Abstract
Overexpression of high mobility group AT-hook 2 (HMGA2) is found in a number of benign and malignant tumors, including the clonal PIGA− cells in 2 cases of paroxysmal nocturnal hemoglobinuria (PNH) and some myeloproliferative neoplasms (MPNs), and recently in hematopoietic cell clones resulting from gene therapy procedures. In nearly all these cases overexpression is because of deletions or translocations that remove the 3′ untranslated region (UTR) which contains binding sites for the regulatory micro RNA let-7. We were therefore interested in the effect of HMGA2 overexpression in hematopoietic tissues in transgenic mice (ΔHmga2 mice) carrying a 3′UTR-truncated Hmga2 cDNA. ΔHmga2 mice expressed increased levels of HMGA2 protein in various tissues including hematopoietic cells and showed proliferative hematopoiesis with increased numbers in all lineages of peripheral blood cells, hypercellular bone marrow (BM), splenomegaly with extramedullary erythropoiesis and erythropoietin-independent erythroid colony formation. ΔHmga2-derived BM cells had a growth advantage over wild-type cells in competitive repopulation and serial transplantation experiments. Thus overexpression of HMGA2 leads to proliferative hematopoiesis with clonal expansion at the stem cell and progenitor levels and may account for the clonal expansion in PNH and MPNs and in gene therapy patients after vector insertion disrupts the HMGA2 locus.
Introduction
Paroxysmal nocturnal hemoglobinuria (PNH) is an acquired hemolytic anemia caused by clonal expansion of a hematopoietic cell that lacks glycosyl phosphatidylinositol (GPI)–linked proteins because of a somatic mutation in the X-linked PIGA gene, which is essential for the synthesis of GPI-anchors (PNH cell).1 The deficiency of GPI-linked proteins accounts for some features of the clinical phenotype such as intravascular hemolysis and hemoglobinuria but a major unanswered question concerns the mechanism underlying the clonal expansion of the mutated hematopoietic stem or early progenitor cell necessary for the development of disease.2-5 A possible clue to the mechanism came from the observation that the HMGA2 gene was rearranged and the mRNA highly expressed, in the PNH cells of 2 patients, suggesting that in addition to the PIGA gene mutation an additional genetic event is required to confer a growth advantage to the PNH clone (two-hit-hypothesis).6 We were thus interested in investigating the consequences of HMGA2 overexpression on hematopoiesis.
The high mobility group AT-hook 2 (HMGA2) protein is a member of the HMGA family of nonhistone chromatin proteins, which also includes HMGA1a, HMGA1b, and HMGA1c.7 Exons 1 to 3 of the HMGA2 gene encode DNA-binding AT-hook domains, which can modulate transcription by affecting the DNA conformation of specific AT-rich regulatory elements promoting transcriptional activity. Exon 4 acts as a linker, and exon 5 encodes the acidic C-terminal domain of the protein and the 3′ untranslated region (UTR) of the mRNA.8,9 The HMGA2 protein is important in a wide spectrum of biologic processes, including cell proliferation, cell-cycle progression, apoptosis, and senescence,10,11 and is thought to play a crucial role in self-renewal and control of differentiation of embryonic stem (ES) cells,12 cancer stem cells,13 and neural stem cells.14
The HMGA2 protein is expressed highly during embryogenesis but only at very low levels in normal adult tissues.8 However, high levels of HMGA2 are found in various benign and malignant tumors, in particular those of mesenchymal origin, and are thought to contribute to transformation in these tumors.7,10,11 In most cases these tumors harbor a rearrangement of chromosome 12q13-15, the location of the HMGA2 gene, causing a deletion of the HMGA2 3′UTR, while sequences encoding the HMGA2 DNA binding domains are intact. The 3′UTR of HMGA2 contains 7 sequences complementary to the let-7-family of micro RNAs (miRNAs). Binding of the complementary sequences by let-7 miRNAs posttranscriptionally and negatively regulates HMGA2 mRNA and protein expression.11 Thus, chromosomal rearrangements within the HMGA2 locus deleting the let-7 binding sites cause overexpression of a full-length or truncated HMGA2 protein with a preserved DNA binding capacity.15,16
Interestingly, a chromosomal rearrangement causing a truncation of the 3′UTR of the HMGA2 gene was also reported in 2 patients with PNH leading to the overexpression of HMGA2 in PNH cells lacking GPI-linked proteins.6 Furthermore, overexpression and/or truncation of HMGA2 have been also found in patients with myelodysplastic syndromes (MDSs) and myelodysplastic syndromes/myeloproliferative neoplasms (MDSs/MPNs).17 These findings suggested the hypothesis that overexpression of HMGA2 may confer a clonal growth advantage to a PNH progenitor cell, thus contributing to pathogenesis in PNH.
The deletion of the Hmga2 gene in mice is associated with a pygmy phenotype18 while overexpression of the full-length or truncated Hmga2 cDNA is associated with increased adipose tissues, a variety of benign mainly mesenchymal tumors, and an increased frequency of breast cancer and hepatocellular carcinomas (reviewed in Ashar et al19 ). However, the consequence of expression of Hmga2 in hematopoiesis has not been investigated.
Here, to study the consequence of overexpression of Hmga2 in hematopoiesis we generated a transgenic mouse line expressing an Hmga2 cDNA with a truncation of its 3′UTR (ΔHmga2), mimicking the truncation seen in the 2 patients with PNH. ΔHmga2 transgenic mice showed increased peripheral blood cell counts in all blood cell lineages, hypercellular bone marrow (BM), splenomegaly, increased colony formation and erythropoietin (EPO)–independent erythroid colony growth, reminiscent of the hematopoiesis seen in MPNs. Hematopoietic cells from ΔHmga2 mice showed a growth advantage over wild-type (WT) cells in competitive repopulation assays and in serial BM transplantation (BMT) experiments, indicating that overexpression of HMGA2 leads to a proliferative growth advantage in hematopoietic cells at both the progenitor and stem cell levels.
Methods
Mice
The construct used in the production of transgenic mice, ΔHmga2, contains the full coding region of Hmga2 cDNA, but lacks 6 of 7 let-7 miRNA complementary sequences in the 3′UTR (Figure 1A). ΔHmga2 was made from mouse Hmga2 cDNA (BC052158; Addgene) by polymerase chain reaction (PCR) using primers with XhoI and ClaI sites. Using these sites, ΔHmga2 was inserted into the pPGKpuro vector (Addgene). The final ΔHmga2 construct with the phosphoglycerate kinase-1 (PGK) promotor and polyadenylate tail (Figure 1A) was obtained by digestion with SalI. Transgenic mice were produced by standard oocyte injection using C57BL6/J mouse-derived fertilized eggs. Founders were identified by PCR. We obtained the ΔHmga2 mouse line by breeding the ΔHmga2 founder mouse with C57BL6/J mice (The Jackson Laboratory). Mice were kept in the transgenic barrier facility at Washington University and the Children's Hospital of Philadelphia (CHOP). Third or later generations of heterozygous ΔHmga2 mice were used for experiments. Two-month-old mice were used for experiments unless otherwise specified. C57BL6/J wild-type mice from the same colony were used as controls. Mice with PIGA− circulating blood cells were produced by breeding Pigalox mice20 with Tg:Tie1-cre mice.21 Pigalox/Tg:Tie1-cre mice express a stable proportion of Piga− blood cells.20 These mice were then crossbred with ΔHmga2 mice to produce ΔHmga2 mice that have a proportion of PNH cells circulating in their peripheral blood (Piga−ΔHmga2+ mice). ΔHmga2 mice or Piga− mice were identified by standard PCR of tail genomic DNA using PCR primers. Information of the oligonucleotides including PCR primers used in this study is available in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). All experiments were approved by the Washington University School of Medicine and CHOP Animal Studies committees.
Identification of ΔHmga2-integrated site and semiquantification of ΔHmga2
To identify the genomic sequence flanking the ΔHmga2 transgene, bubble linker–mediated PCR22 was performed with Platinum Pfx DNA polymerase (Invitrogen). Tail DNA of the ΔHmga2 mouse was digested with ClaI and the fragments were ligated to a double-stranded bubble linker made from 2 oligonucleotides with complementary sequences. The ligation was applied to the primary PCR using a linker-specific primer and a transgene-specific primer. The product of primary PCR was used as the template for secondary PCR with a nested primer pair. The PCR product was purified from a 1.5% agarose gel using QIAquick Gel Extraction kits (QIAGEN) and sequenced using an ABI Big Dye V3.1 analyzer (Applied Biosystems) in the gene sequencing core laboratory at Washington University. The ΔHmga2-integrated site in the chromosome was determined using a BLAST search of the flanking sequences in the Ensemble genome database (http://www.ensembl.org). The analysis showed that the introduced cDNA was integrated into chromosome 9E3 (Figure 1A), where this cDNA is unlikely to affect the expression of any other genes. The copy number of integrated ΔHmga2 was estimated by semiquantitative PCR of genomic DNA from the founder mouse. One hundred nanograms of tail DNA from ΔHmga2 mice was amplified for 25, 30, or 35 cycles. Densities of the bands of these PCR products on 1.5% agarose gel were compared with products of PCR performed using DNA from WT mice mixed with various concentrations of ΔHmga2 cDNA. This analysis indicated the approximately 3 copies of the ΔHmga2 cDNA were integrated.
Quantitative RT-PCR of Hmga2 RNA
Total RNA was extracted from BM, spleen, thymus, and liver using the RNeasy Mini Kit (QIAGEN). After treatment with DNAse I (Invitrogen), reverse transcription (RT) was performed using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative RT-PCR (qRT-PCR) was performed with SYBR Green PCR master mix (Applied Biosystems) and a primer pair specific for Hmga2 cDNA using ABI7500 (Applied Biosystems). Expression of Hprt1 RNA was used as an internal control.
Western blotting of HMGA2 protein
For analysis of HMGA2 protein, total protein was extracted from BM, spleen, thymus, and liver using radioimmunoprecipitation assay buffer containing protease inhibitor cocktail (Sigma-Aldrich). Samples were applied to SDS-PAGE, transferred to a nitrocellulose membrane (GE Healthcare Life Sciences), blocked with Odyssey Blocking Buffer (Licor) and probed with anti-HMGA2 antibody and anti–rabbit secondary antibody. The antibodies used in this study are listed in supplemental Table 2. The membrane was scanned and bands were detected and analyzed using the Odyssey Infra-red Imager (Li-Cor).
Peripheral blood cell counts
One hundered microliters of peripheral blood samples were used for blood counts using a Hemavet 850 blood cell counter (Drew Scientific). Reticulocytes were evaluated by flow cytometry with thiazole orange20 using a FACScan (BD Biosciences).
Cells counts and differential cell populations of BM and spleen
Single-cell suspensions of BM cells were obtained by flushing both femurs and cells were counted using a cell counter (Beckman Coulter). Single-cell suspensions of spleens were obtained by disruption between 2 sterile frosted glass slides. The differential cell populations were determined in these single-cell suspensions by flow cytometry using anti–Gr-1 and anti–Ter-119, anti–T-cell receptor beta, and anti-CD45R/B220 antibodies.
Serum erythropoietin value
To evaluate serum EPO values, serum samples were prepared from peripheral blood using standard procedures. EPO values were measured using a Quantikine Mouse/Rat Epo Immunoassay kit (R&D Systems) according to the manufacturer's protocol.
Histopathology
Hematoxylin and eosin (H&E) staining was performed using paraffin-embedded BM and spleen samples with standard protocols.
Hematopoietic colony assay
BM cells (104) or spleen cells (106) were cultured in 2 wells of a 24-well culture plate supplemented with standard methylcellulose medium containing 3 U/mL human EPO, 10 ng/mL murine IL-3, 10 ng/mL human IL-6, and 50 ng/mL murine stem cell factor (SCF; StemCell Technologies). Numbers of burst forming-unit erythroid (BFU-E) and colony forming-unit granulocyte-macrophage (CFU-GM) were counted after 7-days culture. For the replating culture assay, all BFU-Es in plates were picked and seeded into fresh methylcellulose medium with the same cytokines. To observe EPO-independent formation of CFU erythroid (CFU-E), BM cells were also cultured in the methylcellulose without EPO (StemCell Technologies). Benzidine-positive clusters (benzidine was provided by Sigma-Aldrich) were counted after 3-days culture. Colonies were picked up at random for cytospin and stained with Giemsa solution (Sigma-Aldrich) to confirm morphologic findings.
Competitive repopulation assay
The BM of lethally irradiated recipient mice was reconstituted with mixtures of BM cells from ΔHmga2 mice (glucose phosphate isomeraseb red blood cell [RBC] and CD45.2+ white blood cell [WBC]) and mice with recipient allotypes (glucose phosphate isomerasea RBC in B6.Cg-IgHaThy1aGpia-J and CD45.1+ WBC in B6.SJL-Ptprcapep3b/BoyJ; both from The Jackson Laboratory) at the ratio of 1:1 or 1:9. A total of 5 × 106 BM cells were injected into recipient mice conditioned with 1000 cGy 24 hours before BMT with ratios of 1:1 and 1:9 of ΔHmga2 BM cells to BM cells with the respective recipient allotype. To analyze the chimerism of peripheral RBCs in BMT recipients, we performed cellulose acetate gel electrophoresis of total RBCs lysed with distilled water followed by staining for glucose phosphate isomerase using D-fructose-6-phosphate, β nicotinamide adenine dinucleotide phosphate, glucose-6-phosphate dehydrogenase, phenazine methosulfate, and Thiazolyl blue tetrazolium bromide (all from Sigma-Aldrich).23 Densities of glucose phosphate isomer-asea and glucose phosphate isomeraseb bands were measured using a densitometer (ImageJ 1.24; National Institutes of Health [NIH]). After subtracting a background value, the proportion of the density of glucose phosphate isomeraseb band in the sum of glucose phosphate isomerasea and glucose phosphate isomeraseb bands in the sample was calculated. To analyze the chimerism of WBCs in BMT recipients, we performed flow cytometry using anti-CD45.2 and anti-CD45.1, and lineage-specific antibodies.
Analysis of GPI− cells
Proportions of GPI− cells were determined by flow cytometry20 using antibody to CD24 for RBCs or fluorescently labeled aerolysin (FLAER; Protox Biotech) for granulocytes.
Flow cytometric analysis and sorting of HSC and committed progenitor cells
BM samples were treated with lineage-specific anti-CD3, anti-CD4, anti-CD5, anti-CD8a, anti-B220, anti-Gr1, anti–Ter-119, anti-CD11b, and anti-CD127 antibodies, and conjugated to anti PE-Cy7. Then cells were stained with anti–CD34-PE, anti-CD16/32-APC, anti–CD117-APC-Cy7, and anti–Ly-6A/E (Sca-1)–pacific blue antibodies. Using MoFlo flow cytometer (Beckman Coulter), c-kit+lineage−Sca-1+ (KLS) cells and committed progenitor cell fractions were identified (supplemental Figure 1).24 For microarray analysis, KLS cells and megakaryocyte-erythrocyte progenitor (MEP) cells were directly sorted into Trizol RS reagent (Invitrogen).
Microarray analysis
Eight independent preparations of RNA, which included 2 independent samples of KLS cells and MEP cells from ΔHmga2 mice and 2 of each from WT mice, were generated from sorted cells in Trizol RS by standard methods. Each preparation was made from 4 or 6 mice (2 or 3 males and females, respectively). Amplified cDNA samples were prepared using WT-Ovation Pico System (NuGene) and subjected to analysis with Affymetrix Mouse Gene 1.0 ST GeneChip microarrays. Data were normalized by quantile normalization and analyzed using the Partek Genomic Suite. MetaCore (GeneGo) was used for pathway analysis. Microarray data have been deposited in the Gene Expression Omnibus database (GSE24071).
Analysis of JAK2, STAT3, STAT5, and AKT
Expression of phosphorylated STAT3 (pSTAT3) and STAT5 (pSTAT5) in KLS cells was analyzed by flow cytometry. Lineage-depleted BM cells were prepared by MACS (Miltenyi Biotec) and incubated in IMDM (Invitrogen) containing 2% fetal bovine serum for 20 minutes at 37°C. According to the manufacturer's instructions, cells were fixed and permeabilized using BD Phosflow Buffers (BD Biosciences), and treated with anti-pSTAT3 (pY705) or anti-pSTAT5 (pY694) and anti–C-KIT and anti–SCA-1 antibodies. For Western blots, total BM cells were incubated in the absence or presence of 10 ng/mL IL3 and 3 U/mL EPO for 20 minutes at 37°C, and total protein was extracted. Western blot was performed using antibodies to total or phosphorylated JAK2, STAT3, STAT5, and AKT, and were analyzed using the Odyssey. Anti–β-actin antibody was used as internal control for quantitative analysis.
Statistical analysis
Statistical significance was determined either by a 2-sided Student t test or a 2-way ANOVA analysis. All data represent the mean ± SD.
Results
ΔHmga2 transgenic mice overexpress HMGA2 protein
Transgenic mice were produced by standard techniques by injecting oocytes with a construct with a 3′UTR truncated Hmga2 cDNA containing a Pgk promoter and polyadenylation site. One line was selected for further study and the site of integration of the transgene was determined by bubble PCR22 and the copy number was estimated by semiquantitative PCR. Approximately 3 copies of the transgene were integrated at chromosomal position 9E3, in a large intergenic region between the Clstn2 (calsyntenin-2) and Nmnat3 (nicotinamide nucleotide adenylyltransferase 3) genes (Figure1A). We believe that integration at this position is unlikely to cause any phenotype because of gene disruption. ΔHmga2 mice with a truncation within the 3′UTR (ΔHmga2, Figure 1A) showed increased body weight compared with WT mice (Figure 1B). In transgenic animals the levels of Hmga2 mRNA (Figure 1C) and HMGA2 protein (Figure 1D) were increased in comparison to WT mice in all tissues tested including BM, spleen, and thymus. In BM, spleen, and thymus the Hmga2 mRNA was increased up to 3-fold (Figure 1C) leading to a high level of HMGA2 protein expression, while in WT mice the HMGA2 protein was barely detectable (Figure 1D).
Hmga2 transgenic mice. (A) Diagram of 3′UTR-truncated Hmga2 (ΔHmga2) lacking 6 of 7 complementary sites of let-7-family microRNAs. BP shows the break point as it has been described in patients with PNH.6 The phosphoglycerate kinase-1 promoter (PGK-PROM) and polyadenylate tail (PGK-PA) served to express HMGA2 protein, integrated into chromosome 9E3.3, 5.4 kb from end of Clstn2 and 315.2 kb from end of Nmnat3. (B) Body weight of 3-month-old ΔHmga2 mice (male, n = 5; female, n = 4) compared with that of WT mice (male and female, n = 5 each). (C) Increased expression of Hmga2 mRNA in ΔHmga2 mice (n = 4) compared with WT mice (n = 3) as determined by QRT-PCR analysis. (D-E) Western blots of HMGA2 protein expression. Results of spleen, thymus, and liver derived from a single gel, and a vertical line between WT and ΔHmga2 has been inserted to indicate a repositioned gel lane (D). Results of BM derived from a single gel (E). Representative data of 3 independent experiments are shown. *P < .05.
Hmga2 transgenic mice. (A) Diagram of 3′UTR-truncated Hmga2 (ΔHmga2) lacking 6 of 7 complementary sites of let-7-family microRNAs. BP shows the break point as it has been described in patients with PNH.6 The phosphoglycerate kinase-1 promoter (PGK-PROM) and polyadenylate tail (PGK-PA) served to express HMGA2 protein, integrated into chromosome 9E3.3, 5.4 kb from end of Clstn2 and 315.2 kb from end of Nmnat3. (B) Body weight of 3-month-old ΔHmga2 mice (male, n = 5; female, n = 4) compared with that of WT mice (male and female, n = 5 each). (C) Increased expression of Hmga2 mRNA in ΔHmga2 mice (n = 4) compared with WT mice (n = 3) as determined by QRT-PCR analysis. (D-E) Western blots of HMGA2 protein expression. Results of spleen, thymus, and liver derived from a single gel, and a vertical line between WT and ΔHmga2 has been inserted to indicate a repositioned gel lane (D). Results of BM derived from a single gel (E). Representative data of 3 independent experiments are shown. *P < .05.
Expression of ΔHmga2 results in proliferative hematopoiesis that resembles human MPNs
To investigate the effect of ΔHmga2 overexpression in hematopoietic cells we examined peripheral blood cell counts, BM, and spleen in the transgenic animals. We found a significant increase in peripheral WBCs, neutrophils, monocytes, RBCs, and platelets in ΔHmga2 mice compared with WT mice (Table 1). The increase in RBCs was associated with a decrease in the mean corpuscular volume (MCV) of RBCs and a decrease in serum EPO levels (Table 1). The BM of ΔHmga2 mice was hypercellular with an increased number of nucleated BM cells (Figure 2A) in femurs and increased numbers of megakaryocytes of normal morphology in histologic examinations (Figure 2C). The proportion of Ter-119+ erythroid cells was significantly higher in BM from ΔHmga2 mice (Figure 2D). Furthermore, ΔHmga2 mice showed splenomegaly (Figure 2B) with an expansion of red pulp (Figure 2C), an increase in the proportion of Ter-119+ erythroid cells (Figure 2D), and a perturbation of the normal spleen architecture, suggesting extramedullary erythropoiesis in the spleen of ΔHmga2 mice. The proportion of T and B cells was similar in transgenic and WT mice in BM and spleen. Reticulin fibrosis was absent in BM and spleen.
Peripheral blood cell counts and serum EPO levels
| . | WT . | ΔHmga2 . | P . |
|---|---|---|---|
| WBCs, ×109/L | 8.10 ± 2.11 | 9.55 ± 2.34 | .0315 |
| Neutrophils, ×109/L | 1.31 ± 0.65 | 1.79 ± 0.78 | .0331 |
| Monocytes, ×109/L | 0.37 ± 0.21 | 0.62 ± 0.31 | .0023 |
| Lymphocytes, ×109/L | 6.38 ± 1.60 | 7.09 ± 2.23 | .21 |
| Platelets, ×109/L | 822 ± 81 | 880 ± 94 | .0136 |
| RBCs, ×1012/L | 9.14 ± 0.50 | 9.76 ± 0.44 | .0001 |
| Hemoglobin, g/dL | 13.0 ± 1.1 | 13.7 ± 2.0 | .16 |
| Hematocrit, % | 46.1 ± 3.8 | 46.8 ± 2.7 | .49 |
| MCV, fL | 50.6 ± 4.8 | 47.9 ± 2.5 | .0189 |
| Reticulocytes, ×1012/L | 0.56 ± 0.13 | 0.58 ± 0.11 | .59 |
| EPO, pg/mL* | 202 ± 65 | 125 ± 55 | .0176 |
| . | WT . | ΔHmga2 . | P . |
|---|---|---|---|
| WBCs, ×109/L | 8.10 ± 2.11 | 9.55 ± 2.34 | .0315 |
| Neutrophils, ×109/L | 1.31 ± 0.65 | 1.79 ± 0.78 | .0331 |
| Monocytes, ×109/L | 0.37 ± 0.21 | 0.62 ± 0.31 | .0023 |
| Lymphocytes, ×109/L | 6.38 ± 1.60 | 7.09 ± 2.23 | .21 |
| Platelets, ×109/L | 822 ± 81 | 880 ± 94 | .0136 |
| RBCs, ×1012/L | 9.14 ± 0.50 | 9.76 ± 0.44 | .0001 |
| Hemoglobin, g/dL | 13.0 ± 1.1 | 13.7 ± 2.0 | .16 |
| Hematocrit, % | 46.1 ± 3.8 | 46.8 ± 2.7 | .49 |
| MCV, fL | 50.6 ± 4.8 | 47.9 ± 2.5 | .0189 |
| Reticulocytes, ×1012/L | 0.56 ± 0.13 | 0.58 ± 0.11 | .59 |
| EPO, pg/mL* | 202 ± 65 | 125 ± 55 | .0176 |
Peripheral blood cell counts (n = 29) and serum EPO levels (n = 8) were determined in 2-month-old ΔHmga2 mice and compared with age- and sex-matched WT mice (n = 22 for hematologic parameters and n = 10 for serum EPO values).
WBCs indicates white blood cells; RBCs, red blood cells; MCV, mean corpuscular volume; and EPO, erythropoietin.
Overexpression of HMGA2 promotes hematopoiesis and causes extramedullary erythropoiesis in ΔHmga2 mice. (A) Increased BM cell counts in ΔHmga2 mice (n = 20) compared with WT mice (n = 18), and enlarged spleens (B) in ΔHmga2 mice (n = 8) compared with WT mice (n = 7). Representative examples are shown. (C) H&E stains of BM and spleen demonstrated an increased number of megakaryocytes in BM (left) and expanded red pulp in spleen (right) in ΔHmga2 mice. Representative photographs are shown. (D) Flow cytometry using lineage-specific markers, Gr1 (myeloid), Ter119 (erythroid), TCR (T cell), and B220 (B cell), showed an increased proportion of Ter119+ cells in BM and spleen (n = 3 in each experiment). *P < .05.
Overexpression of HMGA2 promotes hematopoiesis and causes extramedullary erythropoiesis in ΔHmga2 mice. (A) Increased BM cell counts in ΔHmga2 mice (n = 20) compared with WT mice (n = 18), and enlarged spleens (B) in ΔHmga2 mice (n = 8) compared with WT mice (n = 7). Representative examples are shown. (C) H&E stains of BM and spleen demonstrated an increased number of megakaryocytes in BM (left) and expanded red pulp in spleen (right) in ΔHmga2 mice. Representative photographs are shown. (D) Flow cytometry using lineage-specific markers, Gr1 (myeloid), Ter119 (erythroid), TCR (T cell), and B220 (B cell), showed an increased proportion of Ter119+ cells in BM and spleen (n = 3 in each experiment). *P < .05.
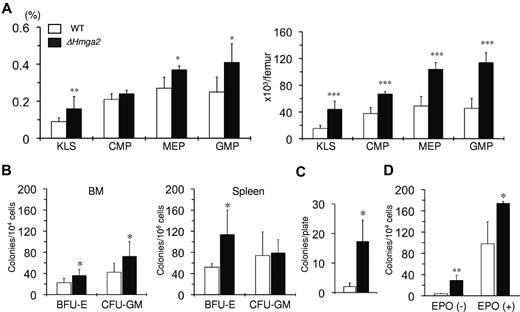
We next evaluated the myeloid progenitor compartment in the BM of ΔHmga2 mice using flow cytometry. We found that the proportion and absolute numbers of cKit+lineage−Sca1+ (KLS) cells, megakaryocyte-erythrocyte progenitor (MEP) cells as well as granulocyte-monocyte progenitor (GMP) cells were significantly increased in ΔHmga2 mice compared with WT (Figure 3A). The common myeloid progenitor cells were increased in absolute numbers but this did not reach statistical significance when the proportion in total BM cells was compared. Clonogenic progenitor assays performed with BM and spleen cells in the presence of SCF, IL-3, IL-6, and EPO resulted in an increased number of BFU-Es from BM and spleen cells and an increased number of CFU-GMs from BM cells from ΔHmga2 mice compared with WT mice (Figure 3B). Furthermore, replating of BFU-Es from BM cells resulted in the growth of new BFU-Es when the cells were obtained from ΔHmga2 mice, but rarely when obtained from WT mice (Figure 3C). In addition, formation of CFU-Es was observed either in the presence or absence of EPO (Figure 3D) from BM cells of ΔHmga2 mice but only in the presence of EPO from WT mice, indicating a self-renewing capacity of BM derived BFU-Es and EPO-independent erythroid colony formation from BM cells from ΔHmga2 mice.
Increased number of HSCs and committed progenitor cells and EPO-independent erythroid colony formation in ΔHmga2 mice. (A) Proportions (left) and absolute numbers (right) of KLS cells and committed progenitor cells in total BM cells as determined by flow cytometry (n = 4 in each). (B) Numbers of BFU-Es and CFU-GMs grown from 1 × 104 BM cells (n = 8 in each) and 1 × 106 spleen cells (n = 4 in each) in the presence of human EPO and IL-6, and murine IL-3 and SCF. (C) New BFU-E formation after replating of primary BFU-Es (n = 3 in each). (D) CFU-E formation from BM cells in the absence (n = 4 in each) and presence (n = 3 in each) of EPO.*P < .05, **P < .01, ***P < .001.
Increased number of HSCs and committed progenitor cells and EPO-independent erythroid colony formation in ΔHmga2 mice. (A) Proportions (left) and absolute numbers (right) of KLS cells and committed progenitor cells in total BM cells as determined by flow cytometry (n = 4 in each). (B) Numbers of BFU-Es and CFU-GMs grown from 1 × 104 BM cells (n = 8 in each) and 1 × 106 spleen cells (n = 4 in each) in the presence of human EPO and IL-6, and murine IL-3 and SCF. (C) New BFU-E formation after replating of primary BFU-Es (n = 3 in each). (D) CFU-E formation from BM cells in the absence (n = 4 in each) and presence (n = 3 in each) of EPO.*P < .05, **P < .01, ***P < .001.
Competitive growth advantage of ΔHmga2 BM cells
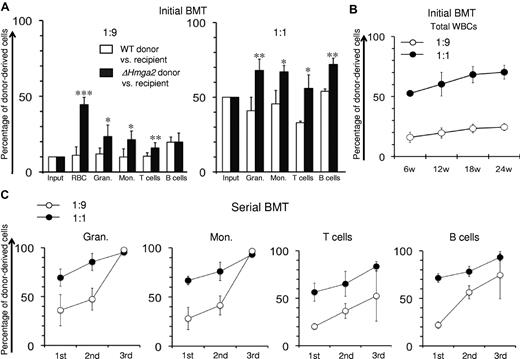
To further investigate the effect of HMGA2 overexpression we performed competitive BMT experiments. BM cells from ΔHmga2 mice were mixed with BM cells from WT mice in 1:1 and 1:9 ΔHmga2:WT ratios and injected into lethally irradiated recipient mice. Twelve weeks after BMT the proportion of ΔHmga2 peripheral blood cells was significantly increased in all lineages in comparison to the ratio used for transplantation (Figure 4A). Over time the proportion of ΔHmga2 mice-derived cells gradually increased (Figure 4B), suggesting a growth advantage of ΔHmga2 expressing BM cells.
Competitive growth advantage of ΔHmga2 BM cells. Chimerism in peripheral blood of lethally irradiated recipient mice (glucose phosphate isomerasea RBCs or CD45.1+ WBCs) after BMT with mixtures of BM cells from donors (WT mice or ΔHmga2 mice with glucose phosphate isomeraseb RBCs and CD45.2+ WBCs) and BM cells from mice with same allotype as recipients at the ratio of 1:1 or 1:9. (A) Donor chimerism in peripheral blood 12 weeks after BMT in primary BMT recipients. (B) Long-term peripheral blood chimerism in primary BMT recipients transplanted with BM cell mixtures consisted of ΔHmga2 donor cells and recipient-allotype cells at the ratios of 1:9 and 1:1 (n = 4-5 in each experiment). (C) Analysis of peripheral blood chimerism in primary, secondary, and tertiary transplantation recipients showed a continuous increase of proportions of ΔHmga2 blood cells in peripheral granulocytes monocytes, T cells, and B cells (n = 4-5 in each experiment). Gran. indicates granulocytes; and Mon., monocytes. *P < .05, **P < .01, ***P < .001.
Competitive growth advantage of ΔHmga2 BM cells. Chimerism in peripheral blood of lethally irradiated recipient mice (glucose phosphate isomerasea RBCs or CD45.1+ WBCs) after BMT with mixtures of BM cells from donors (WT mice or ΔHmga2 mice with glucose phosphate isomeraseb RBCs and CD45.2+ WBCs) and BM cells from mice with same allotype as recipients at the ratio of 1:1 or 1:9. (A) Donor chimerism in peripheral blood 12 weeks after BMT in primary BMT recipients. (B) Long-term peripheral blood chimerism in primary BMT recipients transplanted with BM cell mixtures consisted of ΔHmga2 donor cells and recipient-allotype cells at the ratios of 1:9 and 1:1 (n = 4-5 in each experiment). (C) Analysis of peripheral blood chimerism in primary, secondary, and tertiary transplantation recipients showed a continuous increase of proportions of ΔHmga2 blood cells in peripheral granulocytes monocytes, T cells, and B cells (n = 4-5 in each experiment). Gran. indicates granulocytes; and Mon., monocytes. *P < .05, **P < .01, ***P < .001.
To determine whether this growth advantage occurs at the level of the HSC, serial BMT experiments were performed. In serial transplantation experiments the ΔHmga2 expressing cells continuously increased during 3 rounds of BMT, ultimately accounting for over 90% of peripheral blood granulocytes and monocytes and over 70% of B and T lymphocytes (Figure 4C) indicating that overexpression of HMGA2 confers a clonal growth advantage to hematopoietic cells at the level of the HSC.
Increased expression of pSTAT3 in HSCs and pAKT in total BM cells from ΔHmga2 mice
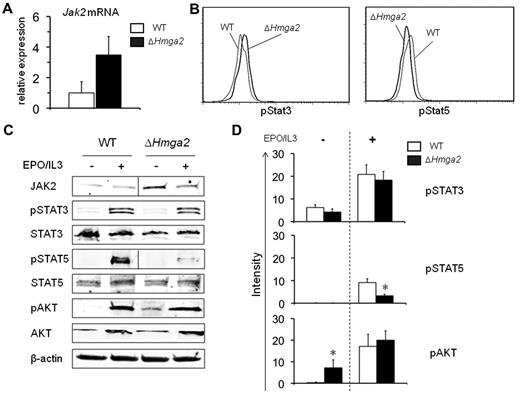
Growth of BM cells from ΔHmga2 mice in colony assays and serial BMT experiments suggested that both HSCs and committed progenitor cells contributed to the increased hematopoiesis in ΔHmga2 mice. HMGA2 is known to alter gene expression by facilitating the accessibility of promoter regions to transcription factors. Therefore, we analyzed altered gene expression because of ΔHmga2 expression, which may be important in cell proliferation or clonality in HSCs and committed progenitor cells. Microarray analysis showed distinct differences between the HSC-enriched KLS and MEP cells (supplemental Tables 3-6, supplemental Figure 2), suggesting different effects of Hmga2 overexpression at different stages of hematopoiesis. Among the differentially expressed genes we noted that up-regulation of Jak2 mRNA was observed only in KLS cells from ΔHmga2 mice. JAK2 is involved in the most significant pathways altered in ΔHmga2 mice, being associated with development, regulation of apoptosis and survival of cells, and immune response (supplemental Table 5, supplemental Figure 2). Therefore, we confirmed up-regulation of Jak2 mRNA in KLS cells by qRT-PCR (Figure 5A). Because JAK2 activation is known to be associated with alterations of the JAK-STAT signaling pathway in hematopoietic cells from patients with MPNs with or without the JAK2V617F mutation,25-27 we next studied whether STAT3 and STAT5, transcription factors downstream of JAK2 in the signaling pathway, are altered in KLS cells. We found significantly higher levels of pSTAT3, but lower levels of pSTAT5 in KLS cells from ΔHmga2 mice compared with WT mice (Figure 5B). We also investigated JAK-STAT and PI3K-AKT pathways in total BM cells because activation of these pathways has been shown in some patients with MPNs with or without the JAK2V617F mutation.27 In total BM cells from ΔHmga2 mice without cytokine stimulation, pAKT expression was significantly higher than in those from WT mice (Figure 5C-D), suggesting an intrinsic phosphorylation of AKT. In total BM cells from ΔHmga2 mice after the stimulation by EPO and IL3, the levels of pAKT and pSTAT3 were comparable to those of WT mice, but that of pSTAT5 was significantly lower compared with WT mice (Figure 5C-D).
ΔHmga2 expression leads to increased EPO-independent phophorylation of AKT. (A) Validation of microarray analysis using qRT-PCR showed similar increase in the expression of Jak2 mRNA in KLS cells from ΔHmga2 mice compared with WT mice. Representative results of ddCT assay using same samples as used in the microarray analysis are shown. (B) Flow cytometry of pStat3 (left) and pStat5 (right) in KLS cells in lineage-depleted BM cells. (C) Western blots of total BM cells incubated in the absence or presence of EPO and IL3. Each protein was examined using a single gel. Vertical lines between WT and ΔHmga2 have been inserted to indicate repositioned gel lanes in JAK2 and pStat5. Data are representative of triplicate experiments in flow cytometry and Western blots. (D) Band intensities of phosphorylated proteins in Western blots measured using infra red imaging (Odyssey). *P < .05.
ΔHmga2 expression leads to increased EPO-independent phophorylation of AKT. (A) Validation of microarray analysis using qRT-PCR showed similar increase in the expression of Jak2 mRNA in KLS cells from ΔHmga2 mice compared with WT mice. Representative results of ddCT assay using same samples as used in the microarray analysis are shown. (B) Flow cytometry of pStat3 (left) and pStat5 (right) in KLS cells in lineage-depleted BM cells. (C) Western blots of total BM cells incubated in the absence or presence of EPO and IL3. Each protein was examined using a single gel. Vertical lines between WT and ΔHmga2 have been inserted to indicate repositioned gel lanes in JAK2 and pStat5. Data are representative of triplicate experiments in flow cytometry and Western blots. (D) Band intensities of phosphorylated proteins in Western blots measured using infra red imaging (Odyssey). *P < .05.
Deficiency in GPI-linked proteins does not increase the clonal advantage of ΔHmga2+ cells
To assess whether the lack of GPI-linked proteins is synergistic with HMGA2 overexpression cells with respect to the growth advantage of hematopoietic cells we monitored the percentage of GPI-linked protein deficient cells in double transgenic mice (Piga−ΔHmga2+ mice). These mice have a proportion of blood cells deficient in GPI-linked proteins,20 so in hematopoiesis HMGA2 overexpressing cells are competing with HMGA2 overexpressing cells that in addition lack GPI-linked proteins. During a 7 month observation period we did not observe a significant difference in the proportion of blood cells that lack GPI-linked proteins, indicating that the lack of GPI-linked proteins does not add an additional growth advantage to the HMGA2 expressing cells (Figure 6).
Lack of GPI-linked proteins is not synergistic to the growth advantage of ΔHmga2+ hematopoietic cells. Proportions of CD24− RBCs and FLAER− granulocytes in Piga−Hmga2WT mice and Piga−ΔHmga2+ mice at the ages of 4 months and 7 months.
Lack of GPI-linked proteins is not synergistic to the growth advantage of ΔHmga2+ hematopoietic cells. Proportions of CD24− RBCs and FLAER− granulocytes in Piga−Hmga2WT mice and Piga−ΔHmga2+ mice at the ages of 4 months and 7 months.
Discussion
Here, we showed that overexpression of HMGA2 protein caused proliferative hematopoiesis and conferred a clonal advantage to hematopoietic cells at the level of the hematopoietic progenitor cell and the HSC. This may explain the proliferative growth advantage of PNH cells in 2 patients with PNH, in which PNH cell clones had acquired a rearrangement on chromosome 12 resulting in truncation of the HMGA2 gene and loss of the 3′UTR.6 Such rearrangements would remove the sites in the 3′UTR that bind the let-7 regulatory microRNAs that normally restrain HMGA2 expression. The causes of clonal expansion in most cases of PNH, however, remain unclear. The frequency of HMGA2 dysregulation in PNH, by gene rearrangement or other means, has yet to be determined.
We cannot formally rule out an influence of the integration site of the ΔHmga2 transgene but we believe this is unlikely because the insertion does not disrupt any genes and the closest gene to the insertion site (Clstn2 at a distance of 5.4 kb) is mainly expressed in neural tissue.28 Furthermore, microarray analysis of KLS cells and MEP cells did not show significant difference in the RNA expression of these 2 genes using the criteria described in supplemental Tables 3 and 4 (≥ 2-fold, P < .01).
The proliferative hematopoiesis we observed in ΔHmga2 mice, is reminiscent of that seen in patients with MPNs including polycythemia vera (PV), essential thrombocythemia (ET), and primary myelofibrosis (PMF). In this context several studies have found overexpression and/or gene disruption of HMGA2 in myeloid neoplasms,17,29,30 suggesting the possible involvement of HMGA2 in the development of these conditions. Although an acquired somatic point mutation in JAK2, JAK2V617F, has been identified in more than 90% of patients with PV, and approximately half of those with ET or PMF,31-34 the precise etiology of these conditions has not been determined. Mutations in other genes, notably MPL, TET2, CBL, and EZH2 have been found in some patients.35-38 In some cases more than one mutation is found in the same clone and in others leukemic transformation takes place in a cell not harboring the mutation.37 HMGA2 overexpression in the MPNs might be an additional factor in MPN etiology.
Hematopoiesis in ΔHmga2 mice was characterized by increased HSCs, committed progenitor cells, and hematopoietic colony forming cells and by EPO-independent erythroid colony formation. Jak2 mRNA was up-regulated and expression of pSTAT3 protein was increased in KLS cells, suggesting that activation of STAT3 may contribute to proliferation of HSCs in ΔHmga2 mice. On the other hand, expression of pSTAT5 was decreased in KLS cells and cytokine-stimulated total BM cells from ΔHmga2 mice. Activation of STAT5 is important in proliferation of erythroid cells in PV.22,24-27,34 However, it appears that expression of pSTAT3 is increased in CD34+ cells from patients with PMF and pSTAT5 is rather decreased in patients with PMF and ET,25,27 as well as ΔHmga2 mice. We also discovered pAKT expression in BM cells from ΔHmga2 mice without the stimulation of cytokines, suggesting the possibility that overexpression of Hmga2 constitutively activates AKT in BM cells. As well as the JAK-STAT signaling pathway involving JAK2, STAT3, and STAT5, the PI3K-AKT pathway is another important pathway implicated in the proliferation of erythroid cells and other myeloid cells in patients with MPNs.27 Thus, AKT activation induced by overexpression of Hmga2 may play a role in proliferative hematopoiesis including formation of EPO-independent erythroid colonies in ΔHmga2 mice.
Serial BMT experiments indicated enhanced self-renewal capacity of HSCs from ΔHmga2 mice which express an Hmga2 mRNA lacking sequences complementary to let-7-family miRNAs. Expression of HMGA2 protein was increased more than that of Hmga2 mRNA compared with WT mice. High expression of the Hmga2 contributes to self-renewal in young neural stem cells in mice, and an increase of let-7b miRNA reduces expression of the Hmga2, leading to decline of self-renewal in old neural stem cells.14 In cancer stem cells, overexpression of Hmga2 is associated with low expression of let-7-family miRNAs and may play a role in controlling differentiation.13 Furthermore, high expression of HMGA2 is also associated with very low expression of let-7-family miRNAs and may contribute to proliferation and maintaining pluripotency in ES cells.12 Together these findings suggest that stem cell function in general is enhanced by robust expression of HMGA2. Interestingly, gene expression analysis in KLS cells from ΔHmga2 mice showed significant increases in the expression of genes involved in pathways associated with repair of DNA damage, which plays a crucial role in the maintenance of HSC function.39 Whether HMGA2 is directly associated with DNA repair pathways, or affects them indirectly through ATM,40,41 also increased in KLS cells from ΔHmga2 mice, remains to be determined.
Several transgenic mouse lines overexpressing Hmga2 have been reported previously,19,42-45 but hematopoiesis in the mice was not investigated in detail. Large cell lymphoma was found in 8 of 67 transgenic mice overexpressing full-length murine Hmga2 cDNA.44 So far, by observing our ΔHmga2 mice until they were 1 year old we have not found development of lymphoma, based on necropsy, but their B cells and T cells showed competitive growth advantage compared with WT. In addition, despite extramedullary erythropoiesis, proportions of B cells and T cells in enlarged spleens of ΔHmga2 mice were similar to those in normal spleens of WT mice, suggesting that numbers of B cells and T cells were increased in the spleen, with the potential for proliferation and lymphoma development. To our knowledge, there has been no detailed investigation of erythropoiesis or granulopoiesis in mice that overexpress Hmga2. Interestingly however, overexpression of HMGA1a, another member of the HMGA family, caused aggressive T cell lymphoma via activation of STAT3 in transgenic mice with HMGA1a.46,47 Blocking of STAT3 function induced apoptosis of the lymphoma cells in these mice.47
The importance of HMGA2 in hematopoietic stem cell biology has recently received independent support from an unexpected source. In several human gene therapy trials using lenti- or retroviral transduction of human genes, insertion of the virus into the HMGA2 locus has been observed as a relatively frequent event.48-50 Insertion into HMGA2, removing suppression by let-7 miRNA, has led to clonal outgrowth of cells with the insertion but with an observation period of up to 8 years has not been associated with malignant transformation,49 reflecting the benign tumor phenotype frequently associated with HMGA2 overexpression. In the recent report by Cavazzana-Calvo and colleagues using lentiviral β-globin gene transfer, an adult patient with severe transfusion dependent β-thalassemia became transfusion independent.48 This patient developed a dominant cell clone in which the vector integrated into the HMGA2 locus, leading to a truncated mRNA which was insensitive to let-7 regulation and expressed HMGA2 highly in erythrocytes under the influence of the β-globin locus control region (LCR). Similarly, in patients whose condition improved after retrovirus mediated gene therapy for X-linked severe-combined immunodeficiency, analysis of vector integrated sites showed multiple integration sites in the HMGA2 locus; when examined these led to HMGA2 mRNA truncation and HMGA2 expression, again invoking HMGA2 involvement in clone expansion.49 Likewise integration into HMGA2 in multiple cell clones was seen in patients successfully treated by retroviral gene therapy for Wiskott-Aldrich syndrome.50 These data suggest that overexpression of HMGA2 may lead to limited expansion of cell clones in gene therapy leading to a positive outcome thus far without the development of malignancy.
In conclusion, the overexpression of HMGA2 caused by a 3′UTR deletion may explain the clonal expansion of PNH cells in at least 2 patients with PNH, and may also contribute to clonal disease in patients with MPNs.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank M. White for generating ΔHmga2 transgenic mice; D. C. Link for helpful discussions; W. Eades and J. Hughes for flow cytometric cell sorting; the Alvin J. Siteman Cancer Center at Washington University School of Medicine, and Barnes-Jewish Hospital in St Louis, MO, for use of the Center for Biomedical Informatics and Multiplex Gene Analysis Genechip Core Facility.
This work was supported by the Research Grant of Aplastic Anemia & Myelodysplastic Syndrome International Foundation (3048-42179) and the Research Fellowship of Japan Society of Blood Transfusion and Cell Therapy to K.I., and NIH grant 2R01 CA105312 to M.B.
National Institutes of Health
Authorship
Contribution: K.I. designed the research, performed experiments, analyzed results, and wrote the manuscript; and P.J.M. and M.B. designed the research, interpreted the results, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Monica Bessler, Division of Hematology, Children's Hospital of Philadelphia, 3615 Civic Center Blvd, Suite 302, Philadelphia, PA 19104; e-mail: besslerm@e-mail.chop.edu.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal