Abstract
Single nucleotide polymorphism arrays (SNP-As) have emerged as an important tool in the identification of chromosomal defects undetected by metaphase cytogenetics (MC) in hematologic cancers, offering superior resolution of unbalanced chromosomal defects and acquired copy-neutral loss of heterozygosity. Myelodysplastic syndromes (MDSs) and related cancers share recurrent chromosomal defects and molecular lesions that predict outcomes. We hypothesized that combining SNP-A and MC could improve diagnosis/prognosis and further the molecular characterization of myeloid malignancies. We analyzed MC/SNP-A results from 430 patients (MDS = 250, MDS/myeloproliferative overlap neoplasm = 95, acute myeloid leukemia from MDS = 85). The frequency and clinical significance of genomic aberrations was compared between MC and MC plus SNP-A. Combined MC/SNP-A karyotyping lead to higher diagnostic yield of chromosomal defects (74% vs 44%, P < .0001), compared with MC alone, often through detection of novel lesions in patients with normal/noninformative (54%) and abnormal (62%) MC results. Newly detected SNP-A defects contributed to poorer prognosis for patients stratified by current morphologic and clinical risk schemes. The presence and number of new SNP-A detected lesions are independent predictors of overall and event-free survival. The significant diagnostic and prognostic contributions of SNP-A–detected defects in MDS and related diseases underscore the utility of SNP-A when combined with MC in hematologic malignancies.
Introduction
Metaphase cytogenetics (MC) has proven to be an extremely valuable clinical tool in the management of hematologic malignancies. It is the “gold standard” for karyotypic analysis in a variety of blood diseases. It can detect balanced chromosomal changes, including translocations or inversions, and unbalanced chromosomal changes, including trisomies, duplications, and deletions.1-3 Specific clonal chromosomal aberrations can be diagnostic, as shown in chronic myelogenous leukemia and acute promyelocytic leukemia.4,5 Some recurrent chromosomal defects can also be prognostic in myelodysplastic syndromes (MDSs), as exemplified by isolated del5q in 5q− syndrome and del20q, which are associated with good outcomes.3 In addition, they can predict drug sensitivity, as exemplified by imatinib in chronic myelogenous leukemia 6 and all-trans retinoic acid in acute promyelocytic leukemia7 or drug resistance.8
MDS is a hematopoietic stem cell disorder typically affecting older persons, characterized by a propensity to leukemic transformation, impaired blood cell production, and unbalanced chromosomal abnormalities.9 Although survival of patients with high-risk disease can be extended with 5-azacytidine,10 only a few curative treatment options currently exist. The clinical success of lenalidomide in patients with 5q− syndrome highlights the diagnostic and prognostic effects of specific chromosomal defects and shows the importance of detailed cytogenetics in disease management. However, because of technical limitations of MC, including its relatively low resolution and especially the need for dividing cells, this technique is not optimally suited for MDS, missing many important defects, thus resulting in detectable genomic aberrations in only 40%-50% of patients with MDS.9,11,12
Although whole-genome scanning technologies with the use of single nucleotide polymorphism arrays (SNP-As) were originally developed for genetic association studies, they are now commonly used as a cytogenetic tool in the research setting.13,14 Hybridization of tumor DNA to arrays containing probes specific for allelic variants of SNPs allows for measurement of gene copy number (hybridization signal intensity) and distinction of individual genotypes for detection of loss of heterozygosity (LOH) at a high resolution. SNP-A–based karyotyping has been applied previously in a range of studies that used a limited number of patients with various hematologic malignancies,12-18 all with similar end-point data, suggesting its potential clinical utility. In addition to a high level of resolution, SNP-A allows for a better appreciation of acquired copy-neutral LOH (aCN-LOH), also referred to as somatic uniparental disomy, a common chromosomal defect in hematologic malignancies, undetectable by conventional MC.13,14 In aCN-LOH, there is a LOH of a genomic region without a concomitant reduction in copy number, as would be expected for a deletion. Recurrent cytogenetic abnormalities identified by SNP-A facilitate the identification of pathogenic mutations.19-27
On the basis of these technical advantages of SNP-A, we hypothesized that identification of new defects with the use of this technology may complement MC and improve the diagnosis and prognostication in MDS and other myeloid malignancies with the potential to change current diagnostic and treatment paradigms.
Methods
Patient population
Informed consent for sample collection was obtained according to protocols approved by the review boards of all participating institutions, in accordance with the Declaration of Helsinki. Diagnosis was assigned according to the World Health Organization (WHO) classification and categorized as lower- and higher-risk groups (Table 1).28 In addition, the International Prognostic Scoring System (IPSS) was used to stratify patients into low-, intermediate-1/2–, and high-risk groups and to define cytogenetic risk group by MC.3 Previously, results performed with the use of 250K were presented on 168 patients with a median follow-up of 13 months.14
Baseline characteristics of 430 patients in study
| Characteristic . | Value . |
|---|---|
| Age, y | |
| Median | 71 |
| Range | 19-92 |
| Sex | |
| Male/female, % | 63/37 |
| Risk stratification by WHO,* n (%) | |
| Low risk | 250 (58) |
| High risk | 180 (42) |
| IPSS risk category, n (%) | |
| Low | 112 (26) |
| Int-1 | 124 (29) |
| Int-2 | 61 (14) |
| High | 44 (10) |
| Unclassified† | 89 (21) |
| WHO classification, n (%) | |
| MDS | |
| RCUD/RCMD | 114 (27) |
| RARS | 36 (8) |
| 5q− syndrome | 19 (4) |
| MDS-U | 8 (2) |
| RAEB1/2 | 73 (17) |
| MDS/MPN overlap | |
| CMML-1 | 36 (8) |
| CMML-2 | 18 (4) |
| MDS/MPN-U‡ | 41 (10) |
| sAML§ | 85 (20) |
| Characteristic . | Value . |
|---|---|
| Age, y | |
| Median | 71 |
| Range | 19-92 |
| Sex | |
| Male/female, % | 63/37 |
| Risk stratification by WHO,* n (%) | |
| Low risk | 250 (58) |
| High risk | 180 (42) |
| IPSS risk category, n (%) | |
| Low | 112 (26) |
| Int-1 | 124 (29) |
| Int-2 | 61 (14) |
| High | 44 (10) |
| Unclassified† | 89 (21) |
| WHO classification, n (%) | |
| MDS | |
| RCUD/RCMD | 114 (27) |
| RARS | 36 (8) |
| 5q− syndrome | 19 (4) |
| MDS-U | 8 (2) |
| RAEB1/2 | 73 (17) |
| MDS/MPN overlap | |
| CMML-1 | 36 (8) |
| CMML-2 | 18 (4) |
| MDS/MPN-U‡ | 41 (10) |
| sAML§ | 85 (20) |
RCUD indicates refractory anemia with unilineage dysplasia; RCMD, refractory cytopenia with multilineage dysplasia; RARS, refractory anemia with ring sideroblasts; MDS-U, myelodysplastic syndromes unclassified; RAEB-1/2, refractory anemia with excess blasts-1/2; CMML, chronic myelomonocytic leukemia; and MDS/MPN-U, myelodysplastic syndromes/myeloproliferative neoplasms unclassifiable.
Low risk includes RCUD, RARS, RCMD, CMML-1, MDS/MPN-U < 5% blasts, RARS-T, MDS-U, and 5q− syndrome. High risk includes RAEB-1/2, CMML-2, MDS/MPN-U ≥ 5% blasts, and AML.
Includes 28 cases of no growth and 61 cases that were not classified because they were CMML with white blood cell count > 12 000/μL, AML by WHO classification, and other factors precluding appropriate classification.
Sixteen cases of MDS/MPN-U are classified as RARS-T.
Thirty-four patients are classified as RAEB-T by French-American-British classification.
Presentation BM biopsies were obtained from 430 patients to help establish the diagnosis (Cleveland Clinic = 330, King's College = 73, Johns Hopkins University = 19, H. Lee Moffitt Cancer Center = 7, and UCLA = 1), and serial samples were obtained in 53 patients. All bioinformatic evaluation was performed at Cleveland Clinic. We also analyzed SNP-A data from 1003 controls (internal controls and Framingham database controls). Patients received treatments that included hematopoietic stem cell transplantation, high-intensity chemotherapy, low-intensity chemotherapy, and supportive treatments (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). MC analysis was performed according to standard procedures, and International System for Cytogenetic Nomenclature29 (ISCN; 2005) was used to define the karyotype. Routinely, 20 metaphase spreads were analyzed for each patient. Patients with no growth of the cell culture for MC are defined as noninformative cases. Clinical parameters studied included age, sex, peripheral blood (PB) counts and morphology, BM morphology, overall survival (OS), and other outcome parameters including event-free survival (EFS), progression-free survival (PFS), and disease-free survival (DFS). The median follow-up of the entire cohort, including deceased and live patients, from diagnosis was 25 months (range, 0-233 months). Among 26% of patients who were still alive at the time of analysis, the median follow-up time by IPSS risk group was low-risk group = 46 months, Int-1 risk group = 43 months, Int-2 risk group = 25 months, and high-risk group = 24 months (supplemental Table 2).
SNP-A analyses
The Affymetrix GeneChip Human Mapping 250K, Genome-Wide Human SNP Array 6.0 and reagents (Affymetrix) were used according to the manufacturer's protocols and have been previously described.13,14 Paired BM and germline (CD3+ cells) DNA were used to distinguish germline lesions.14 The decision to use CD3 lymphocytes for germline confirmation of SNP-A lesions was based on its practical accessibility. Buccal swabs were at risk of contamination with clonal cells, especially in patients with high white blood counts and circulating blast counts, whereas skin punch biopsy specimens were not routinely obtained in patients with myeloid malignancies.
A stringent algorithm was applied in the identification of SNP-A lesions. Lesions identified by both SNP-A and MC were not included in the analysis. Matching aberrations present in our internal or publicly available copy number variant (CNV) databases were considered germline and excluded. CNVs represent losses/gains of genomic material identified in healthy controls and are thought to reflect normal human genomic variation.30,31 All other lesions were confirmed as somatic by analysis of CD3+ cells or serial analyses (n = 76). Previously, we established that nonclonal regions of germline aCN-LOH can be found in 12% of controls with the mean size plus 2 SDs of < 25 Mb.16 If aCN-LOH was detected, the location and size of lesions (telomeric/interstitial and ≥ 25 Mb) were indicative of a somatic abnormality, whereas those interstitial and < 25 Mb were subjected to validation through analysis of CD3+ cells (supplemental Figure 1A-B). Interstitial lesions were defined as those that did not reach the telomere. We confirmed/disproved the somatic nature of lesions by SNP-A analysis in 76 patients (23 with CD3+ cells, 53 with serial samples). In an additional 7 patients, the germline nature of the lesion was confirmed by sequencing of CD3+ cells for selected loci. Some lesions do not have to be confirmed; that is, those present on SNP-A and MC and large aCN-LOH that were never encountered in controls. Previously described recurrent clonal lesions, that is, microdeletions containing TET2, CBL, EZH2 loci, also do not need to be verified. In light of these considerations, validation was performed or not required in 248 cases.
Biostatistical evaluation of SNP-A and clinical data
Signal intensity and SNP genotype data from Affymetrix 250K arrays was determined with the use of Gene Chip Genotyping Analysis software Version 4.0 (GTYPE). Copy number was investigated with the use of a hidden Markov model and copy number Analyzer for Affymetrix GeneChip Mapping 500K arrays (CNAGv.3). Results of Affymetrix 6.0 arrays were analyzed with Genotyping Console Version 4.0 (Affymetrix). Segmental LOH was identified by a statistical assessment of the likelihood that consecutive SNP loci exhibit heterozygosity. For confirmation, 83 samples were analyzed with both Affymetrix 250K and 6.0 SNP-A.
We used the 2006 modified International Working Group (IWG) criteria for evaluating MDS response (supplemental Table 3).32 Patients with secondary AML (sAML) on the basis of the French-American-British classification were analyzed with the revised recommendations of the IWG for AML.33
Although patients with BM blasts > 30% were traditionally classified as AML, IPSS and cytogenetic risk group were assigned to these patients because they evolved from a prior MDS or MDS/myeloproliferative overlap neoplasm (MPN) and therefore belong to the same disease spectrum.
Fisher exact test and χ2 test were applied to compare categorical variables. Clinical outcome parameters analyzed included OS, EFS, PFS, and DFS. OS was measured from day 0 to death from any cause (patients lost to follow-up were censored). Appropriate data censoring was applied to the other outcomes (supplemental Table 4). All 4 outcomes were summarized with the Kaplan-Meier method, and univariable analyses were conducted with the log-rank test and Cox proportional hazards model. The Cox proportional hazards model was also used for multivariable analysis to assess simultaneously the effect of multiple patient factors. A stepwise variable selection algorithm was used to identify independent predictors of EFS and OS. All P values were 2-sided, and P values < .05 indicated statistical significance. Statistical analyses were performed with JMP8.0 software and SAS Version 9.1 (SAS Inc).
Results
SNP-A–based karyotyping as a reliable diagnostic tool
Previously, we established the feasibility of the detection of somatic chromosomal abnormalities by SNP-A karyotyping in a clinical setting.14,18 Our protocol involved exclusion of known nonclonal germline–encoded CNVs and CN-LOH and the confirmation of new somatic defects with the use of paired nonclonal cells (supplemental Figure 1A-B). We performed SNP-A 6.0 arrays on 228 samples, 250K arrays on 200 samples and 50K arrays in 2 samples. Of these, 2 types of SNP-A (6.0 and 250K arrays) were applied in 83 patients, showing 95% concordance for identification of genomic aberrations.13 The 5% difference in the identified genomic aberrations between the 2 arrays is associated with differences in array coverage. Typical data output is shown in Figure 1, comparing higher- and lower-density arrays for an exemplary deletion, CN-LOH, and duplication. MC allowed for detection of 35 balanced translocations (5% of all lesions studied), inherently not detectable by SNP-A. Ninety-three percent of unbalanced chromosomal aberrations found by MC were also detected by SNP-A, excluding those on the Y chromosome; in most instances, defects present in < 30% of metaphases by MC were not identified by SNP-A.
Comparison of exemplary lesions detected by genomewide SNP-A 6.0. (A) Whole-genome view shows the complete diploid set of human chromosomes. A close-up view of chromosome 21 shows the presence of a deletion on its long arm, illustrated by a decrease in the copy number line (blue line and red dots) and LOH (paucity of green bars, indicating the frequency of heterozygous loci [red box]). A closer look at the region shows loci potentially involved in disease pathogenesis. (B) Illustration of the 3 exemplary characteristic lesions seen by 6.0 SNP-A: deletion, duplication, and aCN-LOH. For deletions, LOH as indicated by contraction of the AA and BB alleles (blue lines) and reduction of the copy number (top) marks the affected region. For duplications, the doubling of the heterozygous AB allele and an increase in copy number delineate the abnormality. For aCN-LOH, the loss of the AB allele without contraction of the AA and BB alleles and a normal copy number identify the lesion. In each example, the normal copy number and allele calls for CD3+ cells (green lines) shows the absence of a germline abnormality, thus confirming the somatic nature of the lesions. (C) An exemplary microdeletion in chromosome 7, seen as a drop in the diploid copy number line (blue line in red box) and the absence of this abnormality in the CD3+ lymphocytes, is shown.
Comparison of exemplary lesions detected by genomewide SNP-A 6.0. (A) Whole-genome view shows the complete diploid set of human chromosomes. A close-up view of chromosome 21 shows the presence of a deletion on its long arm, illustrated by a decrease in the copy number line (blue line and red dots) and LOH (paucity of green bars, indicating the frequency of heterozygous loci [red box]). A closer look at the region shows loci potentially involved in disease pathogenesis. (B) Illustration of the 3 exemplary characteristic lesions seen by 6.0 SNP-A: deletion, duplication, and aCN-LOH. For deletions, LOH as indicated by contraction of the AA and BB alleles (blue lines) and reduction of the copy number (top) marks the affected region. For duplications, the doubling of the heterozygous AB allele and an increase in copy number delineate the abnormality. For aCN-LOH, the loss of the AB allele without contraction of the AA and BB alleles and a normal copy number identify the lesion. In each example, the normal copy number and allele calls for CD3+ cells (green lines) shows the absence of a germline abnormality, thus confirming the somatic nature of the lesions. (C) An exemplary microdeletion in chromosome 7, seen as a drop in the diploid copy number line (blue line in red box) and the absence of this abnormality in the CD3+ lymphocytes, is shown.
Effect of SNP-A analysis on precision and diagnostic yield of cytogenetic diagnosis
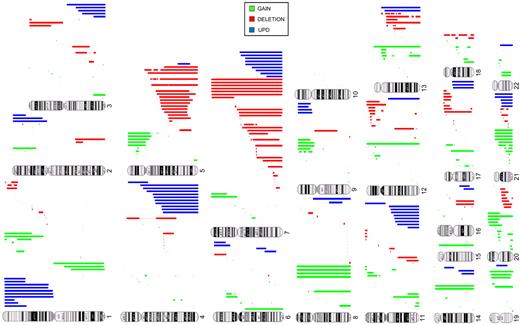
Identification of chromosomal abnormalities by SNP-A karyotyping would greatly enhance the diagnostic evaluation of MDS, particularly in cases in which MC is not informative. In 430 patients analyzed, 189 (44%) showed abnormal MC, 213 (49%) had a normal karyotype, and 28 (7%) had noninformative results. The frequency of abnormal cytogenetic findings for MDS, MDS/MPN, and sAML (defined as AML derived from MDS or MDS/MPN) by MC alone was 46%, 38%, and 45%, respectively, but combining SNP-A and MC increased the diagnostic yield to 74%, 75%, and 74%, respectively (Table 2). When SNP-A was applied together with MC, chromosomal lesions were identified in 319 patients (74%) compared with 44% with MC alone (P ≤ .0001). Importantly, analysis of 28 patients with unsuccessful MC by SNP-A analysis enabled the detection of chromosomal lesions in 16 of these patients (57%) (Table 1). In addition to detecting cryptic aberrations in 130 of 241 patients (54%) with normal/noninformative MC, additional lesions were identified in 117 of 189 patients (62%) with a previously abnormal karyotype (Table 2). Many new abnormalities detected by SNP-A karyotyping were submicroscopic deletions and regions of aCN-LOH involving chromosomes 1, 5, 7, 11, 17, and 21 (Figure 2; supplemental Table 5). Our analysis also identified a subset of 111 cases (65 MDS, 24 MDS/MPN, and 22 MDS/AML) with normal cytogenetics confirmed by both SNP-A and MC. A total of 247 patients had new SNP-A–detected lesions. Sole SNP-A defects were found in 119 patients, with most seen in patients with normal karyotype (n = 69) compared with those with abnormal MC (n = 47) and no growth (n = 3). Single SNP-A abnormalities were most frequently found in chromosomes 4, 7, 8, 9, and 12. The frequency of 2 SNP-A abnormalities were similar between patients with normal (n = 27) and abnormal (n = 28) MC. More complex abnormalities were predominantly seen in patients with previously abnormal MC (n = 42) compared with patients with normal MC (n = 20; supplemental Table 5).
Comparison of cytogenetic detection rate between MC and MC cytogenetics combined with SNA-A karyotyping
| Disease group/MC . | n (%) . | MC + SNP-A . | n (%) . | P* . |
|---|---|---|---|---|
| MDS (n = 250) | ||||
| NI† | 17 (7) | Normal | 65 (26) | |
| Normal | 118 (47) | Abnormal | 70 (28) | < .0001 |
| Abnormal | 115 (46) | No additional | 47 (19) | |
| Abnormal | Additional | 68 (27) | ||
| MDS/MPN (n = 95) | ||||
| NI† | 4 (4) | Normal | 24 (25) | |
| Normal | 55 (58) | Abnormal | 35 (37) | < .0001 |
| Abnormal | 36 (38) | No additional | 10 (11) | |
| Abnormal | Additional | 26 (27) | ||
| AML (n = 85)‡ | ||||
| NI† | 7 (8) | Normal | 22 (26) | |
| Normal | 40 (47) | Abnormal | 25 (29) | .0002 |
| Abnormal | 38 (45) | No additional | 15 (18) | |
| Abnormal | Additional | 23 (27) |
| Disease group/MC . | n (%) . | MC + SNP-A . | n (%) . | P* . |
|---|---|---|---|---|
| MDS (n = 250) | ||||
| NI† | 17 (7) | Normal | 65 (26) | |
| Normal | 118 (47) | Abnormal | 70 (28) | < .0001 |
| Abnormal | 115 (46) | No additional | 47 (19) | |
| Abnormal | Additional | 68 (27) | ||
| MDS/MPN (n = 95) | ||||
| NI† | 4 (4) | Normal | 24 (25) | |
| Normal | 55 (58) | Abnormal | 35 (37) | < .0001 |
| Abnormal | 36 (38) | No additional | 10 (11) | |
| Abnormal | Additional | 26 (27) | ||
| AML (n = 85)‡ | ||||
| NI† | 7 (8) | Normal | 22 (26) | |
| Normal | 40 (47) | Abnormal | 25 (29) | .0002 |
| Abnormal | 38 (45) | No additional | 15 (18) | |
| Abnormal | Additional | 23 (27) |
NI indicates noninformative.
Comparison of cytogenetic detection rate between MC and MC + SNP-A.
Noninformative, defined as no growth by metaphase cytogenetics.
MC results not available (AML = 1 but FISH results were available. The test showed normal findings in all tested loci for del20q, del5/del5q, del7, del7q, +8).
Types and genomic distribution of chromosomal lesions detected by SNA-A karyotyping in MDSs. Gains (green), deletions (red), and acquired somatic uniparental disomy (blue) are shown.
Types and genomic distribution of chromosomal lesions detected by SNA-A karyotyping in MDSs. Gains (green), deletions (red), and acquired somatic uniparental disomy (blue) are shown.
Effect of SNP-A on prediction of outcomes in MDS and related myeloid malignancies
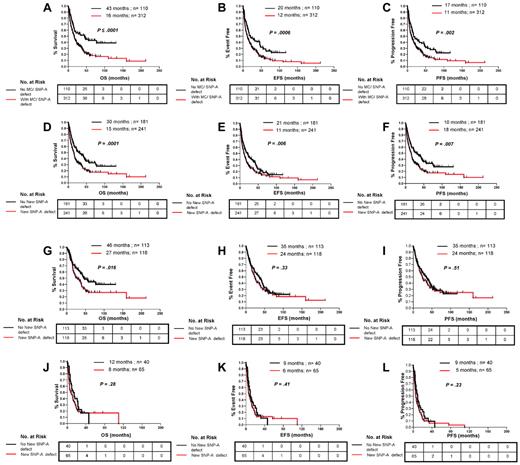
Of utmost importance for the clinical applicability of SNP-A is whether the increased cytogenetic yield translates into an ability to assign prognosis more precisely. Survival of patients with chromosomal defects detected by either MC or SNP-A was worse than in patients in whom no lesions were detected for OS (16 vs 43 months; P ≤ .0001), EFS (12 vs 20 months; P = .0006), and PFS (11 months vs 17; P = .002) (Figure 3A-C). There was no difference in DFS (25 vs 28 months; P = .45; not shown) for patients in whom SNP-A karyotyping detected previously cryptic lesions. We attribute this lack of difference in DFS to the lack of power to assess this survival outcome. Because DFS is analyzed from the time of complete remission to the time of relapse, only 68 patients were left for analysis of DFS, of which 57% (n = 39) were censored. Regardless of prior karyotype, OS (15 vs 30 months; P = .0001), EFS (11 vs 21 months; P = .006), and PFS (10 vs 18 months; P = .007) of patients with new defects uncovered by SNP-A were significantly inferior than for patients with a negative SNP-A examination (Figure 3D-F).
Clinical effect of lesions detected by SNP-A karyotyping analysis in combination with MC and new lesions detected by SNP-A regardless of MC results. Patients with new defects (red line) detected by the combination MC or SNP-A had worse OS (A), EFS (B), and PFS (C) than patients with no defects (black line) detected by both techniques. Patients with new lesions detected by SNP-A regardless of MC results showed worse OS (D), EFS (E), and PFS (F) than patients with an unchanged karyotype. In addition, SNP-A–identified lesions had an effect on OS in patients in the low-risk IPSS group (G) but not on EFS (H) and PFS (I). New SNP-A–detected defects did not affect outcomes in the high-risk IPSS group (J, K, and L). Tables represent the number at risk over time. Characteristics of censored patients have been presented in supplemental Table 4.
Clinical effect of lesions detected by SNP-A karyotyping analysis in combination with MC and new lesions detected by SNP-A regardless of MC results. Patients with new defects (red line) detected by the combination MC or SNP-A had worse OS (A), EFS (B), and PFS (C) than patients with no defects (black line) detected by both techniques. Patients with new lesions detected by SNP-A regardless of MC results showed worse OS (D), EFS (E), and PFS (F) than patients with an unchanged karyotype. In addition, SNP-A–identified lesions had an effect on OS in patients in the low-risk IPSS group (G) but not on EFS (H) and PFS (I). New SNP-A–detected defects did not affect outcomes in the high-risk IPSS group (J, K, and L). Tables represent the number at risk over time. Characteristics of censored patients have been presented in supplemental Table 4.
We also grouped patients by WHO criteria and divided them into low- and high-risk disease defined by morphologic features.28 Patients with low-risk disease for whom MC and SNP-A identified additional lesions showed inferior survival (OS: 35 vs NR, P = .0012; EFS: 25 vs 47 months, P = .0084; PFS: 25 vs 46 months, P = .047) than for patients in whom no additional MC and SNP-A aberrations were detected (supplemental Figure 2A,C,E). Inferior PFS was also observed in the high-risk group of patients (5 vs 9 months, P = .033), but SNP-A did not further resolve other prognostic outcomes (OS: 6 vs 9 months, P = .05; EFS: 5 vs 9 months, P = .07; supplemental Figure 2B,D,F). When WHO categories were analyzed separately, SNP-A karyotyping in conjunction with MC further stratified survival outcomes in patients with MDS, MDS/MPN, and sAML (supplemental Figure 3).
We also analyzed survival outcomes of patients with MDS, MDS/MPN, and AML on the basis of the number of SNP-A–detected defects, and the data showed that patients with ≥ 3 SNP-A–detected defects and those with 1 or 2 detected defects have worse OS, EFS, and PFS than patients without SNP-A–detected defects (supplemental Figure 4).
Effects of chromosomal defects detected by SNP-A on IPSS risk stratification
The IPSS is the most commonly used prognostic system for MDS, which includes variables such as the number of cytopenias, percentage of leukemic blasts, and karyotype, with the karyotype being the most heavily weighted factor. Although the absence of abnormal chromosomes by MC is generally favorable, there is significant variability in the outcomes of patients with normal karyotypes and patients with identical lesions. Consequently, on the basis of the hypothesis that the greater cytogenetic yield by application of SNP-A will contribute to better prognostic resolution, we examined the effect of additional defects detected by SNP-A analyses on outcome parameters within individual IPSS risk groups. Patients were divided into low- (low/intermediate-1) and high-risk (intermediate-2/high; sAML according to French-American-British were excluded) IPSS subtypes. Within the low-risk groups, patients with additional SNP-A–detected defects had worse OS (27 vs 46 months; P = .016; Figure 3G) than patients for whom the cytogenetic diagnosis was unchanged. This difference was not observed in the analysis of EFS and PFS in low-risk patients (Figure 3H-I). No significant difference was found for patients with additional cytogenetic lesions detected by SNP-A within the high-risk groups (Figure 3J-L).
Assignment of prognostic effect to specific recurrent chromosomal defects detected by SNP-A would allow for their incorporation into future prognostic schemes. Such individual analysis is possible only for the most common defects, because most uncovered defects are too heterogeneous and occur at too low a frequency. Analysis of loss and aCN-LOH of chromosomes 7, 11, and 17 as detected by SNP-A shows poor OS, similar to what is seen when corresponding deletions are detected by MC. Similarly, in patients with del5q abnormality regardless of method of detection, SNP-A was able to further delineate a subgroup of patients with abnormalities of 5q only detected with SNP-A that have a different survival in comparison to MDS with a sole del5q abnormality detected by MC and those of MDS with del5q with additional abnormalities when detected by MC (P < .001; supplemental Figure 5).
Univariable and multivariable analyses of clinical and laboratory factors and new prognostic risk groups by MC and SNP-A
Patients with MDS and related myeloid malignancies who had the following clinical features: male sex, increased age, poor risk karyotype by MC defined by IPSS, BM blasts ≥ 5%, ≥ 2 cytopenias as defined by IPSS, presence of new SNP-A–detected defects, increased number of new SNP-A–detected lesions, high risk by IPSS, high risk by WHO risk grouping, and blasts in the PB all have poorer prognosis by univariate analysis for both OS and EFS (supplemental Table 6). More importantly, multivariable analysis showed that the presence of new lesions detected by SNP-A and an increased number of new SNP-A lesions (> 2 vs 1 or 2 vs none) are independent predictors of inferior OS and EFS in patients with MDS and related myeloid malignancies (Table 3). Traditional prognostic factors, including cytogenetic risk by MC as defined by IPSS, WHO risk group, and age, remained important predictors of OS and EFS as did the presence of blasts in the PB.
Mutivariable analysis of clinical and laboratory parameters
| Factor* . | Overall survival . | Event-free survival . | ||
|---|---|---|---|---|
| Hazard ratio (95% CI) . | P† . | Hazard ratio (95% CI) . | P† . | |
| Parameters analyzed | ||||
| Sex | ||||
| Male vs female | 1.25 (0.96-1.63) | .10 | 1.18 (0.92-1.50) | .21 |
| Age at time of sampling | ||||
| ≥ 70 y vs 50-69 y vs < 50 y | 1.82 (1.49-2.23) | < .0001 | 1.68 (1.40-2.01) | < .0001 |
| MC | ||||
| Poor vs intermediate vs good | 1.61 (1.37-1.90) | < .0001 | 1.48 (1.27-1.74) | < .0001 |
| WHO risk group | ||||
| High vs low | 3.24 (2.48-4.23) | < .0001 | 3.11 (2.41-4.01) | < .0001 |
| New lesions by SNP-A | ||||
| Yes or No | 1.64 (1.27-2.12) | .0002 | 1.33 (1.05-1.68) | .02 |
| Parameters analyzed | ||||
| Sex | ||||
| Male vs female | 1.28 (0.98-1.66) | .07 | 1.19 (0.93-1.53) | .17 |
| Age at time of sampling | ||||
| ≥ 70 y vs 50-69 y vs < 50 y | 1.85 (1.51-2.27) | < .0001 | 1.69 (1.41-2.03) | < .0001 |
| MC | ||||
| Poor vs intermediate vs good | 1.58 (1.34-1.86) | < .0001 | 1.47 (1.25-1.72) | < .0001 |
| WHO risk group | ||||
| High vs Low | 3.09 (2.63-4.03) | < .0001 | 3.03 (2.35-3.90) | < .0001 |
| No. of new lesions by SNP-A | ||||
| ≥ 3 vs 1-2 vs 0 | 1.47 (1.24-1.74) | < .0001 | 1.25 (1.06-1.47) | .007 |
| Factor* . | Overall survival . | Event-free survival . | ||
|---|---|---|---|---|
| Hazard ratio (95% CI) . | P† . | Hazard ratio (95% CI) . | P† . | |
| Parameters analyzed | ||||
| Sex | ||||
| Male vs female | 1.25 (0.96-1.63) | .10 | 1.18 (0.92-1.50) | .21 |
| Age at time of sampling | ||||
| ≥ 70 y vs 50-69 y vs < 50 y | 1.82 (1.49-2.23) | < .0001 | 1.68 (1.40-2.01) | < .0001 |
| MC | ||||
| Poor vs intermediate vs good | 1.61 (1.37-1.90) | < .0001 | 1.48 (1.27-1.74) | < .0001 |
| WHO risk group | ||||
| High vs low | 3.24 (2.48-4.23) | < .0001 | 3.11 (2.41-4.01) | < .0001 |
| New lesions by SNP-A | ||||
| Yes or No | 1.64 (1.27-2.12) | .0002 | 1.33 (1.05-1.68) | .02 |
| Parameters analyzed | ||||
| Sex | ||||
| Male vs female | 1.28 (0.98-1.66) | .07 | 1.19 (0.93-1.53) | .17 |
| Age at time of sampling | ||||
| ≥ 70 y vs 50-69 y vs < 50 y | 1.85 (1.51-2.27) | < .0001 | 1.69 (1.41-2.03) | < .0001 |
| MC | ||||
| Poor vs intermediate vs good | 1.58 (1.34-1.86) | < .0001 | 1.47 (1.25-1.72) | < .0001 |
| WHO risk group | ||||
| High vs Low | 3.09 (2.63-4.03) | < .0001 | 3.03 (2.35-3.90) | < .0001 |
| No. of new lesions by SNP-A | ||||
| ≥ 3 vs 1-2 vs 0 | 1.47 (1.24-1.74) | < .0001 | 1.25 (1.06-1.47) | .007 |
Adjusting for the factors in this table, blasts in the peripheral blood, unavailable for 22% of patients, was seen to be an additional independent predictor of overall but not event-free survival (P = .02 and .03, respectively).
Factor with the poorer prognosis is listed first.
Determined by the Wald test.
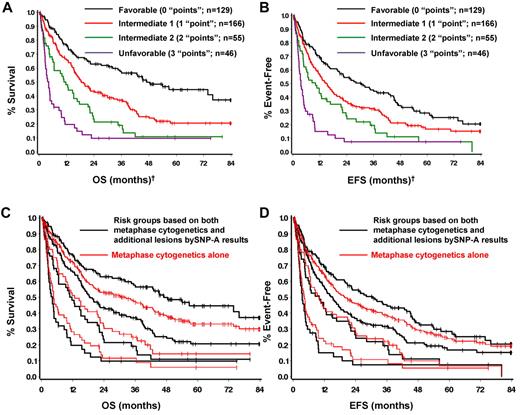
On the basis of the additional prognostic value provided by SNP-A analysis, and the fact that the effect was similar to that of conventional MC, both methods were combined to form 4 prognostic groups by simply counting the number of poor prognostic features present. That is, counting intermediate cytogenetics by MC and new defects by SNP-A as one poor feature or “point” each, and unfavorable cytogenetics by MC as 2 poor features (“points”), 4 patient groups with very different prognoses can be defined on the basis of on the total number of poor prognostic features (or “points”) present. The resulting prognostic groups and outcomes are summarized in Table 4 and Figure 4A-B. Comparing the survival curves of the newly defined risk groups according to SNP-A with those based on MC alone shows that adding SNP-A results in further stratification of each cytogenetic risk group; moving the better prognosis patients up to a more favorable group and the poorer prognosis patients to a less favorable group (Figure 4C-D; supplemental Figure 6). Similar results can be obtained with the use of the number of additional lesions found; therefore, for simplicity the new risk stratification was based simply on the presence or absence of additional lesions by SNP-A.
Prognostic groups based on MC and SNP-A
| Prognostic group . | Points . | MC . | New lesions by SNP-A? . | n . | Median OS, mo . | Median EFS, mo . |
|---|---|---|---|---|---|---|
| Favorable | 0 | Good | No | 129 | 47.0 | 30.4 |
| Intermediate-1 | 1 | Good | Yes | 166 | 18.6 | 15.2 |
| Intermediate | No | |||||
| Intermediate-2 | 2 | Intermediate | Yes | 55 | 11.7 | 10.0 |
| Poor | No | |||||
| Unfavorable | 3 | Poor | Yes | 46 | 4.2 | 3.4 |
| P* | < .0001 | < .0001 |
| Prognostic group . | Points . | MC . | New lesions by SNP-A? . | n . | Median OS, mo . | Median EFS, mo . |
|---|---|---|---|---|---|---|
| Favorable | 0 | Good | No | 129 | 47.0 | 30.4 |
| Intermediate-1 | 1 | Good | Yes | 166 | 18.6 | 15.2 |
| Intermediate | No | |||||
| Intermediate-2 | 2 | Intermediate | Yes | 55 | 11.7 | 10.0 |
| Poor | No | |||||
| Unfavorable | 3 | Poor | Yes | 46 | 4.2 | 3.4 |
| P* | < .0001 | < .0001 |
Determined by Wald test from proportional hazards model.
New prognostic risk groups based on combined MC and SNP-A analyses. SNP-A was able to further delineate cytogenetic risk groups determined by MC as defined by IPSS (A-B). Four prognostic risk groups, namely favorable, intermediate-1, intermediate-2, and unfavorable were defined with the use of the combined karyotypic analysis by MC and SNP-A (C-D). †Favorable indicates good cytogenetics and no additional lesions detected by SNP-A. Intermediate-1 indicates good cytogenetics and additional SNP-A lesions detected, or intermediate cytogenetics and no additional lesions by SNP-A. Intermediate-2 indicates intermediate cytogenetics and additional lesions detected, or poor cytogenetics and no additional lesions detected by SNP-A. Unfavorable indicates poor cytogenetics by MC and additional lesions detected by SNP-A.
New prognostic risk groups based on combined MC and SNP-A analyses. SNP-A was able to further delineate cytogenetic risk groups determined by MC as defined by IPSS (A-B). Four prognostic risk groups, namely favorable, intermediate-1, intermediate-2, and unfavorable were defined with the use of the combined karyotypic analysis by MC and SNP-A (C-D). †Favorable indicates good cytogenetics and no additional lesions detected by SNP-A. Intermediate-1 indicates good cytogenetics and additional SNP-A lesions detected, or intermediate cytogenetics and no additional lesions by SNP-A. Intermediate-2 indicates intermediate cytogenetics and additional lesions detected, or poor cytogenetics and no additional lesions detected by SNP-A. Unfavorable indicates poor cytogenetics by MC and additional lesions detected by SNP-A.
Discussion
Our study shows that SNP-A karyotyping, alone or in combination with routine MC, can affect outcome prediction and improve prognostic stratification in patients with various types of MDS, MDS/MPN, and sAML. Patients with new SNP-A defects had worse survival outcomes when they were stratified by cytogenetic risk with the use of conventional MC, IPSS, and WHO risk schemes. The combined application of MC and SNP-A also increased the yield of chromosomal defects in these diseases which, through the aid of additional molecular techniques, may help better define patients with a truly normal karyotype. These patients appear to have better clinical prognosis and may account for the well-recognized tail on all MDS survival curves. Our studies show that MDS, MDS/MPN, and sAML are closely related cytogenetically and share a similar frequency of aCN-LOH. Similarly, patients with new SNP-A lesions, for example aCN-LOH7q, 17p, or 11q, could be included into higher-risk cytogenetic groups that currently contain MC-detected complex chromosomal defects and partial or numerical losses. For less frequent lesions, such an analysis will be possible when data from large cohorts of patients become available.
Although karyotyping analysis with SNP-A have been used in the past,34 the current study differs in several important and critical aspects compared with the initial report by Gondek et al14 and Mohamedali et al.18 First, this study emphasizes the complementary usage of both MC and SNP-A, whereas the original studies of SNP-A karyotyping were focused on the differences between the techniques. Second, we have now developed and used a refined diagnostic algorithm and have used better analytic methods and array technologies (6.0 Arrays). Third, in the current study, a more extensive analysis of survival outcomes were undertaken, including parameters such as EFS, PFS, and DFS, which were done in accordance with response and survival criteria set by the IWG for both MDS and AML. Finally, the intricate analyses were possible because the median follow-up was longer and the numbers of patients were greater.
Karyotyping tools such as FISH have a higher sensitivity (minimal number of clonal cells in the sample) compared with SNP-A, which is similar to comparative genomic hybridization in that one can detect clonal abnormalities with the use of 250K arrays if present in 20%-25% of cells.13,14 Affymetrix 6.0 arrays primarily differ from 250K arrays in their genomic coverage, the is, the number of SNPs present on the array. Increasing the number of probes will improve the resolution (decreasing the size of detectable lesions) but will not greatly affect sensitivity, which is comparable between 6.0 and 250K arrays. We attribute the improvement in the detection of chromosomal defects to the superior resolution level of SNP-A, the ability to study nondividing cells, and the recognition of aCN-LOH. The dual application of MC and SNP-A is ideal, because MC offsets the inability of SNP-A to detect balanced abnormalities and provides the additional advantage of distinguishing individual clones in the sample. The high concordance rate between MC and SNP-A in the detection of MC-defined unbalanced chromosomal defects also shows the reliability of SNP-A karyotyping. New lesions detected by SNP-A, which were previously not seen by MC, are clinically relevant and prognostic for outcome parameters. Moreover, the presence of SNP-A–detected defects, their number and type can influence survival outcomes in patients with MDS and MDS-related myeloid malignancies. The combination of SNP-A with MC is complementary and will be helpful in the evaluation of patients with MDS and related cancers.
Our study also reaffirms that recurrent areas of aCN-LOH are widespread lesions occurring particularly frequently in MDS/MPN subtypes such as chronic myelomonocytic leukemia (CMML). The new SNP-A–detected chromosomal lesions, including aCN-LOH and microdeletions, have been instrumental in the detection of novel gene mutations in these disorders, including TET2 in MDS/MPN,20,24-26 CBL family in CMML,21,23 MPL in refractory anemia with ring sideroblasts and thrombocytosis,22 and EZH2 in MDS, CMML, and AML35 with further mutations likely to be discovered on the basis of the presence of characteristic lesions. Recurrent areas of aCN-LOH indicate the presence of homozygous mutations in the affected regions, whereas microdeletions facilitate narrowing the search for genes affected by mutations. These new lesions, such as mutations in TET2, CBL, or EZH2, may show an effect on survival, as clearly is the case for p53.20,23,27
The goal of detecting molecular lesions, including invariant chromosomal defects and mutations, is to allow for objective nosologic and prognostic stratification in a heterogenous group of myeloid disorders and ultimately to devise targeted therapies. In general, whether detecting individual recurrent lesions or abnormalities within submorphologic or prognostic categories, the combination of MC and SNP-A added prognostic precision, confirming the clinical utility of this method. For example, application of SNP-A in patients with idiopathic cytopenia of undetermined significance, or those with hypocellular MDS versus aplastic anemia, may allow for improved disease classification and prognostic stratification and distinction of potentially new subsets of patients. Moreover, univariate and multivariate analyses confirmed that the presence of new SNP-A–detected defects, independent of MC and other prognostic factors in MDS, are predictors of poor outcomes in MDS and related malignancies, which allowed for the creation of a new risk stratification system that is relevant for the disease.
In conclusion, our report shows the potential of novel whole-genome scanning technologies, in particular SNP-A–based karyotyping, to improve cytogenetic diagnostics of hematologic malignancies such as MDS and to provide clinically useful diagnostic and prognostic information that cannot be obtained with currently used traditional technologies. On the basis of our data, we recommend the concurrent use of SNP-A and MC in the initial karyotypic evaluation of patients with hematologic conditions, whereby cytogenetics has a clear role in the prognosis of patients, particularly in MDS, MDS/MPN, and AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The work was supported by National Institutes of Health grants R01-HL082983 (J.P.M.), U54-RR019391 (J.P.M. and M.A.S.), and K24-HL077522 (J.P.M.); Department of Defense grant MPO48018 (M.A.M.); and a charitable donation from Robert Duggan Cancer Research Foundation.
The Framingham Heart Study is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with Boston University (contract no. N01-HC-25195). This manuscript was not prepared in collaboration with investigators of the Framingham Heart Study and does not necessarily reflect the opinions or views of the Framingham Heart Study, Boston University, or NHLBI.
National Institutes of Health
Authorship
Contribution: R.V.T. was responsible for overall design, data collection, data analysis, data interpretation, statistical analysis, manuscript preparation, writing and completion and final approval of manuscript; L.P.G. was responsible for overall design, data gathering, data analysis, data interpretation, and final approval of manuscript; C.L.O. analyzed data, interpreted data, wrote the manuscript, and approved the final manuscript; P.E. analyzed statistical data and approved the final manuscript; J.H. and A.K. gathered data and approved the final manuscript; A.M. gathered data, edited the manuscript, and approved the final manuscript; A.S.A., A.F.L., and M.A.S. provided patient samples, edited the manuscript, and approved the final manuscript; R.P. provided patient samples and approved the final manuscript; M.A.M. provided patient samples, gathered data, interpreted data, edited the manuscript, and approved the final manuscript; G.J.M. analyzed data, provided patient samples, interpreted data, wrote the manuscript, and approved the final manuscript; and J.P.M. was responsible for overall design, patient samples, financial support, manuscript preparation, writing and completion, and final approval of manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jaroslaw P. Maciejewski, Department of Translational Hematology and Oncology Research, Taussig Cancer Institute, Cleveland Clinic, 9500 Euclid Ave R40, Cleveland, OH 44195; e-mail: maciejj@ccf.org; or Ghulam J. Mufti, Department of Haematological Medicine, King's College London School of Medicine, The Rayne Institute, 123 Coldharbour Ln, London SE5 9NU, United Kingdom; e-mail: ghulam.mufti@kcl.ac.uk.
References
Author notes
J.P.M. and G.J.M. are joint senior authors of this article.

![Figure 1. Comparison of exemplary lesions detected by genomewide SNP-A 6.0. (A) Whole-genome view shows the complete diploid set of human chromosomes. A close-up view of chromosome 21 shows the presence of a deletion on its long arm, illustrated by a decrease in the copy number line (blue line and red dots) and LOH (paucity of green bars, indicating the frequency of heterozygous loci [red box]). A closer look at the region shows loci potentially involved in disease pathogenesis. (B) Illustration of the 3 exemplary characteristic lesions seen by 6.0 SNP-A: deletion, duplication, and aCN-LOH. For deletions, LOH as indicated by contraction of the AA and BB alleles (blue lines) and reduction of the copy number (top) marks the affected region. For duplications, the doubling of the heterozygous AB allele and an increase in copy number delineate the abnormality. For aCN-LOH, the loss of the AB allele without contraction of the AA and BB alleles and a normal copy number identify the lesion. In each example, the normal copy number and allele calls for CD3+ cells (green lines) shows the absence of a germline abnormality, thus confirming the somatic nature of the lesions. (C) An exemplary microdeletion in chromosome 7, seen as a drop in the diploid copy number line (blue line in red box) and the absence of this abnormality in the CD3+ lymphocytes, is shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/17/10.1182_blood-2010-07-295857/4/m_zh89991168500001.jpeg?Expires=1768371340&Signature=IzoRBI1VK63-I86HMfNfEReHr8r2O-cWRlxvpkpq6DX8Pegdx1nlHI0Ul5UzNhZzy5N5xmIKbJqgnXDby8fs9oJ9yLvBUc0btiYN3rg4qXygvmw8JlZ9GBOUPANkPaR8XAsE7l4UK0trErET5aet0wm0-eMAzSCdjjjLNdpFwDQOWSJzVtB5cUIoldICCSBuDf~qKo2VSnoJhSy6IdEsqqmpptVhNb0FzBVcPjwhEdyEbuoNmIeQ6An0DRzQqanjyixCNEepFpyLJK6BMiUQ1y37QjKd4L~54cexDI0uiBjj7bvpXEvzwYe~vzv1U-zlOGETs-k3tAYBC4Br4ESTDQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal