3/BAK1 (filled circle) cells during G-CSF-treatment. All 32D/shRNA-STAT3 derivates were sorted for low RFP expression before lentiviral transduction with shRNA-BAK1. Mean of 4 experiments and representative Western blots are shown, respectively. *P < .05 compared with control.
Interestingly, reduction of both BAK1 and STAT3 by combinatorial RNAi to similar levels as observed in the presence of enforced miR-125b expression can delay cell death but does not block granulocytic differentiation in the presence of G-CSF (Figure 5C).
Over-expression of miR-125b affects DNA-binding of STAT3 but not induction of tyrosine phosphorylation and nuclear translocation
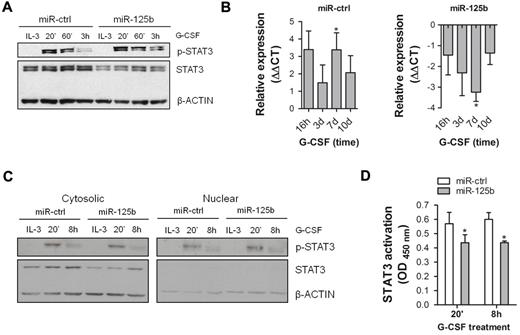
To analyze the impact of miR-125b on STAT3 function as opposed to protein expression, 32D/miR-125b and 32D/miR-ctrl cells were stimulated with G-CSF and tyrosine phosphorylation of STAT3 was evaluated by Western blotting. As shown in Figure 6A, stimulation with G-CSF induced rapid tyrosine phosphorylation that consistently remained at higher levels after 3h in 32D/miR-125b compared with 32D/miR-ctrl cells. Because STAT3 induces transcription of Socs3, a known negative regulator of G-CSF receptor signaling,37,38 we quantified Socs3 mRNA levels by qRT-PCR. As shown in Figure 6B, Socs3 mRNA levels increase in the presence of G-CSF in 32D/miR-ctrl cells (left diagram), whereas 32D/miR-125b cells express significantly less Socs3 mRNA in the presence of G-CSF (right diagram). These data indicate that transcriptional regulatory activity of STAT3 may be reduced in the presence of miR-125b over-expression. Therefore, we evaluated nuclear translocation and DNA-binding of STAT3 in 32D/miR-ctrl and 32D/miR-125b cells. As shown in Figure 6C, tyrosine-phosphorylated STAT3 is equally distributed in nuclear and cytosolic fractions of both 32D/miR-125b and 32D/miR-ctrl cells, although STAT3 protein is reduced in 32D/miR-125b cells. However, DNA-binding of STAT3 upon G-CSF treatment was significantly reduced in 32D/miR-125b compared with 32D/miR-ctrl cells (Figure 6D). Collectively, these results indicate that enforced miR-125b expression does not prohibit STAT3 activation or G-CSF–induced STAT3 shuttling but rather affects its transcriptional activity by diminished DNA-binding.
Enforced expression of miR-125b affects STAT3 signaling at multiple levels. (A) Western blotting of phospho-STAT3 (p-STAT3) and STAT3 in 32D/miR-ctrl and 32D/miR-125b cells in continuously growing culture (IL-3) and after indicated time points of G-CSF stimulation. (B) Expression level of Socs3 mRNA in 32D/miR-ctrl (left graph) and 32D/miR-125b (right graph) cells at indicated time points of G-CSF treatment. Hprt served as an endogenous control. (C) Western blotting of p-STAT3 and STAT3 in cytosolic (left panel) and nuclear (right panel) fractions of 32D/miR-ctrl and 32D/miR-125b cells. Both membranes were exposed at the same time. (D) Analysis of DNA-binding of STAT3 in 32D/miR-ctrl and 32D/miR-125b cells after indicated time points of G-CSF stimulation. Seven μg of nuclear fraction was used. OD/G-CSF values normalized to OD/IL-3 are presented. Mean of 4 (panels B,D) and representative experiments are shown, respectively. *P < .05 compared with corresponding control at indicated time points.
Enforced expression of miR-125b affects STAT3 signaling at multiple levels. (A) Western blotting of phospho-STAT3 (p-STAT3) and STAT3 in 32D/miR-ctrl and 32D/miR-125b cells in continuously growing culture (IL-3) and after indicated time points of G-CSF stimulation. (B) Expression level of Socs3 mRNA in 32D/miR-ctrl (left graph) and 32D/miR-125b (right graph) cells at indicated time points of G-CSF treatment. Hprt served as an endogenous control. (C) Western blotting of p-STAT3 and STAT3 in cytosolic (left panel) and nuclear (right panel) fractions of 32D/miR-ctrl and 32D/miR-125b cells. Both membranes were exposed at the same time. (D) Analysis of DNA-binding of STAT3 in 32D/miR-ctrl and 32D/miR-125b cells after indicated time points of G-CSF stimulation. Seven μg of nuclear fraction was used. OD/G-CSF values normalized to OD/IL-3 are presented. Mean of 4 (panels B,D) and representative experiments are shown, respectively. *P < .05 compared with corresponding control at indicated time points.
miR-125b regulates expression of c-JUN and JUND
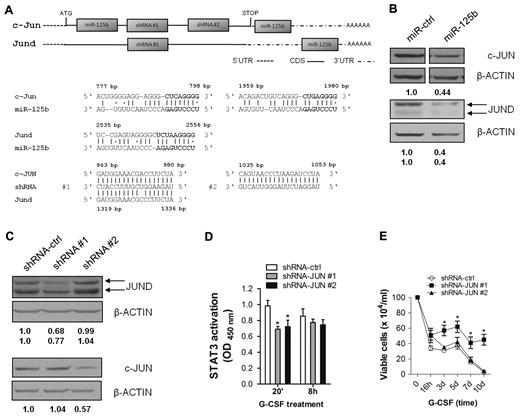
Data presented so far suggest that miR-125b may regulate protein expression of one or more STAT3 cofactors required for optimal STAT3 activity. CBP/p300,39,40 NcoA/SRC1a,41 MITF42 and c-JUN22,23 are known to directly interact with and/or to enhance transcriptional activity of STAT3. However, none of these factors is identified as a potential miR-125b target by DIANA microT v 0.3, miRDB or Target Scan. In contrast, potential miR-125b binding sites within the CDS and UTR regions of c-Jun and Jund mRNAs were identified using RNA2211 (Figure 7A). RNA22 allows identification of miRNA/mRNA binding even in the presence of nucleotide mismatches within the seed sequence. For gene-specific RNAi, we lentivirally expressed published shRNAs28,29 predicted to target both c-JUN and JUND (shRNA#1) or c-JUN only (shRNA#2; Figure 7A). As shown in Figure 7B, miR-125b reduced expression of both c-JUN and JUND by approximately 50% and 60%, respectively. Surprisingly, shRNA#1 reduced JUND protein expression but consistently failed to reduce c-JUN expression in 32D cells (Figure 7C). In contrast, shRNA#2 reduced expression of c-JUN but not JUND, as expected (Figure 7C). In parallel, DNA binding of STAT3 was analyzed in 32D cells with reduced c-JUN (shRNA#2) or JUND (shRNA#1) expression. As shown in Figure 7D, STAT3 binding is impaired upon reduction of both c-JUN and JUND, at least at early time points of G-CSF treatment. Finally, silencing of JUND but not c-JUN expression blocked G-CSF–induced differentiation and enabled G-CSF–dependent expansion of transduced cells, although with some delay (Figure 7E, and supplemental Figure 4).
miR-125b–mediated reduction of c-JUN and JUND protein expression may participate in miR-125b–induced block of granulocytic differentiation. (A) Predicted miR-125b binding sites within the CDS and UTR regions, and predicted shRNA target sites within the CDS regions of c-Jun (NM_010591.1) and Jund (NM_010592.4). (B) Western blotting of c-JUN (top panel) and JUND (bottom panel) in 32D/miR-ctrl and 32D/miR-125b cells. (C) Western blotting of c-JUN and JUND in 32D cells transduced with shRNA-ctrl, shRNA #1, and shRNA #2 lentiviruses. (D) Analysis of STAT3 DNA-binding in 32D/shRNA-ctrl, 32D/shRNA #1 and 32D/shRNA #2 cells after indicated time points of G-CSF stimulation. Fifteen μg of whole cell lysate was used. OD/G-CSF values normalized to OD/IL-3 are presented. (E) Proliferation kinetics of 32D cells expressing shRNA-ctrl (open circle), shRNA #1 (filled squares), and shRNA #2 (filled triangles) during G-CSF treatment. Mean of 4 (panels D-E) and a representative experiments of 4 are shown.
miR-125b–mediated reduction of c-JUN and JUND protein expression may participate in miR-125b–induced block of granulocytic differentiation. (A) Predicted miR-125b binding sites within the CDS and UTR regions, and predicted shRNA target sites within the CDS regions of c-Jun (NM_010591.1) and Jund (NM_010592.4). (B) Western blotting of c-JUN (top panel) and JUND (bottom panel) in 32D/miR-ctrl and 32D/miR-125b cells. (C) Western blotting of c-JUN and JUND in 32D cells transduced with shRNA-ctrl, shRNA #1, and shRNA #2 lentiviruses. (D) Analysis of STAT3 DNA-binding in 32D/shRNA-ctrl, 32D/shRNA #1 and 32D/shRNA #2 cells after indicated time points of G-CSF stimulation. Fifteen μg of whole cell lysate was used. OD/G-CSF values normalized to OD/IL-3 are presented. (E) Proliferation kinetics of 32D cells expressing shRNA-ctrl (open circle), shRNA #1 (filled squares), and shRNA #2 (filled triangles) during G-CSF treatment. Mean of 4 (panels D-E) and a representative experiments of 4 are shown.
Discussion
To study the influence of miRNAs on granulocytic differentiation and miRNA-triggered molecular mechanisms, lentivirus-based gain- and/or loss-of-function phenotypes for 9 miRNAs differentially expressed during G-CSF–induced differentiation of 32D cells have been generated. We found that only over-expression of miR-125b in 32D cells induced a complex phenotype, namely a block in granulocytic differentiation, G-CSF–dependent proliferation, and partial protection of 32D cells from IL-3 withdrawal–induced cell death. However, we cannot rule out the possibility that other miRNAs tested will induce distinct phenotypes under different experimental conditions. Interestingly, miR-125b over-expression phenotype seems to depend on the level of miRNA expression. Mature miR-125b arises from 2 different precursors, 125b-1 and 125b-2, encoded on distinct chromosomes. However, cells transduced with the pre-miR-125b-2 express much higher levels of miR-125b, and only 32D cells transduced with the miR-125b-2 precursor proliferate in the presence of G-CSF and maintain the immature blast cell morphology. The different miR-125b levels are most likely because of differences of pre-miRNA processing from the bicistronic and heterologous pre-miR-125b/miR-30-eGFP transcripts, because lentiviral transduction of both pre-miRNAs resulted in almost equal GFP expression.
Cellular differentiation is regulated by the tightly controlled expression of lineage-specific transcription factors.43 In 32D cells, miR-125b is expressed at very low levels in the presence of IL-3 but its expression increases following G-CSF treatment. On the contrary, enforced miR-125b expression up to much higher levels than that reached during differentiation can block granulocytic differentiation. We therefore speculate that increasing levels of miR-125b may act as a negative regulator of G-CSF receptor signaling in 32D cells. This hypothesis is supported by (1) reduced DNA-binding and transcriptional activity of STAT3 in the presence of elevated miR-125b levels, (2) diminished expression of myeloid-specific genes upon G-CSF stimulation, (3) reduced number of granulocytes in colonies generated by miR-125b-transduced progenitors in G-CSF-supplemented clonogenic assays, and (4) high miR-125b levels required to block granulopoiesis of 32D cells. All together, these data suggest that very high miR-125b levels may inhibit early steps of differentiation induction by interfering with appropriate STAT3 signaling (see next paragraph). However, transgenic and knock-out mouse models are required to definitively address this question. Finally, a similar kinetic during differentiation has been reported for other miRNAs, that is, miR-223 that is increasingly expressed during granulopoiesis and negatively regulates MEF2C expression.44
In primary murine lineage-negative cells enforced miR-125b expression enhances colony formation and changes the cellular composition of progenitor-derived colonies in the presence of G-CSF. In vivo, myeloid cells over-expressing miR-125b show enhanced engraftment in bone marrow and spleen compared with controls, suggesting that over-expression of miR-125b confers a proliferative and/or survival advantage to myeloid cells. This hypothesis may be supported by miR-125b–mediated repression of the proapoptotic BAK1 protein. Furthermore, the described phenotype is myeloid-specific because development of lymphoid cells is not affected. Interestingly, miR-125b over-expression because of t(2;11)(q21;q23) translocation in human AML and MDS results in defective myeloid differentiation.18 Moreover, the positive effects of miR-125b over-expression not only on macrophages but also on granulocytes in chimeric mice may be due to the compensation of defects in G-CSFR–induced STAT3-signaling by other signals in vivo. This may include STAT3 cofactors, such as JUN transcription factors induced by other cytokine receptors. Alternatively, miR-125b over-expression may induce several distinct phenotypes in a cell type–specific manner. Finally, our data are in line with the improved engraftment of miR-125b–transduced bone marrow cells reported by O'Connell et al, published during revision of this manuscript.45
To identify potential miR-125b target genes, we used several established programs that predict approximately 500 genes. Hence, we focused on targets that may functionally contribute to the observed phenotypes that is, block of granulocytic differentiation, G-CSF–dependent cell proliferation, and prolonged survival upon growth factor deprivation. In this context, BCL2 family members involved in regulation of cell survival46 and STAT3 were of particular interest. We confirmed mild but very consistent repression of BAK1 and STAT3 by over-expressed miR-125b in 32D cells but repression of other predicted BCL2 family members such as BMF or PUMA was not detected. This may be a cell lineage–specific effect, since reduced accessibility of miRNA-binding sites or the ratio of miRNA to target mRNAs may prevent effective gene silencing.7 The proapoptotic BAK1 is abundantly expressed in mature neutrophils,47 and repression of BAK1 by miR-125b over-expression may contribute to the delay of 32D cell death upon IL-3 withdrawal.
In addition, we identified STAT3 as a direct target of miR-125b. STAT3 that belongs to the STAT family of transcription factors, is activated following G-CSF treatment48 and is essential for granulocytic differentiation.49,50 Interestingly, the effects of reduced STAT3 expression are dose-dependent, and our data provide experimental evidence that the amount of protein repression by regulatory RNAs can be crucial for different cellular phenotypes. Because si/shRNAs usually induce stronger (and more specific) repression of individual target genes compared with miRNAs, dose-equivalent gene silencing by si/shRNAs is critical for proper evaluation of miRNA target gene function. Finally, because even combinatorial RNAi targeting STAT3 and BAK1 does not mimic the miR-125b–induced phenotype, we might either miss an additional/unknown important target or the miR-125b–induced phenotype requires simultaneous repression of more targets in addition to STAT3 and BAK1.
Biochemical studies on activity and function of STAT3 in response to G-CSF clearly suggested that miR-125b might also reduce one or more STAT3 cofactors. Thus, we analyzed known STAT3 cofactors such as CBP/p300, NcoA/SRC1a, MITF and c-JUN for potential miR-125b binding sites. Standard miRNA target gene prediction programs failed to identify such binding sites but RNA22 identified them within the CDS and UTR regions of c-JUN and JUND. The JUN family components can form either hetero- or homodimers among themselves.51 Consequently, composition of activator protein 1 (AP-1) determines the specific pattern of target gene expression. Although highly conserved within their DNA-binding domains, all JUN proteins differ in their ability to induce AP-1–dependent transcription.51 JUN proteins transactivate unique and overlapping genes or may have competing effects on individual target genes.52 c-JUN has been shown to enhance STAT3 transcriptional regulatory activity by direct binding to STAT3,22,23 whereas interaction between JUND and STAT3 has not been reported so far. miR-125b over-expression results in reduction of c-JUN and JUND proteins by approximately 50% and 60%, respectively. Although sequence alignment predicts silencing of c-JUN and JUND by shRNA#1 and c-JUN only by shRNA#2, we found reduced protein expression of either c-JUN or JUND by shRNA#2 and shRNA#1, respectively. The reasons for this differential gene silencing are currently not known but may include cell line–specific effects as discussed in the preceding paragraph. Gene-specific reduction of both c-JUN and JUND expression impairs STAT3 DNA-binding at least at early time points of G-CSF treatment. Conversely, only silencing of JUND but not c-JUN partially mimics the miR-125b–induced block of granulocytic differentiation in 32D cells. Besides modulating of STAT3 activity, reduction of c-JUN and JUND protein expression may affect other signaling pathways regulated by AP-1, thereby differentially modulating their final regulatory output.
Because over-expression of c-JUN on the other side can partially antagonize the miR-125b phenotype in 32D/miR-125b cells (data not shown), c-JUN and JUND may have overlapping and unique effects on G-CSF–induced granulocytic differentiation. The underlying molecular mechanisms, however, are currently unknown. Finally, our approach demonstrates that new miRNA target genes may be identified based on their assumed function using prediction programs allowing target identification with lower stringency.
In summary, our data identify regulation of JAK-STAT-, BAK1- and c-JUN/JUND-signaling by miR-125b that lead to distinct cellular phenotypes. Protein expression of STAT3, BAK1, c-JUN, and JUND is reduced by approximately 30%-60% in the presence of enforced miR-125b expression. Thereby, silencing of each gene may individually contribute to the observed phenotypes in a coordinated manner. Because miRNAs themselves may be targets for therapeutic intervention either by over-expression or silencing, precise and quantitative analysis of miRNA targets is required to evaluate the safety and benefit of miRNA-based therapeutic strategies to modulate complex cellular phenotypes.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Letizia Venturini and Michael Morgan for scientific discussion and critical reading of the manuscript. We are also grateful to Michael Heuser for help with optimizing and analyzing clonogenic assays and to Denise Hilfiker-Kleiner for discussion of JUN immunoblots. We acknowledge the assistance of the Cell Sorting Core Facility of the Hannover Medical School supported in part by Braukmann-Wittenberg-Herz-Stiftung and Deutsche Forschungsgemeinschaft (DFG).
This work was supported by the DFG excellence initiative REBIRTH, SFB 566 and the DFG Emmy-Noether program (KR2320/2-1).
Authorship
Contribution: E.S. designed research, performed experiments, analyzed data, and wrote the manuscript; M.C. and M.L. performed experiments and analyzed data; I.D. performed experiments; A.K. and A.G. analyzed data; M.S. designed research and analyzed data; and M.E. designed research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michaela Scherr and Matthias Eder, Medizinische Hochschule Hannover, Zentrum Innere Medizin, Abteilung Hämatologie, Hämostaseologie, Onkologie und Stammzelltransplantation, Carl-Neuberg Strasse 1, D-30623 Hannover, Germany; e-mail: M.Scherr@t-online.de and Eder.Matthias@MH-Hannover.de.
References
Author notes
M.S. and M.E. contribute equally to this work.