Abstract
Cumulative evidence indicates that MYC, one of the major downstream effectors of NOTCH1, is a critical component of T-cell acute lymphoblastic leukemia (T-ALL) oncogenesis and a potential candidate for targeted therapy. However, MYC is a complex oncogene, involving both fine protein dosage and cell-context dependency, and detailed understanding of MYC-mediated oncogenesis in T-ALL is still lacking. To better understand how MYC is interspersed in the complex T-ALL oncogenic networks, we performed a thorough molecular and biochemical analysis of MYC activation in a comprehensive collection of primary adult and pediatric patient samples. We find that MYC expression is highly variable, and that high MYC expression levels can be generated in a large number of cases in absence of NOTCH1/FBXW7 mutations, suggesting the occurrence of multiple activation pathways in addition to NOTCH1. Furthermore, we show that posttranscriptional deregulation of MYC constitutes a major alternative pathway of MYC activation in T-ALL, operating partly via the PI3K/AKT axis through down-regulation of PTEN, and that NOTCH1m might play a dual transcriptional and posttranscriptional role in this process. Altogether, our data lend further support to the significance of therapeutic targeting of MYC and/or the PTEN/AKT pathways, both in GSI-resistant and identified NOTCH1-independent/MYC-mediated T-ALL patients.
Introduction
T-cell acute lymphoblastic leukemias (T-ALL) are malignant proliferations of T-cell precursors that represent 10% of pediatric and 25% of adult ALL.1 Although treatment outcome has significantly improved in the last decade, ∼ 30% of patients relapse and remain of dismal prognosis, stressing the critical importance of gaining further insights on the molecular pathways controlling malignant transformation and drug resistance. However, a major obstacle in deciphering such pathways and implementing targeted therapy strategies resides in the fact that T-ALLs constitute a particularly heterogeneous and complex group of disease, resulting from numerous combinations of multigenic aberrations and oncogenic cooperation.2 To date, the deregulation of > 30 distinct oncogenes and tumor suppressors (TS) has been reported, occurring through a large diversity of genomic aberrations and epigenetic deregulations. All such mechanisms are not functionally equivalent,3 and distinct modes of oncogenic activation may drive different oncogenic processes, and generate distinct subtypes of prognostic significance. Some oncogenes (eg, TLX1, TAL1) appear to be mutually exclusive (type A) and delineate distinct subgroups of prognostic significance, correlating with specific stages of thymocyte developmental arrest (immature/DN, intermediate/pre-αβ, and mature/T-cell receptorαβ+, respectively).4,5 By contrast, other deregulations, such as loss of CDKN2A/p14ARF, or constitutive NOTCH1 activation, are found in a large proportion of cases and irrespective of subgroups (type B),2 indicating a more universal role for these alterations in T-ALL pathogenesis, and pointing to attractive therapeutic targets. One such target is NOTCH1 and downstream pathways. Indeed, the key finding that > 50% of T-ALL cases display gain-of-function NOTCH1 mutations (NOTCH1m) initially held great promise for targeted therapy through the use of γ-secretase inhibitors (GSI).6 However, the frequent occurrence of GSI-resistance (GSIR) has revealed an unsuspected complexity of the oncogenic network signaling downstream of NOTCH1.6-8 Among the numerous target genes and pathways triggered by NOTCH1m, MYC has been proposed to represent one of the most critical for tumor growth and maintenance.9-12
MYC is a master transcription factor (recent estimates suggest that it might regulate upto 15% of the genes) governing many critical cell functions such as metabolism, proliferation and survival.13 It is also a very potent oncogene, which might be involved in up to 70% of cancers.14-16 However, MYC is also a particularly complex factor, which, likely because of the critical roles it plays in cell fate decision, is tightly regulated at many levels. In line with this, previous studies have indicated that upregulation of MYC expression is insufficient to induce lymphomagenesis.17,18 First, MYC has a dual functional role: while it promotes cell growth and cell cycle progression, it also enhances apoptosis through the P53/ARF and BIM pathways. So under normal circumstances, the extra cell divisions MYC causes when overexpressed are cancelled out by an increase in cell-death.17,18 Second, the fine tuning of MYC is occurring at the protein level through modulation of its stability: MYC protein has indeed a very short half-life largely determined by the phosphorylation of 2 specific sites (T58 and S62) leading to its degradation by the ubiquitin-proteasome pathway.15,19,20 Mutation of such phosphorylation sites is frequent in Burkitt Lymphoma (BL), and provokes both the inhibition of MYC's degradation, and a failure to induce BIM, releasing all the brakes on aggressive cell proliferation.17,18
In T-ALL, deregulated transcriptional activation of MYC is mostly triggered by NOTCH1m. However, the putative involvement of MYC is less well understood in NOTCH1wt cases, probably because of the fact that direct genomic alterations of MYC are rare in this disease (< 5%).21 Nevertheless, the quasi-systemic CDKN2A/p14ARF loss-of-function in T-ALL constitutes a highly favorable ground for MYC-mediated oncogenesis,22 and MYC direct or indirect activation in transgenic mouse and zebrafish models,23-27 as well as rare MYC/T-cell receptor translocations in human indicate that MYC can also trigger T-ALL initiation and/or maintenance, with or without the acquisition/cooperation of NOTCH1m. Considering the known multiplicity of physiologic pathways acting on MYC transcriptional and posttranscriptional control,28 some of the alternative routes of MYC activation might be shared between NOTCH1-independent and NOTCH1m/GSIR cases. FBXW7 mutations (FBXW7m), which can occur concurrently to (∼ 20%) or in absence of (∼ 5%) NOTCH1m, might represent such an alternative pathway.7,29-31 Indeed, by preventing efficient degradation of several proteins such as Cyclin E, NOTCH1 and MYC, FBXW7m allows NOTCH1-dependent MYC transcriptional activation, but also NOTCH1-independent MYC stabilization. MYC escape from NOTCH1 control might thus constitute a major source of resistance to GSI in such cases.
Because MYC activity has so far mostly been inferred in T-ALL on the basis of its transcriptional status, we sought to provide a more complete picture of MYC deregulation in a comprehensive set of primary diagnosis T-ALL patient samples. Here, we show that high MYC expression levels can be observed in a large number of cases in absence of (known) NOTCH1 and/or FBXW7 mutations. Furthermore, our results indicate that modulation of MYC protein abundance through downregulation of PTEN constitutes a major alternative pathway of MYC activation in T-ALL, operating via the PI3K/AKT axis, both as a result of, and independently from NOTCH1m.
Methods
Patients
Peripheral blood or bone marrow samples from T-ALLs patients (n = 164, 128 adults, 36 children) were collected from Necker Hospital (Paris, France), la Timone Children's Hospital, and Paoli Calmettes Institute (Marseille, France). Details for clinical, immunophenotype, T-cell receptor analysis and oncogene gene expression have been previously described for most cases.31 Diagnosis and classification were defined by expression of specific T-cell markers and negativity for B cells and myeloid markers. Informed consent was obtained from the patients or relatives in accordance with the Declaration of Helsinki, with institutional review board approval of all involved hospitals. Samples were purified by Ficoll-Hypaque centrifugation.
Sequencing
FBXW7 and NOTCH1 mutations were previously reported for most cases, and remaining cases determined accordingly.31 The sequencing of PTEN exon 7 and Myc box I was performed on 116 T-ALL specimens, comprising T-ALL nos. 1 to 28, and 88 additional randomly selected T-ALL cases. Briefly, genomic deoxyribonucleic acid was extracted using QIAamp deoxyribonucleic acid Blood mini kit according to the manufacturer's instructions (QIAGEN) and 50 ng of deoxyribonucleic acid was PCR-amplified (primers are reported in supplemental Table 1, available on Blood Web site; see the Supplemental Materials link at the top of the online article). PCR products were directly sequenced, some amplicons harboring mutations were cloned, and 5 clones randomly selected for sequencing. Sequencing was performed on both strands by BigDye Terminator v1.1 Sequencing Kit (Applied Biosystems) and analyzed on an automated fluorescence-based ABI PRISM 3130XL Genetic analyzer according to the manufacturer's protocol. Sequence analysis was performed by CodonCode Aligner.
Real-time quantitative PCR
RNA was extracted from blood/bone marrow samples using either a TRIzol reagent (Invitrogen), or a column-based system RNAeasy mini kit (QIAGEN) according to the manufacturer's instructions. Reverse-transcription was performed with MMLV reverse transcriptase (Roche) or quantitect RT kit (QIAGEN) and cDNAs analyzed for MYC transcripts by real-time quantitative PCR (RQ-PCR) using Taqman probes on an ABIPRISM 7700 or 7500 (Applied Biosystems) according to guidelines of the EAC program.32 complementary deoxyribonucleic acid quality was checked using ABL housekeeping gene and samples with cycle threshold (Ct) values > 32 or with an ABL copy number < 1000 were excluded from the analysis. Each experiment included 2 negative controls, and all RQ-PCRs were performed in duplicate. MYC/ABL primers and Taqman probes were intron spanning to prevent amplification of genomic deoxyribonucleic acid (listed in supplemental Table 1). Amplification efficiency was assessed by the slope obtained from logarithmic dilutions of calibrator plasmids from 106 to 101 copies for both genes. Efficiency slopes for MYC and ABL were comparable and gave −3.46 and −3.55, respectively (Intercept Ct: 38.9 and 39.5, equivalent to 1 copy/well). To allow comparison between T-ALL samples, transcript quantification was performed after normalization with ABL from previously generated standard curves using the ΔCt method and results calculated according to the following formula 2Δ(CtABL–CtMYC). Analysis of MYC expression was performed using the GraphPad Prism 5 software and statistical significance was calculated as a P value using the nonparametric Mann-Whitney test. Data were considered significant when P < .05.
Immunoblotting analysis
Cells (10 × 106 to 50 × 106) were lysed for 30 minutes at 4°C in urea/thiourea buffer (7M urea, 2M thiourea, 4% CHAPS and 25mM tris(hydroxymethyl)aminomethane pH = 8.5), and samples centrifuged at 75000g for 30 minutes at 4°C. Protein concentrations of supernatants were determined using Bio-Rad protein assay (Bio-Rad). Equivalent protein extract (∼ 80 μg) for each sample was separated by SDS-PAGE and transferred to nitrocellulose membrane using Iblot Gel Transfer stacks and Iblot system (Invitrogen). Membranes were blocked in TTBS (137mM NaCl, 2mM KCl, 25mM tris(hydroxymethyl)aminomethane and 0.1% Tween 20) supplemented with 5% nonfat milk and incubated with primary antibodies against MYC (clone 9E10; Santa Cruz Biotechnology), PTEN (clone 138G6; Cell signaling Technology), phospho-AKT Ser473 (Cell Signaling Technology), phospho-GSK3β Ser9 (Cell Signaling Technology) or actin (clone I-19; Santa Cruz Biotechnology) overnight at 4°C with agitation. The secondary anti–rabbit, –mouse or –goat antibodies (Santa Cruz Biotechnology) conjugated to horseradish peroxidase were added for 1 hour at room temperature. Immunoblots were revealed using enhanced chemiluminescence Western blotting detection reagents (GE Healthcare) and stripped using Restore Western blot stripping buffer (Pierce). Densitometric analyses were performed using TotalLab Quant software Version 2008 (TotalLab Ltd). Each band was first normalized to the β-actin. For inter-gel comparisons, a reference sample, T-ALL no. 5, was systematically loaded on each SDS-PAGE gel, and MYC/actin or PTEN/actin values of the various samples were normalized to the reference TALL no. 5 value, then arbitrarily multiplied by 10.
Array Comparative Genomic Hybridization analysis
Pan-genomic high-density 244K oligonucleotide arrays (Agilent Technologies) were used as described previously,3 and analyzed using the CGH Analytics 3.2 software (Agilent Technologies). Loss of heterozygosity (LOH) analysis was performed using the GeneChip Human Mapping 250K SNP-array (Affymetrix), and analyzed using the GeneChip Genotyping Analysis Software GTYPE 4.0.
Cell lines and modulation of PTEN expression
Human T-ALL DND41 and Jurkat Tet-on (Clontech) cell lines were cultured at 37°C with 5% CO2 in RPMI media supplemented with 10% FBS (Lonza). 106 Jurkat Tet-on cells (Clontech) were transiently transfected with 5 μg of PTRE-PTEN vector; the transfection was performed by electroporation using the Amaxa system and solution V according to the manufacturer's protocol. Six hours after transfection, doxycycline (1 μg/mL) was added to the culture to induce PTEN expression. After an additional 12 hours, cells were harvested for Western blotting. PTRE-PTEN vector was obtained by cloning in the EcoRI site of the PTRE-Tight plasmid (Clontech) the human PTEN complementary deoxyribonucleic acid amplified with PTEN1 and PTEN2 primers (supplemental Table 1). To generate TRIP-shPTEN, a PTEN shRNA under the control of the U6 promoter was cloned into TRIPΔU3EFIαEGFP, (referred to as TRIP in this study), a third generation HIV-1–derived lentiviral vector that expresses EGFP.33 Viral particles were produced using standard procedures. For transduction, 2×105 DND41 cells were incubated with lentiviral particles for 30 minutes at 37°C, then cells were centrifuged at 800g for 60 minutes at 33°C. Five days after transduction, cells were harvested for Western blot and RQ-PCR.
Results
T-ALLs display a wide spectrum of MYC expression in both NOTCH1/FBXW7m and NOTCH1/FBXW7wt subgroups
We first examined MYC transcription levels by real-time PCR in a large collection of adult and pediatric samples (n = 164). As previously reported for pediatric cases,34,35 a remarkably large spectrum of relative MYC expression levels (> 2000-fold) could be observed overall (Figure 1A), consistent with the possibility that some samples displaying very low transcript levels might have built oncogenic circuits not critically dependent on MYC. Notably, median MYC expression was slightly higher in pediatric cases compared with adults (supplemental Figure 1A). We then compared the distribution of MYC transcripts between the different T-ALL differentiation subgroups (immature vs pre-αβ vs mature/cortical). No significant difference could be found between subgroups, either as a whole (Figure 1A) or when splitting children and adults (supplemental Figure 1B). This is in line with the previous suggestion that MYC activation occurs within all T-ALL sub-types,4 and that its oncogenic cooperation operates in a similar diverse fashion and in a nonexclusive manner (type B oncogene) in all subtypes. The effect of activating mutations of NOTCH1 on MYC transcription was next evaluated (Figure 1B). As expected, the lower 25% quartile of expression levels clearly differed overall between NOTCH1/FBXW7wt and NOTCH1/FBXW7m cases, in line with the reported activation of MYC transcription34 by NOTCH1. However, the NOTCH1/FBXW7m group still displayed a surprisingly large spectrum of MYC expression (> 50-fold). Interestingly, while a similar proportion of NOTCH1/FBXW7m cases were found in pediatric (66%) and adults (64%), only the adult group showed a significant difference in the median MYC expression between NOTCH1/FBXW7wt and NOTCH1/FBXW7m (supplemental Figure 1B). This difference seems owing to higher MYC expression in our pediatric NOTCH1/FBXW7WT series compared with adults, rather than to a difference in the NOTCH1/FBXW7m subset. Although this observation should be taken with caution as our pediatric series is small and contrasts with other pediatric series of equal size,34,35 it is in line with recent data from the large series from Zuurbier et al.36 We also tested the possibility that the distinct NOTCH1 mutations might exert various activation potential on the MYC target gene. However, no significant difference in MYC expression could be found in adults between PEST, HD, or combinations thereof, although there was a small trend for higher MYC expression in HD/PEST (supplemental Figure 1C). Most importantly, the upper 25% quartile did not differ between the NOTCH1/FBXW7wt and NOTCH1/FBXW7m groups, indicating that high MYC expression can be generated in a significant number of cases in absence of (known) NOTCH1 and/or FBXW7 mutations, or direct genomic alterations of MYC.
MYC expression in T-ALL. (A) Sample distribution according to differentiation subgroups. (B) Sample distribution according to NOTCH1/FBXW7 status (further splitting in pediatric vs adult subsets is shown in supplemental Figure 1). MYC expression is normalized to ABL. Dots represent individual samples; median values are indicated by horizontal bars. The nonparametric Mann-Whitney test was used to calculate P values.
MYC expression in T-ALL. (A) Sample distribution according to differentiation subgroups. (B) Sample distribution according to NOTCH1/FBXW7 status (further splitting in pediatric vs adult subsets is shown in supplemental Figure 1). MYC expression is normalized to ABL. Dots represent individual samples; median values are indicated by horizontal bars. The nonparametric Mann-Whitney test was used to calculate P values.
Altogether, and in keeping with the known multiplicity of indirect signaling pathways controlling MYC at the transcriptional level (including Sonic hedgehog, Wnt, NOTCH1, TGF-β),14,28 our results suggest that the transcriptional activation of MYC might be controlled in T-ALL by several pathways, and potentially in additive/synergistic fashion, both in presence or absence of NOTCH1/FBXW7 mutations.
Posttranscriptional deregulation represents an alternative route of MYC activation
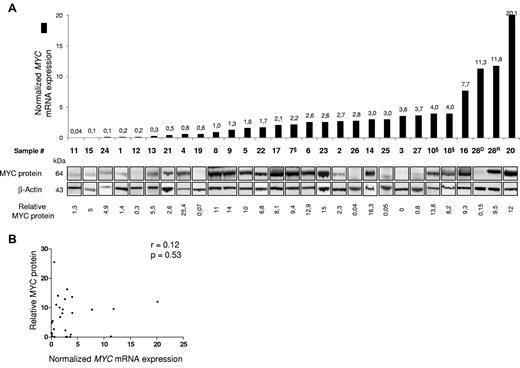
MYC is a complex transcription factor, which has been shown to be finely regulated at both posttranscriptional and posttranslational levels, both in physiologic and pathologic settings.7,15,28,29,37 To evaluate if MYC expression provided a reliable measure of MYC activity in T-ALL samples, we assessed protein levels by Western blot in a panel of 29 primary T-ALL samples for which enough material was available. As found for transcripts, a large spectrum of protein levels was seen (Figure 2A botttom panel). Strikingly, however, no significant correlation was apparent between transcript level (Figure 2A histograms) and protein abundance (Figure 2B), as estimated by quantification of relative MYC and β-actin band intensities (see “Immune blotting analysis” for details on relative quantification between sample). Indeed, high (eg, no. 23), medium (eg, no. 2), or low (eg, no. 26) levels of MYC protein could be found within all ranges of transcripts (Figure 2A), with high levels of MYC protein being produced in some cases with minimal transcript levels (compare for example samples 4 and 19), and conversely quite modest levels of protein being seen in samples with relatively high expression levels (compare for example samples 27 and 10, or 28D and 28R). This discrepancy between transcript levels and protein abundance is in line with the frequent deregulation of factors controlling MYC protein turn-over in tumor cells. In BL, mutations in the MYC-box (in and around the critical T58 phosphorylation site) impede efficient addressing to the proteasome, and results in oncogenic MYC protein stabilization.19,20 Our sequencing analysis of the MYC-box region in 116 primary T-ALL samples (including the subset used for proteomic analysis), revealed the absence of MYC-box mutation, suggesting that BL and T-ALL use different routes to achieve oncogenic MYC levels. Inactivating mutations of FBXW7 have also been shown to impair MYC degradation, and are estimated to be present in 10%-25% of T-ALL cases.7,29,31,34,38 However, only 3/29 samples (∼ 10%, cases 7, 10, and 18, Figure 2A) were found to harbor such mutations in our protein subset (FBXW7m were underrepresented in this subset as it represented ∼ 20% of the 139 cases screened from the whole T-ALL collection, not shown). Although all 3 cases were associated with high MYC protein levels as expected, FBXW7m alone could not account for the numerous cases showing MYC protein accumulation in our series, thus implying the presence of other, alternative mechanisms.
Relationship between MYC transcript levels and MYC protein abundance in primary T-ALL samples. (A) Samples are ordered according to increasing MYC mRNA levels (histograms in top panel, values are indicated above each bar), and the corresponding Western blots are shown below (bottom panel, relative values of MYC protein levels are indicated below each sample). Immunoblotting was performed using anti-MYC and anti–β-actin antibodies. § indicate cases with FBXW7m. (B) Plot showing relative MYC protein versus MYC transcript. Data were analyzed using the Spearman non parametric correlation test; the correlation coefficient (r) is indicated.
Relationship between MYC transcript levels and MYC protein abundance in primary T-ALL samples. (A) Samples are ordered according to increasing MYC mRNA levels (histograms in top panel, values are indicated above each bar), and the corresponding Western blots are shown below (bottom panel, relative values of MYC protein levels are indicated below each sample). Immunoblotting was performed using anti-MYC and anti–β-actin antibodies. § indicate cases with FBXW7m. (B) Plot showing relative MYC protein versus MYC transcript. Data were analyzed using the Spearman non parametric correlation test; the correlation coefficient (r) is indicated.
PTEN inactivation constitutes a likely candidate of MYC stabilization
To uncover alternative routes of MYC accumulation in T-ALL, we focused on a case which was analyzed at diagnosis and relapse (samples 28D and 28R), and which displayed striking differences in protein abundance despite very similar high transcript levels (Figure 2A). As both diagnosis and relapse samples were devoid of FBXW7 (and NOTCH1) mutations, we performed in-depth molecular characterization of this case. Karyotype and multicolor FISH (M-FISH) revealed the occurrence of 2 translocations in the same blast cells: a t(1;14)(p32;q11) involving TAL1, and a previously undescribed cryptic t(7;8)(q34;q24) (Figure 3A). FISH mapping, cloning, and breakpoint sequencing confirmed that the der8 chromosome juxtaposed MYC upstream of the T-cell receptorβ enhancer, at a propitious orientation, distance and timing39,40 for sustained MYC expression (Figure 3A and supplemental Figure 2). Breakpoint and transcription analysis further confirmed the occurrence of both translocations at diagnosis and relapse, and presence of similar TAL1 and MYC expression levels at both time points. Using high-resolution Array Comparative Genomic Hybridization analysis we further uncovered 7 additional chromosomal alterations in this case (not shown), 3 of which are recurrently observed in T-ALL oncogenesis (del 9p21, 10q23, and 4q25). Strikingly, only one difference could be observed between diagnosis and relapse: the del10q23 alteration, which evolved from a mono- to a bi-allelic deletion (Figure 3B) because of loss of heterozygosity of a large 10q(q22.2-qter) region including the phosphatase and tensin homolog deleted on chromosome 10 (PTEN) gene (supplemental Figure 3). Accordingly, Western blot analysis confirmed the presence of PTEN protein in the diagnosis sample, but its absence at relapse, in anti-correlation with MYC protein levels (Figure 3C). This points out to the major role played by PTEN in the emergence of the early relapse which occurred in this case, and the potential key role of MYC protein accumulation in this process.
Analysis of T-ALL case no.28 at diagnosis and relapse. (A) Top panel, identification of the t(1;14)(p32;q11) and a new cryptic t(7;8)(q34;q24) by R-Banding and M-FISH. M-FISH chromosome painting is shown in false color view with fluorescence profiles. Middle panel, chromosomal mapping of the t(7;8)(q34;q24) around MYC by FISH. (i) Interphase nucleus hybridized with the dual color MYC break apart fluorescence in situ hybridization probe (the 5′ probe is located immediately 5′ of MYC, and the 3′ probe ∼ 1.5 Mb 3′); (ii) metaphase FISH analysis with BAC clone RP11-440N18. The leukemic nuclei are identified by the presence of the der1 in DAPI image; (iii) metaphase fluorescence in situ hybridization analysis with BAC clone RP11-125A17. Bottom panel, metaphase FISH mapping with BAC clone RP11-701D14 (T-cell receptorβ) and CEP8 (left image), and BAC clones RP11-701D14 and RP11-125A17 (3′ MYC; right image), confirming reciprocal fusion between T-cell receptorβ and the 3′region of MYC. (B) Array Comparative Genomic Hybridization profiles of chromosome 10 at diagnosis and relapse. The arrow indicates the anomaly at the 10q23 region. Right, a zoom on the 10q23 region is shown. Homozygous and heterozygous deletions had ratios of −1 and −4 respectively. (C) Western blot analysis of MYC and PTEN proteins at diagnosis and relapse. The MYC transcript levels normalized to ABL are indicated on the top of the blots.
Analysis of T-ALL case no.28 at diagnosis and relapse. (A) Top panel, identification of the t(1;14)(p32;q11) and a new cryptic t(7;8)(q34;q24) by R-Banding and M-FISH. M-FISH chromosome painting is shown in false color view with fluorescence profiles. Middle panel, chromosomal mapping of the t(7;8)(q34;q24) around MYC by FISH. (i) Interphase nucleus hybridized with the dual color MYC break apart fluorescence in situ hybridization probe (the 5′ probe is located immediately 5′ of MYC, and the 3′ probe ∼ 1.5 Mb 3′); (ii) metaphase FISH analysis with BAC clone RP11-440N18. The leukemic nuclei are identified by the presence of the der1 in DAPI image; (iii) metaphase fluorescence in situ hybridization analysis with BAC clone RP11-125A17. Bottom panel, metaphase FISH mapping with BAC clone RP11-701D14 (T-cell receptorβ) and CEP8 (left image), and BAC clones RP11-701D14 and RP11-125A17 (3′ MYC; right image), confirming reciprocal fusion between T-cell receptorβ and the 3′region of MYC. (B) Array Comparative Genomic Hybridization profiles of chromosome 10 at diagnosis and relapse. The arrow indicates the anomaly at the 10q23 region. Right, a zoom on the 10q23 region is shown. Homozygous and heterozygous deletions had ratios of −1 and −4 respectively. (C) Western blot analysis of MYC and PTEN proteins at diagnosis and relapse. The MYC transcript levels normalized to ABL are indicated on the top of the blots.
Posttranscriptional PTEN downregulation is a major pathway of MYC activation in T-ALL
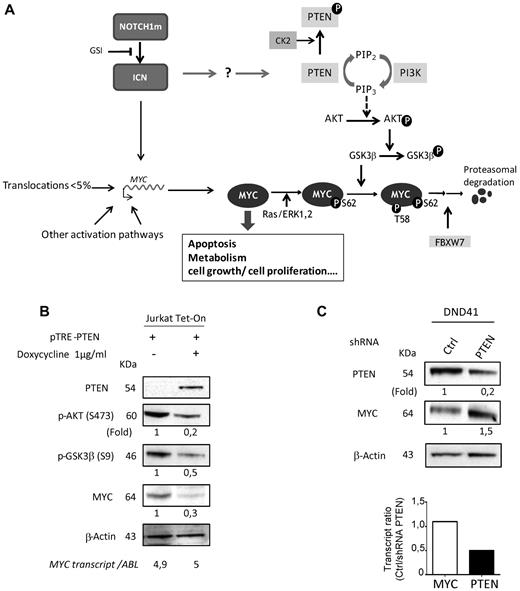
PTEN is a powerful tumor suppressor, and has been associated with GSI resistance in T-ALL,8 although this is debated.41 PTEN is considered the main negative regulator of the PI3K-AKT signaling. By dephosphorylating phosphatidylinositol-3,4,5-triphosphate (PIP3), PTEN antagonizes PI3K and dampens down AKT activation.42 As AKT inactivates by phosphorylating GSK3β, the serine-threonine kinase involved in MYC phosphorylation at T58,14,19,20 PTEN has been proposed to indirectly contribute to MYC degradation37 (Figure 4A). To test this signaling circuitry in T-ALL, we performed PTEN gain-of-function (using doxycyclin-inducible recombinant PTEN) and loss-of-function (using PTEN shRNA) experiments, and monitored the effect on MYC and upstream effectors. As shown in Figure 4B, PTEN re-expression in the Jurkat PTENneg T-ALL cell line resulted in significant reduction of MYC, phospho-AKT, and phospho-GSK3β protein levels. Conversely, PTEN knock-down (even if partial) led to a significant increase of MYC protein levels in the PTENpos DND41 T-ALL cell (Figure 4C and supplemental Figure 4B). Most importantly, this modulation chiefly occurred at the posttranscriptional level as MYC transcript was barely affected (Figure 4, see also supplemental Figure 5). Considering that this same circuitry has been found in other cell types,14,19,20,37 it is likely that a similar mechanism of MYC regulation is also happening in patient-derived cells.
Modulation of MYC protein abundance occurs via the PTEN phosphatidylinositol 3-kinase/AKT axis. (A) Schematic representation of upstream effectors of MYC activity. Efficient addressing of MYC to the proteasome degradation pathway is performed through an ordered pattern of phosphorylation, starting at serine 62 (S62) via the RAS/extracellular signal-regulated kinase/mitogen-activated protein kinase pathway,19 which enhances both MYC activity and phosphorylation at threonine 58 (T58) via the GSK3β serine-threonine kinase. The double S62T58 phosphorylated form of MYC is then recognized by the PIN1 phosphatase (not depicted for clarity), which rapidly dephosphorylates S62, and allows the T58 mono-phosphorylated form to be recognized and poly-ubiquitinylated by the UbE3 ligase FBXW7 for final degradation.14,19,20 For clarity, only few regulators/effectors are displayed; notably the multiple targets of the major AKT hub (such as mTOR) are not shown. (B) Effect of PTEN gain-of-function in PTENneg T-ALL. Jurkat Tet-On cells were transiently transfected with the doxycycline-inductible expression vector PTRE-PTEN, then PTEN, MYC, p-AKT, and p-GSK3β protein levels monitored by Western blot with or without doxycycline induction (in absence of PTRE-PTEN, doxycycline does not induce a decrease of MYC protein level, see supplemental Figure 4A). MYC transcript levels were monitored by RQ-polymerase chain reaction and normalized to ABL transcripts. (C) Effect of PTEN loss-of-function in PTENpos T-ALL. DND41 cell line was transduced either with TRIP-shPTEN, a lentiviral vector that expresses EGFP and a PTEN shRNA, or with TRIP, the native lentiviral vector that only expresses EGFP used here as control. PTEN and MYC proteins and transcripts levels were then monitored by Western blot and RQ-PCR, respectively. MYC and PTEN transcript levels were normalized to ABL transcripts. (B-C) Bands intensities were quantified using TotalLab Quant software, normalized to β-actin and reported below the blots as relative values to the control assays. Data shown are representative of at least 3 independent experiments.
Modulation of MYC protein abundance occurs via the PTEN phosphatidylinositol 3-kinase/AKT axis. (A) Schematic representation of upstream effectors of MYC activity. Efficient addressing of MYC to the proteasome degradation pathway is performed through an ordered pattern of phosphorylation, starting at serine 62 (S62) via the RAS/extracellular signal-regulated kinase/mitogen-activated protein kinase pathway,19 which enhances both MYC activity and phosphorylation at threonine 58 (T58) via the GSK3β serine-threonine kinase. The double S62T58 phosphorylated form of MYC is then recognized by the PIN1 phosphatase (not depicted for clarity), which rapidly dephosphorylates S62, and allows the T58 mono-phosphorylated form to be recognized and poly-ubiquitinylated by the UbE3 ligase FBXW7 for final degradation.14,19,20 For clarity, only few regulators/effectors are displayed; notably the multiple targets of the major AKT hub (such as mTOR) are not shown. (B) Effect of PTEN gain-of-function in PTENneg T-ALL. Jurkat Tet-On cells were transiently transfected with the doxycycline-inductible expression vector PTRE-PTEN, then PTEN, MYC, p-AKT, and p-GSK3β protein levels monitored by Western blot with or without doxycycline induction (in absence of PTRE-PTEN, doxycycline does not induce a decrease of MYC protein level, see supplemental Figure 4A). MYC transcript levels were monitored by RQ-polymerase chain reaction and normalized to ABL transcripts. (C) Effect of PTEN loss-of-function in PTENpos T-ALL. DND41 cell line was transduced either with TRIP-shPTEN, a lentiviral vector that expresses EGFP and a PTEN shRNA, or with TRIP, the native lentiviral vector that only expresses EGFP used here as control. PTEN and MYC proteins and transcripts levels were then monitored by Western blot and RQ-PCR, respectively. MYC and PTEN transcript levels were normalized to ABL transcripts. (B-C) Bands intensities were quantified using TotalLab Quant software, normalized to β-actin and reported below the blots as relative values to the control assays. Data shown are representative of at least 3 independent experiments.
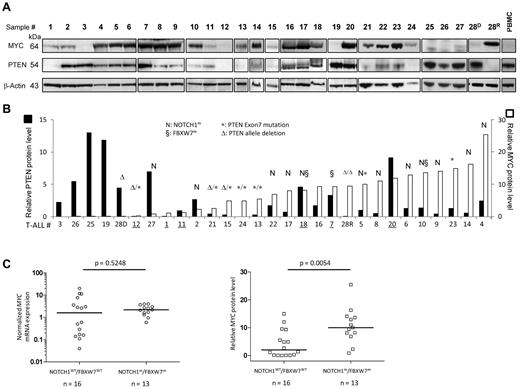
We next sought to assess to which extent MYC abundance relates to PTEN loss/haplo-insufficiency in primary T-ALL samples. PTEN (exon 7) sequencing was performed, and inactivating mutations were found in 12% of cases (14/116, supplemental Table 2), with 2 cases also bearing 1 deleted allele (analysis performed by aCGH analysis in 16/29 available cases from the protein subset). We then probed PTEN on stripped Western blots from the 29 primary T-ALL samples (Figure 5A), and evaluated relative PTEN and MYC levels using TotalLab Quant software (see “Immunoblotting analysis”). A surprisingly good anti-correlation between PTEN and MYC protein levels could be observed (Figure 5B and supplemental Figure 6), especially when considering the anticipated diversity of additional factors potentially acting on AKT hyper-activation downstream of PTEN in T-ALL.43 The inverse inter-relationship was particularly clear in cases with abundant PTEN protein levels (eg, nos. 3, 26, 25, 19, 28D), where only faint MYC could be found despite relatively high transcription levels (see histograms in Figure 2A). Conversely, high MYC protein levels were in most cases associated with rather low relative PTEN protein levels (eg, nos. 6, 10, 9, 23, 14, 4). Few notable exceptions (underlined in Figure 5B) pointed out the multiplicity of factors potentially additionally involved in MYC protein abundance. Case no. 20, for example, displayed both abundant PTEN and MYC proteins. However, an unusually high transcriptional activity of MYC was present in this case (ΔCt = 20.1; Figure 2A), raising the possibility that above a certain threshold, abundant transcription might compensate for protein accumulation, potentially by saturating one or several steps of the degradation machinery. Conversely, substantial MYC stabilization through PTEN loss would obviously first require a minimal sustained MYC transcription level (eg, case no. 11, PTENlow, where MYC is low but detectable despite a transcription level of 0.04, Figure 2A; compare with sample 25, PTENhigh, where a similar low MYC is associated with a ∼ 100-fold higher transcription level). Most interestingly, the 2 other samples with both abundant PTEN and MYC (nos. 18 and 7) corresponded to cases with FBXW7m. That 2 of the 3 cases with FBXW7m displayed abundant MYC protein levels and did not downregulate PTEN is in keeping with the idea that FBXW7m might substitute for PTEN loss, and that increased MYC abundance is a major oncogenic consequence of PTEN modulation among the many other oncogenic effects provoked by the activation of the key AKT hub. We also observed that a large fraction of the NOTCH1m cases was associated with MYChigh /PTENlow protein levels, and occurred in absence of detected PTEN genomic alteration (to one exception, Figure 5B no. 5). Similar to Larson et al,34 no transcriptional down-regulation of PTEN was observed in these cases (not shown), suggesting a posttranscriptional regulation distinct from the NOTCH/HES1 axis.8 Because a significant correlation could be found in this subset between NOTCH1m and MYC at the protein level (P = .0054) but not at the transcriptional level (P = .52; Figure 5C), our data suggest that downregulation of PTEN protein may represent yet an additional pathway of posttranscriptional MYC activation by NOTCH1.
Western blot analysis of PTEN and MYC in primary T-ALL samples and cell lines. (A) Western blot analysis of PTEN and MYC in 29 primary T-ALL samples. The same immunoblots were used to probe MYC, β-actin and PTEN. (B) Quantification of the relative MYC and PTEN protein amount (see “Immunoblot analysis” for details). Samples were ranked according to increasing MYC protein abundance. (C) Relative MYC transcript levels (left) and relative MYC protein levels (right) of the 29 T-ALL subset used for proteomic analysis were plotted according to NOTCH1/FBXW7 status and analyzed using nonparametric Mann-Whitney test.
Western blot analysis of PTEN and MYC in primary T-ALL samples and cell lines. (A) Western blot analysis of PTEN and MYC in 29 primary T-ALL samples. The same immunoblots were used to probe MYC, β-actin and PTEN. (B) Quantification of the relative MYC and PTEN protein amount (see “Immunoblot analysis” for details). Samples were ranked according to increasing MYC protein abundance. (C) Relative MYC transcript levels (left) and relative MYC protein levels (right) of the 29 T-ALL subset used for proteomic analysis were plotted according to NOTCH1/FBXW7 status and analyzed using nonparametric Mann-Whitney test.
Altogether our results suggest that, in addition to the multiple transcriptional pathways of MYC activation, PTEN downregulation constitute a major alternative and/or complementary pathway of posttranscriptional MYC activation in T-ALL.
Discussion
Cumulative evidence indicates that MYC is a critical component of T-ALL oncogenesis,7,9-12,23,25-27,29,37 and a potential candidate for targeted therapy.14,44 However, oncogenic MYC deregulation is a subtle and complex process, involving both fine protein dosage and cell-context dependency, and detailed understanding of MYC-mediated oncogenesis in T-ALL is still lacking. To further understand how MYC is interspersed in the complex and likely numerous oncogenic networks underlying T-ALL tumorigenesis,2 we underwent a thorough analysis of MYC activation in a comprehensive collection of primary adult and pediatric patient samples. Importantly, we show that high MYC expression levels could be found in a large fraction of the samples in absence of known NOTCH1/FBXW7 mutations or direct MYC alteration, suggesting the occurrence of important additional pathway(s) of MYC transcriptional activation, which still remain to be precisely defined. Interestingly, the large spectrum of MYC expression also found within the NOTCH1m/FBXW7m group goes along with the idea that such pathways might act in an integrative fashion, a scenario which could have dire consequences in the set up of targeted therapy strategies. This scenario is not unexpected considering the known multiplicity of pathways controlling MYC transcription,28 and reports of frequent indirect sustained induction of MYC by upstream oncogenic signals such as Ras or Wnt/βcatenin in both human neoplasia44 and recent mouse models of T-ALL.26,27 Transcriptional deregulation is however not the sole factor governing MYC activity. MYC is indeed also very tightly regulated at the posttranscriptional level, partly through the control of protein stability: while MYC in normal resting cells displays a particularly short half-life (∼ 12 minutes.), it typically increases to 20 minutes in cycling cells, and has been shown to reach considerable higher levels in cancer cells (5- to 10-fold increase).29,37 Here we add further support that posttranscriptional control is also frequently at work in T-ALL, providing yet another alternative and/or synergistic route of MYC activation. Most importantly, we show that very low transcriptional levels can be rescued by posttranscriptional control of MYC protein levels, and can in some cases give rise to more abundant MYC proteins than cases displaying considerably higher transcription levels (including a case resulting from a MYC/T-cell receptor translocation). This spectacular compensation suggests that physiologic MYC transcription levels resulting from normal thymocyte differentiation (in particular during the clonal expansion phase after β-selection)45 might be sufficient to trigger oncogenic MYC levels in the context of cooperating mutations allowing protein accumulation. Conversely, the absence of stabilization in cases with deregulations leading to very high transcript levels might avoid converting “low” MYC levels to “acute” MYC, which has been shown to trigger activation of P53-mediated tumor suppression.44 Such a phenomenon would allow a low MYC-mediated oncogenic initiation to progress stepwise through gradual acquisition of P53 pathway inhibition (eg, through P14ARF loss-of-function), allowing to release all brakes to the conversion of acute MYC and aggressive tumorigenesis through a subsequent step of MYC stabilization. In this context, the identification of the mechanisms underlying such posttranscriptional deregulation is essential to the understanding of MYC-mediated oncogenesis and drug resistance. In BL, oncogenic MYC stabilization mainly results from mutations in MYC itself, in or around the critical T58 phosphorylation site, preventing efficient addressing to the proteasome. We show that such MYC mutations are absent in primary T-ALL. Rather, our results indicate that PTEN plays a major role in modulating the abundance of MYC protein. A role of PTEN loss-of-function on MYC stabilization via the up-regulation of AKT is in line with previous findings in B-cell ALL37 and pancreatic cancer,46 and with the signaling circuitry of the AKT axis identified in T-ALL8 (Figure 4A). One of the cases reported here (no. 28), showing a fulminant early relapse associated with the progression from mono- to bi-allelic PTEN deletion and the conversion from low to acute MYC, adds to the mounting evidence that acute PTEN loss plays a major role in the emergence of chemoresistance,8 and of a potential key role of MYC stabilization in this process.7,29 Interestingly, PTEN, like MYC, is very dose-responsive,47 and on acute loss, will also trigger P53-mediated tumor suppression, through induction of senescence.48 This illustrates the critical importance of “locking” the P53 checkpoint for tumor progression in both PTEN- and MYC-mediated oncogenic processes. In line with the “one-by-one” model proposed by Pandolfi and colleagues in prostate cancer,48 the interdependency of MYC and PTEN shown here suggests that in T-ALL, subtle variations of PTEN (and/or MYC) might similarly allow gradual loss of P53-mediated tumor suppression (mostly via P14ARF loss-of-function), driving subsequent acquisition of acute loss of PTEN and/or acute MYC toward the generation of a highly refractory disease.
As the effect of PTEN on MYC would be indirect via activation of the PI3K-AKT-GSK3β pathway, the many mutations recently described in components from this pathway downstream43 or upstream8,49 of PTEN might provide alternative and/or synergistic routes to modulate low versus acute MYC activity. Recent estimates indicate that up to 47% of T-ALL cases contain mutations in the PTEN/PI3K/AKT pathway, a figure likely underestimated due the many upstream factors controlling this key oncogenic hub (eg, via CK2, RAS).43,49 Interestingly, while displaying a large spectrum of MYC expression levels, the large majority of NOTCH1m cases was associated with high MYC and low PTEN protein levels in absence of PTEN lesions. This suggests that NOTCH1-mediated downmodulation of PTEN protein constitutes an important pathway of MYC stabilization, and adds to the evidence that NOTCH1m/FBXW7m (∼ 60% cases) would play a major dual role in both activating MYC transcription and stabilizing MYC via PTEN repression. Although the sum of all these alterations might amount to most if not all T-ALL cases, we do not mean to imply that all such cases are MYC-addicted. The fact that MYC can rescue some, but not all human T-ALL cell lines from GSI-induced growth arrest,9 and the large spectrum of MYC protein found across samples in our series including cases with undetectable levels of MYC both in NOTCH1wt and (rare) NOTCH1m cases are consistent with the possibility that some leukemia cases might have built oncogenic circuits not critically dependent on MYC. Understanding the global impact of this plethora of mutations on the many critical pathways triggered by AKT (including mTOR, NOTCH1, MYC), as well as how it integrates in the distinct oncogenic networks imprinted in the various T-ALL subtypes, is key to the development of therapeutic targeting of MYC14 and/or the PTEN/AKT pathways,43,50 both in GSIR and NOTCH1-independent/MYC-mediated T-ALL patients. However, considering the amazing complexity of MYC target genes and pathways, combined with the subtle effect of MYC protein dosage and the particularly diverse oncogenic circuitry in T-ALL, precisely defining MYC addiction in T-ALL patients will be a challenging task that warrants further study on a comprehensive cohort of protocolar patients.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr E. Clappier for the generous gift of the TRIP-shPTEN vector, Drs B. Malissen, E. Soucie, and J. Nunez for critical reading of the manuscript, and all the clinicians of the French adult ALL GRAALL cooperative group for providing patient samples, which were conserved in the AP-HP and the Institut National du Cancer (INCa) accredited tumorothèque at Necker-Enfants Malades. Work in B.N.'s laboratory was supported by grants from INCa, from la Fondation de France, and institutional grants from Inserm and CNRS. M.B. was supported by a fellowship from the Ministère de l'Enseignement Supérieur et de la Recherche and the Lique Nationale Contre le Cancer (LNCC); B.M. was a recipient of a fellowship from the LNCC, and the French Society of Hematology (SFH); W.A.D., and A.W.L. were supported by grant EMCR 2002-2707 from the Dutch Cancer Society.
Authorship
Contribution: M.B., M.L., and V.A. designed, performed, and analyzed experiments; J.D.C. and C.P. performed and analyzed sequencing data; B.Q., C.F., C.F.T., B.M., J.M.N., C.P., W.A.D., A.W.L., J.G., S.R., and E.M. deciphered and analyzed case T-ALL no. 28; J.S., L.H., V.A., and E.M. performed Array Comparative Genomic Hybridization/SNP data; C.B. produced lentiviral particles; V.A., E.A.M., N.V., T.P., and G.M. provided patient samples; and D.P.B. and B.N. conceived and supervised research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Bertrand Nadel and Dominique Payet-Bornet, Centre d'Immunologie de Marseille-Luminy (CIML), Université de la Méditerranée, 13288 Marseille cedex 9, France; e-mail: nadel@ciml.univ-mrs.fr and payet@ciml.univ-mrs.fr.