To the editor:
Loss of transmembrane lipid asymmetry in apoptotic cells or in activated platelets is thought to be catalyzed by a specific membrane protein named phospholipid scramblase.1 Recently, the transmembrane protein TMEM16F was shown to be required for Ca2+-induced lipid scrambling and exposure of phosphatidylserine at the cell surface.2 The TMEM16F gene is located on chromosome 12 (12q12) and comprises 20 exons encoding a 910–amino acid protein with 8 transmembrane segments.2,3 A patient with Scott syndrome, an inherited bleeding disorder caused by defective scramblase activity, was reported to be homozygous for a TMEM16F mutation (IVS12–1G→T) causing exon 13 skipping, frame shift, and premature termination of translation.2
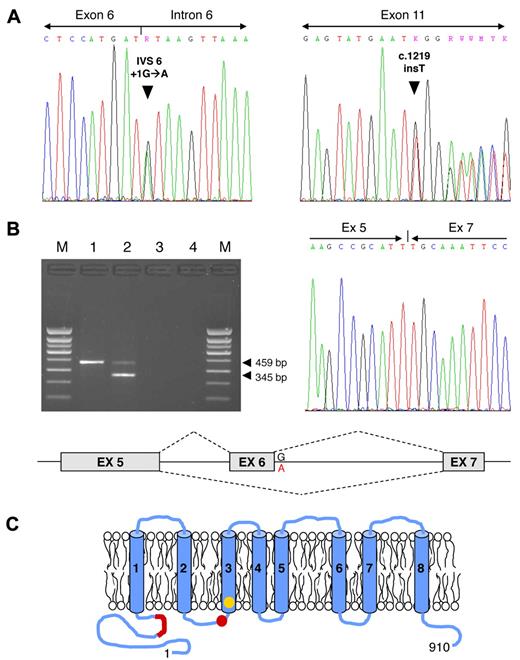
After obtaining informed consent, we investigated another patient with Scott syndrome, a 64-year-old Welsh female with a moderate bleeding tendency. The scramblase defect in her platelets, erythrocytes, and B lymphocytes has been previously characterized.4 The patient's genomic DNA was isolated from peripheral blood leukocytes and all TMEM16F exons (including splicing junctions) were amplified and sequenced. Two different mutations were identified (Figure 1A): a transition at the first nucleotide of intron 6 (IVS6 + 1G→A), disrupting the donor splice site consensus sequence of intron 6, and a single-nucleotide insertion in exon 11 (c.1219insT, cDNA numbering from the ATG), predicting a frame shift and premature termination of translation at codon 411 (Figure 1C).
TMEM16F mutations. (A) TMEM16F mutation screening in the Scott patient under study. Identification of the TMEM16F IVS6 + 1G→A (left) and c.1219insT mutations (right) in the patient's genomic DNA. (B) TMEM16F cDNA analysis. (Top left) Amplification of a TMEM16F cDNA amplicon spanning exons 5-8 yielded only the expected 459-bp fragment in the normal control (lane 1), but also a shorter 345-bp fragment in the patient (lane 2). Lane 3, reverse transcription blank; lane 4, PCR blank; M, molecular weight marker. (Top right) Sequencing of the 345-bp fragment showed that it corresponds to TMEM16F mRNA molecules lacking exon 6 (because of the IVS6 + 1G→A mutation). (Bottom) Schematic representation of the splicing aberration (exon 6 skipping) in the patient's TMEM16F pre-mRNA. (C) Schematic representation of the TMEM16F protein. The positions of the previously described TMEM16F mutation2 (IVS12–1G→T, predicting the truncation of the protein in the third transmembrane domain, shown in yellow) and of the mutations identified in this study (IVS6 + 1G→A, predicting the in-frame deletion of 38 amino acids in the N-terminal cytoplasmic tail, and c.1219insT, predicting the truncation of the protein between the second and third transmembrane domains, both shown in red) are indicated.
TMEM16F mutations. (A) TMEM16F mutation screening in the Scott patient under study. Identification of the TMEM16F IVS6 + 1G→A (left) and c.1219insT mutations (right) in the patient's genomic DNA. (B) TMEM16F cDNA analysis. (Top left) Amplification of a TMEM16F cDNA amplicon spanning exons 5-8 yielded only the expected 459-bp fragment in the normal control (lane 1), but also a shorter 345-bp fragment in the patient (lane 2). Lane 3, reverse transcription blank; lane 4, PCR blank; M, molecular weight marker. (Top right) Sequencing of the 345-bp fragment showed that it corresponds to TMEM16F mRNA molecules lacking exon 6 (because of the IVS6 + 1G→A mutation). (Bottom) Schematic representation of the splicing aberration (exon 6 skipping) in the patient's TMEM16F pre-mRNA. (C) Schematic representation of the TMEM16F protein. The positions of the previously described TMEM16F mutation2 (IVS12–1G→T, predicting the truncation of the protein in the third transmembrane domain, shown in yellow) and of the mutations identified in this study (IVS6 + 1G→A, predicting the in-frame deletion of 38 amino acids in the N-terminal cytoplasmic tail, and c.1219insT, predicting the truncation of the protein between the second and third transmembrane domains, both shown in red) are indicated.
To clarify the impact of the IVS6 + 1G→A mutation on TMEM16F pre-mRNA splicing, total RNA was isolated from the patient's blood cells using TRIzol Reagent (Invitrogen) and reverse-transcribed with the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems). A TMEM16F cDNA amplicon spanning exons 5-8 was amplified and analyzed by agarose gel electrophoresis (Figure 1B). Whereas the normal control showed only the expected 459-bp fragment, the patient showed both the normal fragment (likely the product of the c.1219insT allele) and a shorter (345-bp) fragment. The low intensity of the patient's 459-bp band suggests that the mRNA transcribed from the c.1219insT allele is partially degraded in vivo. Sequencing of the shorter fragment showed that it lacked exon 6 (Figure 1B). Exon 6 skipping, which is attributable to the IVS6 + 1G→A mutation, predicts the in-frame deletion of 38 amino acids (residues 212-249) from the N-terminal cytoplasmic domain of the protein (Figure 1C). The normal intensity of this band suggests that the stability of the mRNA transcribed from this allele is normal.
To definitely exclude the possibility that the identified mutations are polymorphisms present in the general population, 100 unrelated white individuals were genotyped for all 3 mutations by restriction analysis with BstY I (IVS12–1G→T), Rsa I (IVS6 + 1G→A, restriction site introduced with a mutagenic reverse primer) and Tsp509 I (c.1219insT), respectively (New England Biolabs). No instance of any of these mutations was found.
In conclusion, we have identified 2 novel mutations in the TMEM16F gene in a patient with Scott syndrome. This finding confirms the recent report2 that the product of this gene is required for Ca2+-dependent phospholipid scrambling and that loss-of-function mutations in this gene can give rise to Scott syndrome.
Authorship
Acknowledgments: The authors thank Mr N. Deckers for technical support, and the anonymous patient who donated blood for the study.
This study was supported by VIDI grant 917-76-312 (to E.C.) from the Dutch Organisation for Scientific Research (NWO).
Contribution: E.C. performed research, analyzed data, and wrote the manuscript; P.W.C. recruited the patient, took care of blood sampling, and provided intellectual contribution; P.L.W. provided intellectual contribution and wrote the manuscript; and E.M.B. designed and coordinated the study and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Edouard Bevers, Department of Biochemistry, Maastricht University, PO Box 616, 6200 MD Maastricht, The Netherlands; e-mail: em.bevers@maastrichtuniversity.nl.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal