Abstract
Antigen presentation by mature dendritic cells (DCs) is the first step for initiating adaptive immune responses. DCs are composed of heterogeneous functional subsets; however, the molecular mechanisms that regulate differentiation of specific DC subsets are not understood. Here, we report that the basic leucine zipper transcription factor NFIL3/E4BP4 is essential for the development of CD8α+ conventional DCs (cDCs). Nfil3−/− mice specifically lack CD8α+ cDCs but not CD8α− cDCs or plasmacytoid DCs in lymphoid tissues. Flt3 ligand–dependent generation of CD8α+ cDCs in lymphoid tissues and CD8α+-equivalent cDCs from Nfil3−/− bone marrow cells was also impaired. NFIL3 regulates CD8α+ cDC development in part through Batf3 expression. Importantly, Nfil3−/− mice exhibited impaired cross-priming of CD8+ T cells against cell-associated antigen, a process normally performed by CD8α+ cDCs, and failed to produce IL-12 after TLR3 stimulation. Thus, NFIL3 plays an essential role in the development of CD8α+ cDCs.
Introduction
Dendritic cells (DCs) are effective antigen-presenting cells that both initiate antigen-specific immune responses and generate and maintain self-tolerance.1 DCs can develop from bone marrow (BM) progenitor cells and reside in the peripheral lymphoid tissues. Several DC subsets were identified by their cell-surface phenotypes, residential location, and functional differences.2 Splenic DCs are divided into 2 major populations: plasmacytoid DCs (pDCs) and conventional DCs (cDCs). The cDCs are further divided into CD8α+ and CD8α− DCs. CD8α+ DCs predominantly produce IL-12 in response to microbial antigens, tumor cells, and virus-infected cells.1-3 Importantly, CD8α+ DCs are specialized cells for the cross-presentation of antigens by MHC class I molecules.4,5 Analyses of mice lacking transcription factor genes such as Irf4, Irf8, and Batf3 revealed their essential roles in the development of distinct subsets of DCs.6-9
NFIL3 (nuclear factor, IL-3 regulated; also called E4BP4) was originally identified as an adenovirus E4 promoter– and human IL3 gene promoter–binding protein, and its role in circadian regulation has been well studied.10-12 However, the role of NFIL3 in the immune system was not largely elucidated until Nfil3−/− mice were established recently. We have demonstrated that Nfil3−/− mice have defects in IgE class switching by regulating Ig-ϵ germline transcription in B cells.13 Nfil3−/− mice also lack natural killer (NK) cells, which suggests a role for NFIL3 in NK cell development.13-15 Here, we demonstrate impaired CD8α+ DC development and impaired cross-priming activity of CD8+ T cells in response to cell-mediated antigen in Nfil3−/− mice. Our results provide compelling evidence that NFIL3 is a key regulator of CD8α+ DC development.
Methods
Mice
Nfil3−/− mice13 (C57BL/6 background) were used for all experiments. All experimental mouse protocols were approved by the Institutional Animal Care and Use Committee of University of Iowa.
Real-time RT-PCR
cDNA was prepared from RNA isolated from cells by the SuperScript First-Strand Synthesis System (Invitrogen) according to the manufacturer's instructions and was subjected to real-time PCR with SYBR Green PCR Master Mix (Applied Biosystems) as described previously.13 Primers used are listed in the supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Flow cytometry
Cell suspensions from spleen, thymus, blood, and BM culture were stained with the antibodies listed in supplemental Methods. Cells were analyzed with LSR II (BD Biosciences), and data were interpreted with CellQuest (BD Biosciences) or FlowJo (TreeStar).
Isolation and culture of DCs
Spleen cDCs were isolated as described previously.9 For BM-derived DCs, BM cells were cultured with mouse Flt3 ligand (FL; 50 ng/mL) for 9 days.
Viral transduction
FL-activated BM cells were spin-infected with retrovirus as described previously13 and were grown for another 7 days before analysis.
In vivo cross-priming assay
OVA257-264-specific CD8+ T-cell responses in mice injected with irradiated splenocytes from act-mOVA/Kb−/− mice were determined by MHC class I–peptide tetramer staining.16 Detailed materials and methods can be found in the online supplemental Methods.
Results and discussion
Selective developmental defect of CD8α+ DCs in Nfil3−/− mice
Nfil3 expression is high in NK cells and IL-4–stimulated B and T cells (Figure 1A).14,15 Nfil3 expression in sorted splenic pDC and cDC subsets was similar to NK cells and IL-4–stimulated B and T cells, which suggests a potential role of NFIL3 in DC populations. Therefore, we examined DC populations in Nfil3−/− mice. The percentages of both the cDC (CD11chiPDCA-1−) and pDC (CD11cintPDCA-1+) populations of Nfil3−/− mice were slightly increased compared with that of Nfil3+/+ mice (Figure 1B). Strikingly, the CD8α+ cDC population of Nfil3−/− mice was significantly reduced compared with Nfil3+/+ mice (Figure 1B,D). CD8α+ cDCs also express DEC-205, CD24, and SIRP-α,17 and we confirmed the lack of CD8α+ cDCs in Nfil3−/− mice by costaining for expression of these markers (Figure 1B,D). Furthermore, the frequency of the precursor cells (CD8α−CD24hiSIRP-αlo) of CD8α+ cDCs in the spleen18 is also considerably reduced in Nfil3−/− mice. CD8α+ cDCs can develop from T-cell precursors in the thymus; therefore, we determined whether a lack of CD8α+ cDCs was observed in the thymus.2,19 The frequency of pDCs was increased in the thymus of Nfil3−/− mice but that of cDCs was decreased considerably compared with Nfil3+/+ mice (Figure 1C). Moreover, Nfil3−/− mice lacked CD8α+DEC-205+CD24hiSIRP-αlo cDC cells in their thymus (Figure 1C-D). Because CD8α+ cDCs constitute approximately 70% of the cDC population in the thymus,20 the lack of CD8α+ cDCs likely reflects the reduction of the cDC population in Nfil3−/− mice. These results suggest that NFIL3 regulates the development of both splenic and thymic CD8α+ cDCs in vivo.
Selective loss of CD8α+ DCs in Nfil3−/− mice and impaired Flt3-dependent development of CD8α+ DCs in the absence of NFIL3. (A) Nfil3 is highly expressed in DC subsets. Quantitative real-time RT-PCR analysis of Nfil3 mRNA in purified cells (NKs and DCs) or in vitro stimulated cells (B and T cells) was performed. Data show the mean and SD from 3 experiments. (B) Splenocytes from Nfil3+/+ or Nfil3−/− mice were stained for CD11c and PDCA-1 to identify pDCs and cDCs (left) and for CD11c, CD8, CD4, DEC-205, SIRP-α, and CD24 to detect CD8α+ DCs and CD8α − DCs (middle) or to detect CD8α+ DC precursors in CD8α−CD4− DCs (right). Data are representative of 5 experiments with similar results. (C) Thy1.2− thymocytes from Nfil3+/+ or Nfil3−/− mice were stained for CD11c and PDCA-1 to identify pDCs and cDCs (left) and for CD11c, CD8, DEC-205, SIRP-α, and CD24 to detect CD8α+ DCs (right). Data are representative of 5 experiments with similar results. (D) Number of CD8α+DEC-205+ cDCs in spleen and thymus from Nfil3+/+ (○) and Nfil3−/− (●) mice. Cell numbers were calculated on the basis of analyses by flow cytometry (n = 5; *P < .001, **P < .0001). (E) Impaired IL-12 production by splenic DCs in response to polyinosinic:polycytidylic acid [Poly (I:C)] in Nfil3−/− mice. CD11chi cDCs were enriched by a MACS system with anti-Thy1.2 and anti-B220 beads for depletion of T cells, B cells, and pDCs, followed by anti-CD11c beads for isolation of cDCs. Enriched cDCs were unstimulated or stimulated with CpG (0.5 μg/mL), lipopolysaccharide (LPS; 100 μg/mL), or Poly (I:C) (100 μg/mL) for 4 hours. One hour after stimulation, brefeldin A was added to the culture. After stimulation, cells were stained for CD11c and CD45RA, followed by intracellular staining for IL-12. Data are representative of 4 experiments with similar results. (F) Developmental defect of Nfil3−/− BM progenitor cells to CD8α+-equivalent cDCs. BM cells from Nfil3+/+ or Nfil3−/− mice were cultured with FL (50 ng/mL) for 9 days and stained for CD11c, CD45RA, SIRP-α, and CD24. Data are representative of 3 experiments with similar results. (G) FL-DCs from Nfil3+/+ or Nfil3−/− mice were stimulated with Poly (I:C) (100 μg/mL) for 4 hours and stained for IL-12. Data are representative of 3 experiments with similar results. (H) Lack of FL-dependent development of CD8α+ cDC in vivo. Nfil3−/− and Nfil3+/+ mice were injected with B16-FL cells subcutaneously, and spleens were harvested 8 days after injection. Splenocytes were analyzed for CD11c+ (left), and CD8α+ cDCs (right) by flow cytometry. Data are representative of 3 independent experiments. (I) Equal numbers of BM progenitor cells between Nfil3−/− and Nfil3+/+ mice. BM cells from Nfil3−/− and Nfil3+/+ mice were stained for LSK (Lin−c-kithiSca-1+CD127−), CLPs (common lymphoid progenitors; Lin−c-kitintSca-1intCD127+), CMP/GMP/MEPs (myeloid progenitors; Lin−c-kithiSca-1−CD127−), CDPs (common DC progenitors; Lin−Flt3+c-kitloCD127-CD115+), and pre-cDCs (Lin−CD11c+MHCII-Flt3+SIRP-αlo), and cell numbers of each population per 1 × 106 BM cells were calculated. Each dot represents an individual mouse, and gray lines indicate mean cell number. (J) Relative expression of Batf3, Irf8, Id2, Irf4, and Nfil3 genes in Nfil3−/− and Nfil3+/+ BM pre-cDCs was determined by quantitative real-time RT-PCR. Mean expression of RNA in wild-type cells was set as 1. The mean and SD of 2 independent experiments are shown. (K) Rescue of CD8α+-equivalent cDCs by BATF3 and NFIL3 transduction. Nfil3−/− BM cells stimulated with FL were transduced with vector or Batf3- or Nfil3-carrying viruses and cultured for 9 days, then stained for CD11c, CD45RA, SIRP-α, and CD24. Virus-infected cells (green fluorescent protein–positive cells) were analyzed. Data are representative of 3 experiments with similar results.
Selective loss of CD8α+ DCs in Nfil3−/− mice and impaired Flt3-dependent development of CD8α+ DCs in the absence of NFIL3. (A) Nfil3 is highly expressed in DC subsets. Quantitative real-time RT-PCR analysis of Nfil3 mRNA in purified cells (NKs and DCs) or in vitro stimulated cells (B and T cells) was performed. Data show the mean and SD from 3 experiments. (B) Splenocytes from Nfil3+/+ or Nfil3−/− mice were stained for CD11c and PDCA-1 to identify pDCs and cDCs (left) and for CD11c, CD8, CD4, DEC-205, SIRP-α, and CD24 to detect CD8α+ DCs and CD8α − DCs (middle) or to detect CD8α+ DC precursors in CD8α−CD4− DCs (right). Data are representative of 5 experiments with similar results. (C) Thy1.2− thymocytes from Nfil3+/+ or Nfil3−/− mice were stained for CD11c and PDCA-1 to identify pDCs and cDCs (left) and for CD11c, CD8, DEC-205, SIRP-α, and CD24 to detect CD8α+ DCs (right). Data are representative of 5 experiments with similar results. (D) Number of CD8α+DEC-205+ cDCs in spleen and thymus from Nfil3+/+ (○) and Nfil3−/− (●) mice. Cell numbers were calculated on the basis of analyses by flow cytometry (n = 5; *P < .001, **P < .0001). (E) Impaired IL-12 production by splenic DCs in response to polyinosinic:polycytidylic acid [Poly (I:C)] in Nfil3−/− mice. CD11chi cDCs were enriched by a MACS system with anti-Thy1.2 and anti-B220 beads for depletion of T cells, B cells, and pDCs, followed by anti-CD11c beads for isolation of cDCs. Enriched cDCs were unstimulated or stimulated with CpG (0.5 μg/mL), lipopolysaccharide (LPS; 100 μg/mL), or Poly (I:C) (100 μg/mL) for 4 hours. One hour after stimulation, brefeldin A was added to the culture. After stimulation, cells were stained for CD11c and CD45RA, followed by intracellular staining for IL-12. Data are representative of 4 experiments with similar results. (F) Developmental defect of Nfil3−/− BM progenitor cells to CD8α+-equivalent cDCs. BM cells from Nfil3+/+ or Nfil3−/− mice were cultured with FL (50 ng/mL) for 9 days and stained for CD11c, CD45RA, SIRP-α, and CD24. Data are representative of 3 experiments with similar results. (G) FL-DCs from Nfil3+/+ or Nfil3−/− mice were stimulated with Poly (I:C) (100 μg/mL) for 4 hours and stained for IL-12. Data are representative of 3 experiments with similar results. (H) Lack of FL-dependent development of CD8α+ cDC in vivo. Nfil3−/− and Nfil3+/+ mice were injected with B16-FL cells subcutaneously, and spleens were harvested 8 days after injection. Splenocytes were analyzed for CD11c+ (left), and CD8α+ cDCs (right) by flow cytometry. Data are representative of 3 independent experiments. (I) Equal numbers of BM progenitor cells between Nfil3−/− and Nfil3+/+ mice. BM cells from Nfil3−/− and Nfil3+/+ mice were stained for LSK (Lin−c-kithiSca-1+CD127−), CLPs (common lymphoid progenitors; Lin−c-kitintSca-1intCD127+), CMP/GMP/MEPs (myeloid progenitors; Lin−c-kithiSca-1−CD127−), CDPs (common DC progenitors; Lin−Flt3+c-kitloCD127-CD115+), and pre-cDCs (Lin−CD11c+MHCII-Flt3+SIRP-αlo), and cell numbers of each population per 1 × 106 BM cells were calculated. Each dot represents an individual mouse, and gray lines indicate mean cell number. (J) Relative expression of Batf3, Irf8, Id2, Irf4, and Nfil3 genes in Nfil3−/− and Nfil3+/+ BM pre-cDCs was determined by quantitative real-time RT-PCR. Mean expression of RNA in wild-type cells was set as 1. The mean and SD of 2 independent experiments are shown. (K) Rescue of CD8α+-equivalent cDCs by BATF3 and NFIL3 transduction. Nfil3−/− BM cells stimulated with FL were transduced with vector or Batf3- or Nfil3-carrying viruses and cultured for 9 days, then stained for CD11c, CD45RA, SIRP-α, and CD24. Virus-infected cells (green fluorescent protein–positive cells) were analyzed. Data are representative of 3 experiments with similar results.
CD8α+ DCs produce IL-12 in response to stimulation of TLR3, TLR4, and TLR9 by their specific ligands.17,21 Although TLR4 and TLR9 stimulation induced IL-12 in Nfil3−/− splenic cDCs, TLR3 stimulation did not induce IL-12 in Nfil3−/− cDCs (Figure 1E). These results suggest the lack of functional TLR3-expressing CD8α+ DCs in Nfil3−/− mice.
Nfil3 deficiency caused impaired FL-dependent CD8α+ cDC development
In vitro BM culture with FL generates cDCs that are phenotypically and functionally equivalent to splenic CD8α+ and CD8α− cDCs.17 We next determined whether the Nfil3 deficiency in BM cells affected FL-dependent DC development in vitro. Although both cDCs (CD11c+CD45RA−) and pDCs (CD11c+CD45RA+) were generated from Nfil3+/+ and Nfil3−/− BM cells, NFIL3 deficiency resulted in significantly fewer CD8α+-equivalent FL-cDCs (CD24hiSIRP-αlo) compared with Nfil3+/+ BM cells (Figure 1F). FL-cDCs generated from Nfil3−/− BM cells also failed to produce IL-12 by stimulation with TLR3, which suggests the impaired development of CD8α+-equivalent FL-cDCs in the absence of NFIL3 (Figure 1G).
Administration of recombinant FL or a B16 melanoma cell line expressing FL (B16-FL) to mice resulted in the strong induction of pDCs and CD8α+ cDCs in lymphoid tissues.22,23 Despite the strong induction of splenic CD11c+ cells, B16-FL–injected Nfil3−/− mice failed to produce CD8α+DEC-205+ cDCs (Figure 1H). Taken together, these results suggest that NFIL3 is required for FL-dependent CD8α+ cDC development in vivo and in vitro.
CD8α+ DCs can arise from different types of progenitor cells in BM and T-cell precursors in the thymus.2,24,25 Interestingly, Nfil3−/− mice have normal numbers of both cDC progenitors in BM (Figure 1I) and T-cell precursors in the thymus,13 which suggests that lack of CD8α+ cDCs is not due to a defect in progenitor cells. In contrast, Nfil3−/− mice are deficient in the immediate splenic precursors of CD8α+ cDCs (Figure 1B). Therefore, NFIL3 may function during the development or expansion of pre-cDCs to the CD8α+-committed precursor stage.
To elucidate the mechanisms by which NFIL3 regulates CD8α+ DC development, pre-cDCs from Nfil3+/+ and Nfil3−/− mice were purified, and gene expression of transcription factors essential for CD8α+ DC development was examined. Expression of Batf3 but not Irf8 and Id2 was significantly reduced in Nfil3−/− pre-cDCs, which suggests that BATF3 could be downstream of NFIL3 function (Figure 1J). Consistent with this interpretation, BATF3 transduction into Nfil3−/− BM progenitors was able to substantially restore CD8α+ DC development (Figure 1K). These results suggest that NFIL3 regulates CD8α+ DC development in part through BATF3 expression, although other factors appear to be required to fully rescue CD8α+ DC deficiency in the absence of NFIL3.
Impaired cross-priming by Nfil3−/− DCs
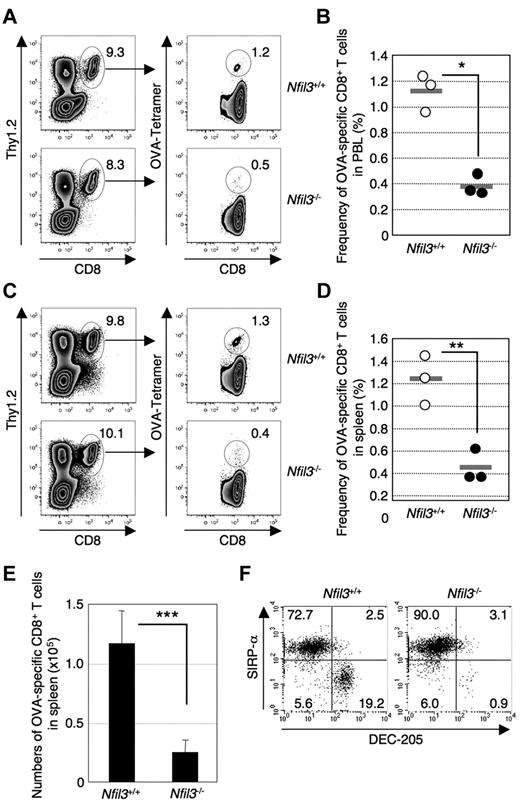
An important function of CD8α+ DCs is the cross-presentation of cell-associated antigens to CD8+ T cells.26 Therefore, we examined whether Nfil3−/− mice exhibit an impaired ability to cross-prime antigen-specific CD8+ T cells. Injection of irradiated act-mOVA/Kb−/− splenocytes, which cannot directly present the OVA257-264 epitope, into Nfil3+/+ mice but not Nfil3−/− mice resulted in induction of OVA257-264-specific CD8+ T cells in both the blood (Figure 2A-B) and spleen (Figure 2C-E). These results suggest that Nfil3−/− mice, which lack a CD8α+ DC population (DEC-205+SIRP-αlo) in the spleen (Figure 2F), are defective in cross-priming antigen-specific CD8+ T cells against cell-associated antigens. Taken together, these results indicate that Nfil3−/− mice lack functional CD8α+ DCs in the spleen.
Impaired cross-priming in Nfil3−/− mice. Nfil3−/− and Nfil3+/+ mice were immunized with irradiated Kb−/−/mOva splenocytes intravenously. Detection of OVA257-specific CD8+ T cells in PBLs and spleen by tetramer (Kb/OVA257) staining was performed at day 5 after immunization. (A) Representative FACS profiles for OVA257-specific CD8+ T cells in PBLs. Similar results were obtained from 3 independent mice in each group. (B) Reduced frequency of OVA257-specific CD8+ T cells among total circulating CD8+ T cells in Nfil3−/− PBLs at day 5 after immunization. Each dot represents an individual Nfil3+/+ mouse (○) or Nfil3−/− (●) mouse, and gray lines indicate mean frequency (*P = .0016). (C) Representative FACS profiles for OVA257-specific CD8+ T cells in spleen. Similar results were obtained from 3 independent mice in each group. (D) Reduced frequency of OVA257-specific CD8+ T cells of Nfil3−/− spleen CD8+ T cells at day 5 after immunization. Each dot represents an individual Nfil3+/+ mouse (○) or Nfil3−/− mouse (●), and gray lines indicate mean frequency (**P = .0067). (E) Reduced numbers of OVA257-specific CD8+ T cells in Nfil3−/− spleen at day 5 after immunization. Data represent 1 of 2 independent experiments with similar results (***P = .035). (F) Detection of CD8α+ cDCs (CD11chiCD8α+DEC-205+SIRP-alo) in Nfil3−/− and Nfil3+/+ mice immunized with irradiated Kb−/−/mOva splenocytes by flow cytometry. Data are representative of 3 independent mice from each group.
Impaired cross-priming in Nfil3−/− mice. Nfil3−/− and Nfil3+/+ mice were immunized with irradiated Kb−/−/mOva splenocytes intravenously. Detection of OVA257-specific CD8+ T cells in PBLs and spleen by tetramer (Kb/OVA257) staining was performed at day 5 after immunization. (A) Representative FACS profiles for OVA257-specific CD8+ T cells in PBLs. Similar results were obtained from 3 independent mice in each group. (B) Reduced frequency of OVA257-specific CD8+ T cells among total circulating CD8+ T cells in Nfil3−/− PBLs at day 5 after immunization. Each dot represents an individual Nfil3+/+ mouse (○) or Nfil3−/− (●) mouse, and gray lines indicate mean frequency (*P = .0016). (C) Representative FACS profiles for OVA257-specific CD8+ T cells in spleen. Similar results were obtained from 3 independent mice in each group. (D) Reduced frequency of OVA257-specific CD8+ T cells of Nfil3−/− spleen CD8+ T cells at day 5 after immunization. Each dot represents an individual Nfil3+/+ mouse (○) or Nfil3−/− mouse (●), and gray lines indicate mean frequency (**P = .0067). (E) Reduced numbers of OVA257-specific CD8+ T cells in Nfil3−/− spleen at day 5 after immunization. Data represent 1 of 2 independent experiments with similar results (***P = .035). (F) Detection of CD8α+ cDCs (CD11chiCD8α+DEC-205+SIRP-alo) in Nfil3−/− and Nfil3+/+ mice immunized with irradiated Kb−/−/mOva splenocytes by flow cytometry. Data are representative of 3 independent mice from each group.
Here, we demonstrated that Nfil3 expression is essential for FL-dependent CD8α+ cDC development in vivo and in vitro and that Nfil3−/− mice exhibited an impaired capacity to cross-prime CD8+ T cells against cell-associated antigens. Thus, we conclude that NFIL3 plays a critical role in the development of CD8α+ cDCs, in addition to NK cells.13-15 Both CD8α+ cDCs and NK cells can develop from T-cell precursors in the thymus, which are different lineages from BM-derived CD8α+ cDCs and NK cells.19,27 Therefore, the lack of CD8α+ cDCs and NK cells in Nfil3−/− mice suggests that NFIL3 could be important for the development of common progenitors of these cells. Control of the expression of the Nfil3 gene or its target genes in progenitor cells for CD8α+ cDCs and NK cells may provide new avenues to dissect the role of DC in regulating immune responses.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants R01 AI54821 (P.B.R) and AI46653 and AI50073 (J.T.H) from the National Institutes of Health.
National Institutes of Health
Authorship
Contribution: M.K., N.-L.L.P., and L.L.P. performed the experiments and analyzed data; M.K., J.T.H., and P.B.R. designed the experiments; and M.K. and P.B.R. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Paul B. Rothman, Roy J. and Lucille A. Carver College of Medicine, University of Iowa, 212 CMAB, 451 Newton Rd, Iowa City, IA 52242; e-mail: paul-rothman@uiowa.edu.

![Figure 1. Selective loss of CD8α+ DCs in Nfil3−/− mice and impaired Flt3-dependent development of CD8α+ DCs in the absence of NFIL3. (A) Nfil3 is highly expressed in DC subsets. Quantitative real-time RT-PCR analysis of Nfil3 mRNA in purified cells (NKs and DCs) or in vitro stimulated cells (B and T cells) was performed. Data show the mean and SD from 3 experiments. (B) Splenocytes from Nfil3+/+ or Nfil3−/− mice were stained for CD11c and PDCA-1 to identify pDCs and cDCs (left) and for CD11c, CD8, CD4, DEC-205, SIRP-α, and CD24 to detect CD8α+ DCs and CD8α − DCs (middle) or to detect CD8α+ DC precursors in CD8α−CD4− DCs (right). Data are representative of 5 experiments with similar results. (C) Thy1.2− thymocytes from Nfil3+/+ or Nfil3−/− mice were stained for CD11c and PDCA-1 to identify pDCs and cDCs (left) and for CD11c, CD8, DEC-205, SIRP-α, and CD24 to detect CD8α+ DCs (right). Data are representative of 5 experiments with similar results. (D) Number of CD8α+DEC-205+ cDCs in spleen and thymus from Nfil3+/+ (○) and Nfil3−/− (●) mice. Cell numbers were calculated on the basis of analyses by flow cytometry (n = 5; *P < .001, **P < .0001). (E) Impaired IL-12 production by splenic DCs in response to polyinosinic:polycytidylic acid [Poly (I:C)] in Nfil3−/− mice. CD11chi cDCs were enriched by a MACS system with anti-Thy1.2 and anti-B220 beads for depletion of T cells, B cells, and pDCs, followed by anti-CD11c beads for isolation of cDCs. Enriched cDCs were unstimulated or stimulated with CpG (0.5 μg/mL), lipopolysaccharide (LPS; 100 μg/mL), or Poly (I:C) (100 μg/mL) for 4 hours. One hour after stimulation, brefeldin A was added to the culture. After stimulation, cells were stained for CD11c and CD45RA, followed by intracellular staining for IL-12. Data are representative of 4 experiments with similar results. (F) Developmental defect of Nfil3−/− BM progenitor cells to CD8α+-equivalent cDCs. BM cells from Nfil3+/+ or Nfil3−/− mice were cultured with FL (50 ng/mL) for 9 days and stained for CD11c, CD45RA, SIRP-α, and CD24. Data are representative of 3 experiments with similar results. (G) FL-DCs from Nfil3+/+ or Nfil3−/− mice were stimulated with Poly (I:C) (100 μg/mL) for 4 hours and stained for IL-12. Data are representative of 3 experiments with similar results. (H) Lack of FL-dependent development of CD8α+ cDC in vivo. Nfil3−/− and Nfil3+/+ mice were injected with B16-FL cells subcutaneously, and spleens were harvested 8 days after injection. Splenocytes were analyzed for CD11c+ (left), and CD8α+ cDCs (right) by flow cytometry. Data are representative of 3 independent experiments. (I) Equal numbers of BM progenitor cells between Nfil3−/− and Nfil3+/+ mice. BM cells from Nfil3−/− and Nfil3+/+ mice were stained for LSK (Lin−c-kithiSca-1+CD127−), CLPs (common lymphoid progenitors; Lin−c-kitintSca-1intCD127+), CMP/GMP/MEPs (myeloid progenitors; Lin−c-kithiSca-1−CD127−), CDPs (common DC progenitors; Lin−Flt3+c-kitloCD127-CD115+), and pre-cDCs (Lin−CD11c+MHCII-Flt3+SIRP-αlo), and cell numbers of each population per 1 × 106 BM cells were calculated. Each dot represents an individual mouse, and gray lines indicate mean cell number. (J) Relative expression of Batf3, Irf8, Id2, Irf4, and Nfil3 genes in Nfil3−/− and Nfil3+/+ BM pre-cDCs was determined by quantitative real-time RT-PCR. Mean expression of RNA in wild-type cells was set as 1. The mean and SD of 2 independent experiments are shown. (K) Rescue of CD8α+-equivalent cDCs by BATF3 and NFIL3 transduction. Nfil3−/− BM cells stimulated with FL were transduced with vector or Batf3- or Nfil3-carrying viruses and cultured for 9 days, then stained for CD11c, CD45RA, SIRP-α, and CD24. Virus-infected cells (green fluorescent protein–positive cells) were analyzed. Data are representative of 3 experiments with similar results.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/23/10.1182_blood-2010-07-295873/4/m_zh89991172140001.jpeg?Expires=1764980233&Signature=e9etCPy0wjNDSaifilD7kCNjF15bnkfCModRqx~hfBSDoIXqolde0uRUPxxSo2WHGzNyhFTUTF8qe1zEIwnAm~34CoGAOnYsIHwF4Ogsarsl5j26GyRMNObfvkLzoiNtPbRsb3XWXQNdbGXvIMkLTDOrq-XE1uNxadxLJK6px50iqhKjsh7P8qmel09dhWNprcVQkFZw4v~OJeiDy54iwMnktq4umqYb6jcNNt2A6YB0QgqBBzpbiJbvXcnsiBodcOkKhU1hkLH0YO-nQB4JOx9hBgGXV6ierKERRDyq38kYE6WTXN7Ob8nftOMfQkYS9ZVfWUvksAQtvkXQ2pdfvw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal