Abstract
DNA methylation plays an important role in the self-renewal of hematopoietic stem cells and in the commitment to the lymphoid or myeloid lineages. Using purified CD34+ hematopoietic progenitor cells and differentiated myeloid cell populations from the same human samples, we obtained detailed methylation profiles at distinct stages of hematopoiesis. We identified a defined set of differentiation-related genes that are methylated in CD34+ hematopoietic progenitor cells but show pronounced DNA hypomethylation in monocytes and in granulocytes. In addition, by comparing hematopoietic progenitor cells from umbilical cord blood to hematopoietic progenitor cells from peripheral blood of adult donors we were also able to analyze age-related methylation changes in CD34+ cells. Interestingly, the methylation changes observed in older progenitor cells showed a bimodal pattern with hypomethylation of differentiation-associated genes and de novo methylation events resembling epigenetic mutations. Our results thus provide detailed insight into the methylation dynamics during differentiation and suggest that epigenetic changes contribute to hematopoietic progenitor cell aging.
Introduction
DNA methylation is a major epigenetic modification that has essential roles in gene regulation and chromatin organization.1,2 Significant changes in DNA methylation patterns have been observed during differentiation processes, in particular comparing pluripotent stem cells or multipotent progenitors with lineage-committed cells.3-7 Consequently, stem cells and differentiated cells are characterized by specific epigenetic modification patterns that are correlated with active and repressed chromosomal regions.8,9 The hallmark of exit from pluripotency and the onset of differentiation is the reorganization of these epigenetic patterns, leading to the repression of stem cell–specific factors, the activation of lineage-specific genes, and the continued silencing of effector genes of other lineages.10,11
The formation of the various hematopoietic cell types is a highly hierarchical process, originating from a unique HSC population in the BM that gives rise to a series of well-characterized progenitors and terminally differentiated cells.12 HSCs are defined on the basis of their multipotency and self-maintaining capacity and by specific expression patterns of cell surface markers.12,13 On the basis of the expression of these surface proteins, populations of multipotent HSC-like progenitor cells can be isolated and separated from lineage-committed cells.12 Hematopoiesis thus provides a well-defined system to study DNA methylation changes during differentiation, for example, by comparing epigenetic maps of HSCs with those of lineage-restricted progenitors and differentiated cells.
A recent landmark study has used this approach to establish comprehensive methylome maps of various mouse hematopoietic cell populations, which led to the identification of new genes and pathways involved in lineage choice and differentiation.14 This work also showed that myelopoiesis and lymphopoiesis result in different terminal DNA methylation patterns and that myeloid commitment is characterized by less global DNA methylation than lymphoid commitment.14 These findings are in agreement with earlier reports showing that DNA methylation, in particular by DNA methyltransferase 1 (DNMT1), has a direct role in regulating the self-renewal of HSCs and the commitment to lymphoid versus myeloid lineages.15,16
Adult HSCs give rise to all blood cells for the entire lifespan and reside in specialized stem cell niches in the BM that create the microenvironment essential for long-term self-renewal.17 Accumulating evidence suggests that aging is associated with a reduction in HSC numbers and functionality.18,19 For example, purified adult HSCs can show a reduced potential to differentiate toward the lymphoid lineage,18,20 which might be related to a loss of DNA methylation.15 Recent studies that used whole blood samples from human donors identified specific sites of age-dependent hypermethylation, which correlated with sets of genes found hypermethylated in various cancers.21,22 These age-related patterns could also be observed in partially purified lineage-committed cells, which suggests that aging is accompanied by small but significant changes in DNA methylation patterns, which are not related to changes in the cellular composition of the examined tissue.21-23
To analyze dynamic DNA methylation changes in the human hematopoietic system, we isolated CD34+ HPCs from umbilical CB (CB-HPCs) and compared their genomic DNA methylation patterns with granulocytes and monocytes, which were purified from the same blood samples. Additional comparisons were made with CD34+ HPCs from mobilized peripheral blood of adult donors (mPB-HPCs), with an average age of 35 years. Our data identify specific epigenetically regulated pathways associated with HPC differentiation and also suggest that epigenetic changes could underpin the altered phenotype of aged HPCs.
Methods
Isolation and sampling of human HPCs, monocytes, and granulocytes
Hematopoietic progenitors were isolated with the use of the CD34+ cell fraction from CB-HPCs or from mPB-HPCs of healthy voluntary adult donors with an average age of 35 years. Mobilization was achieved by treatment with G-CSF before leukapheresis for allogeneic stem cell transplantation. All samples were collected after obtaining informed consent in accordance with the Declaration of Helsinki, following the guidelines approved by the Ethics Committee on the Use of Human Subjects at the University of Heidelberg.
After density centrifugation on Ficoll-Hypaque (Biochrom KG) HPCs and monocytes were enriched from the mononuclear cell fraction. CD34+ cells were isolated with the use of monoclonal anti-CD34 antibody labeled with magnetic beads on an autoMACS system (Miltenyi Biotec). Monocytes were isolated from the CD34− cell fraction and stained with anti–CD14-PE (clone MϕP9; Becton Dickinson). Granulocytes were isolated from the density gradient cell pellet after removal of red blood cells by lysis with Pharm Lyse Buffer (Becton Dickinson) and stained with anti–CD16-FITC antibody (clone NKP15; Becton Dickinson). Finally, cells were sorted by flow cytometry on a FACSVantage SE cell sorter (Becton Dickinson) with the use of a combination of either CD14 (monocytes) or CD16 (granulocytes) expression, as well as cell size and granularity. These parameters were analyzed by flow cytometry with forward and side scatter characteristics and the respective fluorescence channel. Nonviable cells were excluded by additional staining with propidium iodide. Reanalysis of all cell types showed purities > 92% for all samples.
DNA isolation
Genomic DNA was isolated from all samples with the use of the QiAmp DNA Micro Kit (QIAGEN) according to the manufacturer's instruction. DNA quality was controlled by agarose gel electrophoresis and quantified by a NanoDrop ND-1000 Spectrometer (peqLab Biotechnologies).
Array-based DNA methylation profiling and data analysis
Array-based gene-specific DNA methylation analysis was performed with Infinium HumanMethylation27 bead chip technology (Illumina) according to the manufacturer's instructions. Genomic DNA (500 ng) from each sample was bisulfite converted with the EZ-96 DNA Methylation Kit (Zymo Research Corporation) according to the manufacturer's recommendations. Bisulfite-treated genomic DNA was whole-genome amplified and hybridized to HumanMethylation27 BeadChips. The methylation status of a specific cytosine is indicated by average β (AVB) values where 1 corresponds to full methylation and 0 to no methylation. Raw array data were quantile normalized, and P values for comparisons between different datasets were statistically adjusted with the Benjamini-Hochberg correction, as described previously.23 Data from allosomal loci were excluded from further analysis to eliminate the influence of sex-specific methylation differences. Individual loci were scored as differentially methylated if the AVB difference was ≥ 0.15.24 For comparisons, methylation profiles from mesenchymal stem cells25 and from keratinocytes and fibroblasts23 were also included. Functional categories of differentially methylated genes were identified by Ingenuity Pathway Analysis (www.ingenuity.com). The Infinium methylation data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress), under accession no. E-MTAB-487.
454 DNA bisulfite sequencing
Deep DNA bisulfite sequencing was performed from equimolar sample pools, as described previously.23 For 454 sequencing, bisulfite-treated genomic DNA was amplified with sequence-specific primers containing cell type–specific barcodes and 454 linker sequences (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Analysis of transcription factor binding motifs
To identify enriched transcription factor binding motifs we used PScan26 and 130 transcription factor-binding profiles (Homo sapiens) from the JASPAR database.27 The analysis of genes was focused on the region from −450bp to 50bp with respect to their transcription start site. To summarize the results of the PScan analysis, a heatmap displaying the natural logarithm of the P values was generated.
Results
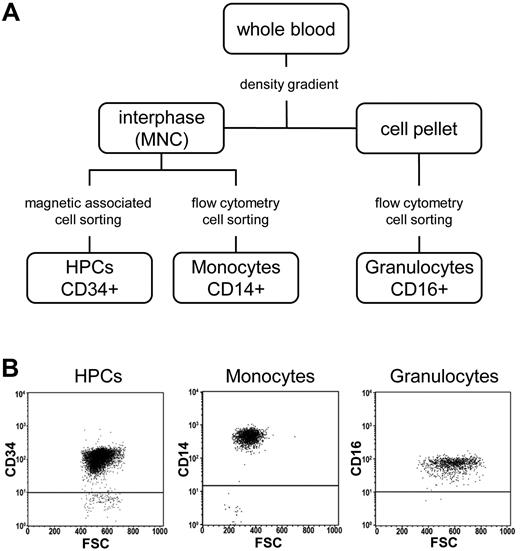
The availability of purified CD34+ HPCs and their differentiated derivatives (see “Isolation and sampling of human HPCs, monocytes, and granulocytes”) provided an opportunity to address several key questions related to epigenetic changes during HSC differentiation and aging. To this end we obtained 18 primary cell samples representing distinct cell populations (7× HPCs from CB-HPCs, 5× HPCs from peripheral blood of G-CSF–treated adult donors [mPB-HPCs], 3× monocytes, and 3× granulocytes; Figure 1A). The purity in all samples was > 92%, as validated by flow cytometry (Figure 1B). Genomic DNA of all 18 samples was analyzed by Illumina Infinium arrays to determine the methylation status of 27 578 CpG dinucleotides within 14 495 promoters of the human genome.24 This analysis generated 18 million methylation scores, with β values for individual markers ranging from 0 (unmethylated) to 1 (completely methylated). Raw data were normalized and statistically corrected as described in “Methods.”
Isolation of primary cell samples. (A) Flow chart showing the isolation strategy for CD34+ HPCs, CD14+ monocytes, and CD16+ granulocytes. (B) Flow cytometric dot plots showing the purity of cell populations. MNC indicates mononuclear cells; FSC, forward scatter.
Isolation of primary cell samples. (A) Flow chart showing the isolation strategy for CD34+ HPCs, CD14+ monocytes, and CD16+ granulocytes. (B) Flow cytometric dot plots showing the purity of cell populations. MNC indicates mononuclear cells; FSC, forward scatter.
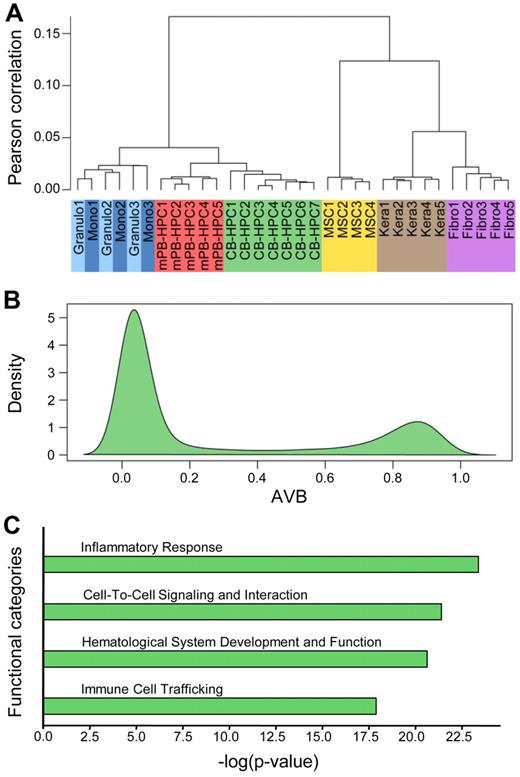
Unsupervised hierarchical clustering of the 18 newly obtained methylation profiles together with 14 published profiles from other tissues showed a high similarity in the methylation patterns of defined cell types (Figure 2A). This very high degree of interindividual similarity allowed us to establish average methylation profiles for each cell type. We also observed that the methylation patterns of differentiated cells from the myeloid lineage appeared closely related. However, granulocytes and monocytes could clearly be separated from HPCs and from the previously published methylation profiles derived from mesenchymal stem cells, fibroblasts, and keratinocytes (Figure 2A). A closer analysis of the distribution of methylation scores (β values) showed that most of the analyzed sites were unmethylated in CB-HPCs (Figure 2B). However, a density plot also showed a pronounced peak of hypermethylated loci, consisting of 4950 markers with β values ≥ 0.8 (Figure 2B). The corresponding genes probably contribute to the specific epigenetic program of HPCs. Indeed, genes associated with hypermethylated markers in HPCs were strongly enriched in functional categories associated with general characteristics of differentiated hematopoietic cells, including inflammatory response, cell-to-cell signaling, hematologic system development, and immune cell trafficking (Figure 2C). These findings are consistent with the notion that DNA methylation in HPCs restricts the functions of differentiated hematopoietic cells.
Methylation patterns of pluripotent CB-HPCs. (A) Unsupervised hierarchical clustering of the 18 newly obtained methylation profiles together with 14 published profiles from other tissues. Defined cell types show a high overall similarity in the methylation patterns and thus cluster together. (B) Density plot of Illumina methylation scores of CB-HPCs. Although most markers are unmethylated, the plot also shows a pronounced peak of hypermethylated loci, consisting of 4950 markers with β values ≥ 0.8. (C) Ingenuity Pathway Analysis of markers with AVB ≥ 0.8 in CB-HPCs. The plot shows the 4 functional categories most significantly overrepresented in the hypermethylated loci in CB-HPCs. These categories are commonly associated with differentiated hematopoietic cells.
Methylation patterns of pluripotent CB-HPCs. (A) Unsupervised hierarchical clustering of the 18 newly obtained methylation profiles together with 14 published profiles from other tissues. Defined cell types show a high overall similarity in the methylation patterns and thus cluster together. (B) Density plot of Illumina methylation scores of CB-HPCs. Although most markers are unmethylated, the plot also shows a pronounced peak of hypermethylated loci, consisting of 4950 markers with β values ≥ 0.8. (C) Ingenuity Pathway Analysis of markers with AVB ≥ 0.8 in CB-HPCs. The plot shows the 4 functional categories most significantly overrepresented in the hypermethylated loci in CB-HPCs. These categories are commonly associated with differentiated hematopoietic cells.
To further analyze the data, we compared the HPC methylation profile with the methylation profiles of granulocytes and monocytes. These cells were purified in parallel to the HPCs and from the same blood samples. Pairwise comparison between HPCs and the 2 differentiated cell types showed a clear hypomethylation effect in both granulocytes (Figure 3A) and monocytes (Figure 3B), which affected > 600 markers. Notably, a highly overlapping set of markers was identified in both differentiated cell types (Figure 3C). To validate the array-based predictions, 4 genes were selected on the basis of their predicted hypomethylation and annotated gene functions, and methylation patterns were determined by deep bisulfite sequencing from equimolar sample pools. We obtained several thousand high-quality reads for 12 amplicons (4 genes in 3 cell types) with a total coverage ranging from 74× to 1644×. As shown in Figure 3D, the 454 sequencing profiles were essentially identical to the Infinium-based results and confirmed significant differentiation-dependent DNA hypomethylation for all 4 genes. For example, S100A8 is known to be highly expressed in cells of the myeloid lineage and has been shown to mediate differentiation-dependent inhibition of telomerase activity.28 Our methylation data show very high levels of homogeneous methylation across the S100A8 promoter in HPCs and complete loss of methylation in granulocytes and monocytes (Figure 3D). Similarly, MPO, a specific marker for the myeloid lineage, becomes hypomethylated and activated during myeloid differentiation.29 In agreement with these findings, our sequencing results also show that the MPO upstream region, which harbors the probe from the Infinium array, becomes demethylated in differentiated myeloid cells (Figure 3D). Finally, array-based predictions could also be validated for SLAMF7 and TREM2, 2 additional myeloid genes30,31 (Figure 3D). Interestingly, the differentiation-dependent demethylation of SLAMF7 was restricted to the promoter region, whereas the gene body remained methylated, which is consistent with a specific role of promoter DNA methylation in gene silencing. Our results thus confirm the differentiation-dependent demethylation of myeloid genes by an independent approach.
Analysis of differentiation-dependent methylation changes in CB-HPCs. Comparison of genomic DNA methylation profiles of granulocytes (A) and monocytes (B) with those of CB-HPCs. Red dots indicate differentially methylated sites. (C) Venn diagram showing an overlapping set of 490 hypomethylated CpG dinucleotides (core set). (D) Validation of selected markers by 454 bisulfite sequencing. Schematic outline of validation genes. Gray lines represent the sequenced regions, arrows indicate transcriptional start sites, vertical lines represent individual CpG dinucleotides, and triangles indicate CpG markers interrogated on the array. Sequencing results are shown as heatmaps in which each row represents one sequence read. Individual red boxes indicate methylated and green boxes indicate unmethylated CpG dinucleotides. Sequencing gaps are shown in white. Panels below heatmaps summarize the results from 454 bisulfite sequencing (454) and from the array-based methylation analysis (Inf). Sequencing coverage ranged from 74 to 1644 reads, as indicated.
Analysis of differentiation-dependent methylation changes in CB-HPCs. Comparison of genomic DNA methylation profiles of granulocytes (A) and monocytes (B) with those of CB-HPCs. Red dots indicate differentially methylated sites. (C) Venn diagram showing an overlapping set of 490 hypomethylated CpG dinucleotides (core set). (D) Validation of selected markers by 454 bisulfite sequencing. Schematic outline of validation genes. Gray lines represent the sequenced regions, arrows indicate transcriptional start sites, vertical lines represent individual CpG dinucleotides, and triangles indicate CpG markers interrogated on the array. Sequencing results are shown as heatmaps in which each row represents one sequence read. Individual red boxes indicate methylated and green boxes indicate unmethylated CpG dinucleotides. Sequencing gaps are shown in white. Panels below heatmaps summarize the results from 454 bisulfite sequencing (454) and from the array-based methylation analysis (Inf). Sequencing coverage ranged from 74 to 1644 reads, as indicated.
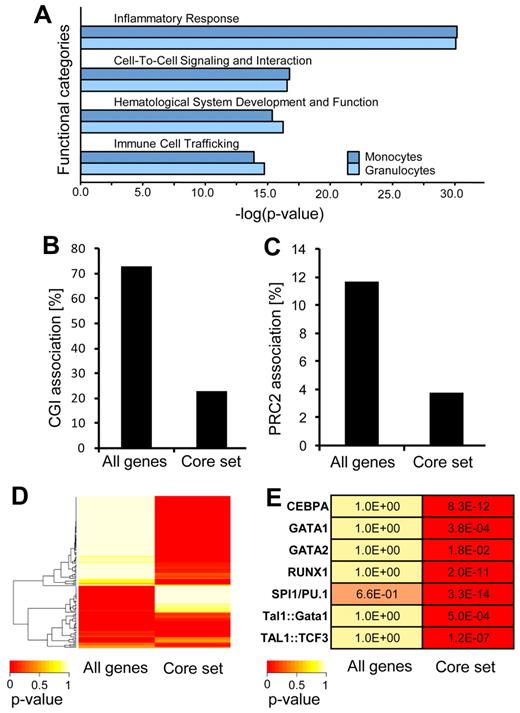
To further support the notion that the differentiation of HPCs is associated with the demethylation of a specific epigenetic program, we subjected all markers with differentiation-dependent hypomethylation to pathway analysis. The results showed a strong enrichment in functional categories associated with general characteristics of differentiated hematopoietic cells, including inflammatory response, cell-to-cell signaling, hematologic system development, and immune cell trafficking (Figure 4A). Notably, these were the same functional categories that we previously showed to be affected by DNA hypermethylation in HPCs (Figure 2C). In conclusion, our results thus uncover a specific DNA methylation program that is probably used to silence differentiation-specific genes in HPCs.
Mechanisms regulating differentiation-dependent demethylation. (A) Ingenuity Pathway Analysis (IPA) of markers with differentiation-dependent demethylation. (B) Percentage of genes showing differentiation-dependent methylation changes in CpG islands (CGIs). (C) Association of genes showing differentiation-dependent methylation changes with known PRC2 targets. (D) Analysis of hypomethylated promoters in differentiated myeloid cell populations for specific transcription factor binding motifs with the use of PScan and transcription factor-binding motifs from the JASPAR database (see “Methods”). A specific set of transcription factor binding shows a high probability of binding near promoters of genes derived from the hypomethylated core set. (E) Motifs of transcription factors involved in hematopoiesis are highly enriched near promoters derived from the hypomethylated core set.
Mechanisms regulating differentiation-dependent demethylation. (A) Ingenuity Pathway Analysis (IPA) of markers with differentiation-dependent demethylation. (B) Percentage of genes showing differentiation-dependent methylation changes in CpG islands (CGIs). (C) Association of genes showing differentiation-dependent methylation changes with known PRC2 targets. (D) Analysis of hypomethylated promoters in differentiated myeloid cell populations for specific transcription factor binding motifs with the use of PScan and transcription factor-binding motifs from the JASPAR database (see “Methods”). A specific set of transcription factor binding shows a high probability of binding near promoters of genes derived from the hypomethylated core set. (E) Motifs of transcription factors involved in hematopoiesis are highly enriched near promoters derived from the hypomethylated core set.
We then further analyzed the data to determine the molecular characteristics of the genes that showed differentiation-dependent hypomethylation. This showed that the effect was distinctly more pronounced in CpG dinucleotides outside of CpG islands (Figure 4B). In addition, known target genes of the PRC2 (Polycomb Repressive Complex 2) were substantially underrepresented among the hypomethylated genes (Figure 4C). This is consistent with the previously reported association of PRC2 chromatin and de novo DNA methylation during differentiation.6 Furthermore, because transcription factors have been implied in the establishment and maintenance of DNA methylation patterns,32 we analyzed whether hypomethylated promoters in differentiated myeloid cell populations are characterized by specific transcription factor binding sites. With the use of PScan26 and 130 transcription factor-binding motifs from the JASPAR database,27 we observed a significant enrichment of specific transcription factor binding sites in promoters of genes contained in our hypomethylated core set, that is, the set of genes that became demethylated both in granulocytic and in monocytic differentiation (Figure 4D). This suggests that genes losing DNA methylation during myeloid differentiation are regulated by a specific set of transcription factors. By comparing these enriched sites with binding motifs of known hematopoietic transcription factors,33 such as TAL1, RUNX1, GATA1, we found many well-established motifs significantly overrepresented (Figure 4E). Together, these data suggest that specific chromatin structures and sets of hematopoietic transcription factors regulate DNA demethylation and the concomitant activation of myeloid genes necessary for myeloid differentiation.
Having established differentiation-dependent DNA methylation changes, we also sought to analyze the DNA methylation patterns of HPCs in the context of stem cell aging. This represented an important extension of this study, because aging of HPCs has been associated with reduced differentiation capacity, which possibly reflects reduced phenotypic plasticity.17,34 To obtain “old” HPCs, CD34+ cells were purified from G-CSF–mobilized peripheral blood of donors with an average age of 35 years (mPB-HPCs), and their methylation profiles were compared with the profiles of CB-HPCs. Methylation profiles from 5 mPB-HPC samples again showed little overall methylation changes between individual samples (Figure 2A), thus confirming the presence of an HPC-specific methylation pattern.
The comparison of the mPB-HPC methylation profiles with the CB-HPC methylation profiles showed a bimodal pattern with 350 markers becoming hypomethylated and 192 markers that were hypermethylated in mPB-HPCs (Figure 5A). To analyze these changes in detail and to validate the array-based predictions, 4 genes were selected on the basis of their predicted methylation changes and/or their annotated gene functions and were subjected to methylation analysis by deep bisulfite sequencing. We obtained several thousand high-quality reads for 8 amplicons (4 genes in 2 age groups) with the total coverage ranging from 243× to 1657×. As shown in Figure 5B, the 454 sequencing profiles are highly consistent with the Infinium-based results. For example, the KCNAB3 upstream region was almost completely unmethylated in CB-HPCs but showed high levels of homogeneous methylation across the entire amplicon in mPB-HPCs (Figure 5B). For SOCS1, the methylation changes were more restricted to the region around the CpG represented on the array, but the gene still showed clear hypermethylation in mPB-HPCs (Figure 5B). Finally, the 454 sequencing results also confirmed the array-predicted hypomethylation effect in mPB-HPCs. Both ICAM2 and GBP1 showed substantially reduced methylation in mPB-HPCs across the entire amplicon (Figure 5B), thus further showing the differences in methylation between HPCs isolated from young and from old donors.
Analysis of age-dependent methylation patterns in HPCs. (A) Comparison of DNA methylation profiles of CB-HPCs and mPB-HPCs. Red dots indicate differentially methylated sites. (B) Validation of selected markers by 454 bisulfite sequencing. PCR amplification was performed on equimolar sample pools, and sequencing results are shown as in Figure 3D. Sequencing coverage ranged from 243 to 1657 reads, as indicated. Triangles indicate CpG dinucleotides that were interrogated on the array.
Analysis of age-dependent methylation patterns in HPCs. (A) Comparison of DNA methylation profiles of CB-HPCs and mPB-HPCs. Red dots indicate differentially methylated sites. (B) Validation of selected markers by 454 bisulfite sequencing. PCR amplification was performed on equimolar sample pools, and sequencing results are shown as in Figure 3D. Sequencing coverage ranged from 243 to 1657 reads, as indicated. Triangles indicate CpG dinucleotides that were interrogated on the array.
Age-related methylation changes have been investigated in a variety of human tissues, and we therefore sought to analyze the molecular characteristics of age-dependent methylation changes in greater detail. Pathway analysis failed to identify an enrichment for specific cellular pathways among the genes with age-related methylation changes (data not shown). However, we observed a substantial overlap between the genes that are hypomethylated in mPB-HPCs with genes that are hypomethylated during the myeloid differentiation of CB-HPCs (Figure 6A). This overlap was highly significant (P = 2.2 × 10−16, Fisher exact test) and indicates the existence of related DNA hypomethylation signatures for myeloid differentiation and aging of HPCs.
Mechanisms regulating age-related methylation changes. (A) Venn diagram showing overlapping hypomethylated markers in granulocytes, monocytes, and mPB-HPCs. (B) Correlation of genes showing age-related methylation changes with known PRC2 targets. (C) Transcription factor binding motif analysis of hypermethylated promoters in mPB-HPCs was performed as in Figure 4D and compared with the hypomethylated core dataset of differentiated myeloid cell populations (Figure 4D). A substantial difference in the transcription factor binding profiles for differentiation- and age-related methylation changes was observed.
Mechanisms regulating age-related methylation changes. (A) Venn diagram showing overlapping hypomethylated markers in granulocytes, monocytes, and mPB-HPCs. (B) Correlation of genes showing age-related methylation changes with known PRC2 targets. (C) Transcription factor binding motif analysis of hypermethylated promoters in mPB-HPCs was performed as in Figure 4D and compared with the hypomethylated core dataset of differentiated myeloid cell populations (Figure 4D). A substantial difference in the transcription factor binding profiles for differentiation- and age-related methylation changes was observed.
To further investigate DNA hypermethylation in mPB-HPCs we analyzed the distribution of Polycomb target sites among the differentially methylated genes. Our results showed that PRC2 target genes were substantially overrepresented among genes with DNA hypermethylation in mPB-HPCs (Figure 6B), whereas a reduced PRC2 association was observed among hypomethylated genes. A similar association has previously been identified as an important mechanism underlying age-related methylation changes in human peripheral blood samples21 and thus supports the notion that the observed methylation differences between CB-HPCs and mPB-HPCs are indeed age related.
Finally, we also analyzed the transcription factor binding motifs of genes that became hypermethylated in mPB-HPCs. The results failed to show any major differences to the control set (all genes). This is in marked contrast to the genes that became hypomethylated during differentiation, which showed a highly specific transcription factor binding motif pattern that is almost complementary to the control set (Figure 6C). These findings further show the underlying differences between the observed DNA hypomethylation and de novo methylation changes and suggest that both effects represent independent epigenetic changes associated with HPC aging.
Discussion
DNA methylation is extensively reprogrammed during mammalian development,35 but little is known about sequence specific methylation changes. We took advantage of an array-based approach to identify genome-wide DNA methylation changes at a single base pair resolution to analyze differentiation- and age-dependent DNA-methylation dynamics in the human hematopoietic system. Our results show that genomic DNA methylation profiles define distinct cell types and that the profiles of hematopoietic cells of the same type are closer related to each other than to other cell types.
Two recent studies described an unexpected reduction of promoter DNA methylation during mouse embryogenesis and during lineage commitment of mouse hematopoietic progenitors.14,36 In addition, it was shown that a conditional loss of mouse DNMT1 leads to the rapid death of hematopoietic stem cells, whereas reduced DNMT1 activity inhibited HPC self-renewal.15 Hypomorphic DNMT1 mice showed normal myelopoiesis but drastically reduced lymphopoiesis, suggesting that low levels of DNA methylation are sufficient to control genes of the lymphoid lineage, whereas DNA methylation is essential to repress genes of the myeloid lineage.15 Our results suggest that DNA methylation has a similar role during the differentiation of human HPCs. Genes that showed differentiation-dependent hypomethylation were strongly enriched in functional categories which are characteristic of differentiated hematopoietic cells, suggesting that HPCs use DNA methylation to silence the myeloid differentiation program.15,16
Pluripotency of embryonic stem cells relies on Polycomb-group proteins to reversibly repress genes required for differentiation.37-39 It has been suggested that Polycomb-group repression through the PRC2 complex is also needed to keep hematopoietic progenitors multipotent because enforced expression of the PRC2 proteins BMI1 and EZH2 enhances self-renewal of HPCs.40-42 However, the overexpressed PRC2 proteins targeted a set of genes that was distinct from those defined in embryonic stem cells.43 In agreement with these findings, our data show myeloid differentiation-dependent DNA hypomethylation predominantly at loci that are not associated with Polycomb target genes. Furthermore, we found that differentially hypomethylated genes are enriched for binding motifs of hematopoietic transcription factors. A close association between transcription factor binding, DNA methylation changes, and altered cellular phenotypes has also been found in other studies.44,45 The mechanistic details of this interaction, that is, whether transcription factor binding promotes DNA demethylation or whether DNA hypomethylation allows transcription factor binding, remain to be elucidated.
Epigenetic processes have been suggested to underpin the reduced phenotypic plasticity of pluripotent cells during aging.34 To identify methylation differences between young and old HPCs we compared methylation patterns between HPCs isolated from newborns with HPCs isolated from G-CSF–treated adult donors with an average age of 35 years (range from 28 to 41 years). Although we cannot exclude the possibility that the G-CSF–induced mobilization has an effect on the methylation pattern, the methylation changes identified in these cells are consistent with age-related epigenetic changes observed in other studies.21-23,46
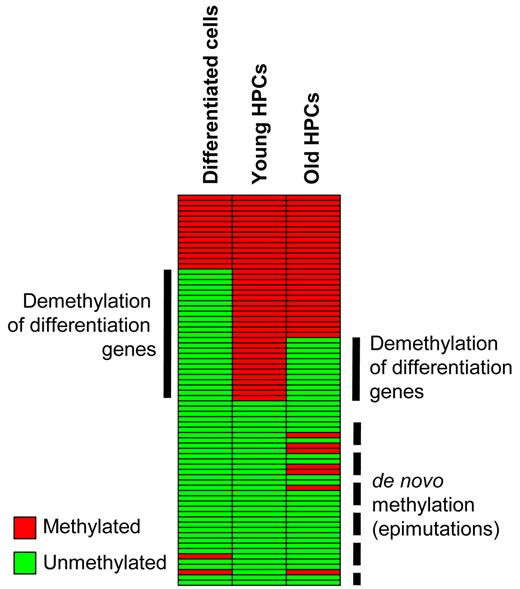
Importantly, our analysis of methylation patterns from G-CSF–mobilized HPCs identified epigenetic changes that might be relevant for understanding previously reported phenotypic changes in aging hematopoietic stem cells (Figure 7). (1) We observed hypomethylation of a subset of genes that showed a highly significant overlap with the genes hypomethylated during myeloid differentiation. These epigenetic alterations might explain the diminished pluripotency and the reduced differentiation potential of HPCs from older donors.20,47 (2) Furthermore, we observed de novo methylation of a subset of genes that are associated with Polycomb chromatin, an important factor in establishing de novo DNA methylation during aging and tumorigenesis.21,22,48 These de novo methylation events are usually associated with gene silencing and could thus contribute to the reduced phenotypic plasticity of aged stem cells.34 A prominent example is provided by the age-related hypermethylation of SOCS1 (Figure 5B), which has also been described in patients with multiple myeloma.49 SOCS1 acts as a negative regulator of cytokine signaling, is important for lymphoid differentiation,50 and could thus be an important factor for maintaining pluripotency in young HPCs. Further studies will be required to analyze comprehensively the functional significance of age-related epigenetic changes.
Schematic representation of DNA methylation changes during differentiation and aging. Promoter DNA methylation of a set of myeloid-specific differentiation genes in hematopoietic progenitor cells is lost during myeloid differentiation. A similar, albeit reduced, effect is also observed during HPC aging, in addition to de novo methylation. Both changes might contribute to the reduced phenotypic plasticity of aged hematopoietic progenitor cells.
Schematic representation of DNA methylation changes during differentiation and aging. Promoter DNA methylation of a set of myeloid-specific differentiation genes in hematopoietic progenitor cells is lost during myeloid differentiation. A similar, albeit reduced, effect is also observed during HPC aging, in addition to de novo methylation. Both changes might contribute to the reduced phenotypic plasticity of aged hematopoietic progenitor cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Tanja Musch for technical assistance and Oliver Heil, Cassandra Falckenhayn, and Bodo Brueckner for their valuable support in the analysis of methylation data.
This work was supported by grants from the Ministerium für Wissenschaft, Forschung und Kunst Baden-Württemberg (Forschungsverbund Molekulare und biomedizinische Grundlagen von Alterungsvorgängen; F.L. and A.D.H.) and by grants from the Deutsche Forschungsgemeinschaft (Priority Programmes 1356 and 1463; A.B. and F.L.).
Authorship
Contribution: M.T.B., I.H., A.B., A.D.H., and F.L. designed the research; M.T.B. and I.H. performed experiments and collected and analyzed the data; I.H., V.E., and A.D.H. contributed material; and M.T.B., A.B., and F.L. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Frank Lyko, Deutsches Krebsforschungszentrum, Im Neuenheimer Feld 580, 69120 Heidelberg, Germany; e-mail: f.lyko@dkfz.de.
References
Author notes
M.T.B. and I.H. contributed equally to this study.