Abstract
Pediatric mixed-lineage leukemia (MLL)–rearranged acute monoblastic leukemia with t(9;11)(p22;q23) has a favorable outcome compared with other MLL-rearranged AML. The biologic background for this difference remains unknown. Therefore, we compared gene expression profiles (GEPs; Affymetrix HGU133 + 2.0) of 26 t(9;11)(p22;q23) patients with 42 other MLL-rearranged AML patients to identify differentially expressed genes. IGSF4, a cell-cell adhesion molecule, was found to be highly expressed in t(9;11)(p22;q23) patients, which was confirmed by real-time quantitative polymerase chain reaction and Western blot. IGSF4 expression within t(9;11)(p22;q23) patients was 4.9 times greater in French-American-British morphology classification (FAB)–M5 versus other FAB-types (P = .001). Methylation status investigation showed that high IGSF4-expressing t(9;11)(p22;q23) patients with FAB-M5 have no promoter hypermethylation, whereas all other cases do. Cell-line incubation with demethylating agent decitabine resulted in promoter demethylation and increased expression of IGSF4. Down-regulation of IGSF4 by siRNA did not affect proliferation or drug sensitivity. In a cohort of 79 MLL-rearranged AML cases, we show significant better overall survival for cases with high IGSF4 expression (5-year overall survival 0.70 vs 0.37, P = .03) In conclusion, we identified IGSF4 overexpression to be discriminative for t(9;11)(p22;q23) patients with FAB-M5, regulated partially by promoter methylation and resulting in survival benefit.
Introduction
Pediatric acute myeloid leukemia (AML) is a heterogeneous disease. Currently, apart from response to treatment, the most important prognostic factor is cytogenetic aberrations. Well-known cytogenetic abnormalities that predict differences in survival are t(15;17)(q22;q21) (PML-RARα), t(8;21)(q22;q22) (RUNX1-RUNX1T1), inv(16)(p13q22) (CBF-MYH11), and mixed-lineage leukemia (MLL)-rearranged AML.1-3 Intensive chemotherapy has improved survival rate during the past few decades (5-year event-free survival [EFS] 60%). Future therapeutic strategies should be directed toward outcome as well as toward limitation of short- and long-term toxicity.4 It is anticipated that such strategies can be determined by the molecular targeting of abnormally expressed genes in specific genetic types of pediatric AML.5
In recent years, more than 60 different translocation partners of the MLL gene have been described.6 In pediatric MLL-rearranged AML, the most common translocations are t(9;11)(p21;q23) (MLL-AF9), which occurs in approximately 50% of patients; t(10;11)(p12;q23) (MLL-AF10), t(6;11)(q27;q23) (MLL-AF6); and t(11;19)(q23;p13.3) (MLL-ENL).1,2 Of interest is that t(9;11) has been linked with favorable outcome.7-9 Recently, we identified that superior prognosis in the t(9;11) cases was restricted to those with French-American British morphology classification (FAB) M5 phenotype.10
So far, the underlying biologic factors that determine the differences in clinical outcome of MLL-rearranged AML cases on the basis of translocation partner are not known because there is little information available on the molecular aberrations. Therefore, the aim of this study was to investigate the biologic background of t(9;11)(p22;q23) AML with and without FAB M5 compared with AML with other MLL-translocation partners.
Methods
Patients
Viably frozen diagnostic bone marrow or peripheral blood samples from 269 de novo and 8 secondary pediatric AML cases were provided by the Dutch Childhood Oncology Group, the AML “Berlin-Frankfurt-Münster” Study Group, the Czech Pediatric Hematology Group, and the St Louis Hospital in Paris, France. Samples were chosen to represent all common cytogenetic groups and were selected on the basis of availability of high-quality RNA. Each study group performed central morphologic reviews according to the FAB classification. Informed consent was obtained from all patients in accordance with the Declaration of Helsinki, after Erasmus MC Institutional Review Board approval according to national law and regulations.
The samples included 33 pediatric MLL-rearranged cases with t(9;11)(p22;q23) and 52 with other MLL rearrangements, the other 192 samples represented all other common AML cytogenetic groups (Table 1). Among the 8 secondary AML cases, 3 harbored a t(9;11)(p22;q23). These 3 cases were all classified as FAB-M5. The 5 other secondary AML cases did not harbor an MLL rearrangement.
Clinical characteristics of GEP cohort of initial pediatric AML samples
| . | Original GEP cohort (n = 245) . | Additional cases (n = 32) . | Total (n = 277) . |
|---|---|---|---|
| Sex | |||
| Male | 137 (56) | 13 (45) | 150 |
| Female | 108 (44) | 16 (55) | 124 |
| Age, y, median (range) | 9.8 (0-18.8) | 3.0 (0.4-17.3) | |
| WBC × 109/L, median (range) | 41.3 (0.0-483) | 117.5 (1.8-475) | |
| FAB | |||
| M0 | 14 (6) | 3 (9) | 17 (6) |
| M1 | 25 (10) | 1 (3) | 26 (9) |
| M2 | 55 (22) | 1 (3) | 56 (20) |
| M3 | 20 (8) | 20 (7) | |
| M4 | 56 (23) | 8 (25) | 64 (23) |
| M5 | 53 (22) | 18 (56) | 71 (26) |
| M6 | 3 (1) | 3 (1) | |
| M7 | 8 (3) | 8 (3) | |
| Unknown | 11 (4) | 1 (3) | 12 (4) |
| Cytogenetics | |||
| MLL rearrangements | 53 (23) | 32 (100) | 85 (31) |
| t(9;11)(p22;q23) | 21 (9) | 12 (38) | 33 (12) |
| Other MLL rearrangements | 32 (13) | 20 (63) | 52 (19) |
| t(8;21) (q22;q22) | 28 (11) | 28 (10) | |
| inv(16)(q13q22) | 27 (11) | 27 (10) | |
| t(15;17)(q22;q21) | 18 (7) | 18 (6) | |
| Cytogenetically normal (CN)-AML | 41 (17) | 41 (15) | |
| AML-other/unknown | 78 (32) | 78 (28) | |
| Successful GEP | 245 | 14 | 259 |
| Successful RT-qPCR | 76 | 19 | 95 |
| . | Original GEP cohort (n = 245) . | Additional cases (n = 32) . | Total (n = 277) . |
|---|---|---|---|
| Sex | |||
| Male | 137 (56) | 13 (45) | 150 |
| Female | 108 (44) | 16 (55) | 124 |
| Age, y, median (range) | 9.8 (0-18.8) | 3.0 (0.4-17.3) | |
| WBC × 109/L, median (range) | 41.3 (0.0-483) | 117.5 (1.8-475) | |
| FAB | |||
| M0 | 14 (6) | 3 (9) | 17 (6) |
| M1 | 25 (10) | 1 (3) | 26 (9) |
| M2 | 55 (22) | 1 (3) | 56 (20) |
| M3 | 20 (8) | 20 (7) | |
| M4 | 56 (23) | 8 (25) | 64 (23) |
| M5 | 53 (22) | 18 (56) | 71 (26) |
| M6 | 3 (1) | 3 (1) | |
| M7 | 8 (3) | 8 (3) | |
| Unknown | 11 (4) | 1 (3) | 12 (4) |
| Cytogenetics | |||
| MLL rearrangements | 53 (23) | 32 (100) | 85 (31) |
| t(9;11)(p22;q23) | 21 (9) | 12 (38) | 33 (12) |
| Other MLL rearrangements | 32 (13) | 20 (63) | 52 (19) |
| t(8;21) (q22;q22) | 28 (11) | 28 (10) | |
| inv(16)(q13q22) | 27 (11) | 27 (10) | |
| t(15;17)(q22;q21) | 18 (7) | 18 (6) | |
| Cytogenetically normal (CN)-AML | 41 (17) | 41 (15) | |
| AML-other/unknown | 78 (32) | 78 (28) | |
| Successful GEP | 245 | 14 | 259 |
| Successful RT-qPCR | 76 | 19 | 95 |
Values reflect number of cases (%) unless otherwise specified.
AML indicates acute myeloid leukemia; FAB, French-American-British morphology classification; GEP, gene expression profiling array; MLL, mixed-lineage leukemia; RT-qPCR, quantitative real-time polymerase chain reaction; and WBC, white blood cell count.
Materials
Leukemic cells were isolated and enriched as previously described.11,12 All resulting samples contained 80% or more leukemic cells, as determined morphologically by May-Grünwald-Giemsa (Merck)–stained cytospins. A minimum of 5 × 106 leukemic cells was lysed in Trizol reagent (Gibco BRL/Life Technologies) and stored at −80°C. Isolation of genomic DNA and total cellular RNA was performed as described previously.13 Leukemic samples were routinely investigated for MLL rearrangements by standard chromosome-banding analysis and/or fluorescence in situ hybridization. If translocation with one of the common partners (MLL-AF9, MLL-AF10, MLL-AF6, MLL-ENL, and MLL-ELL) was suspected, reverse transcriptase polymerase chain reaction (PCR) was performed. (Primers are described in supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article,) Of the 85 MLL-rearranged cases, 33 harbored a t(9;11)(p22;q23), 19 a t(10;11)(p12;q23), and 15 a t(6;11)(q27;q23). The remaining 18 cases were confirmed with long-distance inverse PCR as MLL other.
Gene expression profiling
Gene expression profiling (GEP) was performed on the RNA of a cohort of 237 de novo and 8 secondary pediatric AML samples. We included 14 additional cases of MLL-rearranged AML (5 of which carried a t(9:11)) for GEP to increase group size. Integrity of total RNA was checked with the use of the Agilent 2100 Bio-analyzer (Agilent). cDNA and biotinylated cRNA was synthesized, hybridized, and processed on the Affymetrix Human Genome U133 Plus 2.0 Array (Affymetrix) according to the manufacturer's guidelines. Data acquisition was performed by the use of Expresso (Bioconductor package Affy), and probe-set intensities were normalized by the use of the variance stabilization normalization (Bioconductor package VSN) in the statistical data analysis environment R, version 2.7.0.14,15 Expression levels were log-transformed during this normalization. An empirical Bayes linear regression model was used to compare the signatures for the t(9;11) cases to all other MLL-rearranged AML cases.16 Moderated T-statistics P values were corrected for multiple testing by use of the false discovery rate method defined by Benjamini and Hochberg.17 IGSF4 was identified from a top-50 differentially expressed gene list. For the expression analysis of IGSF4, probe set 209031_at was used.
Quantitative real-time PCR
Quantitative real-time PCR (RT-qPCR) was performed on cDNA of 95 pediatric AML patient samples, selected on availability of remaining cDNA, produced as previously described.18 Within this group, 57 were classified as MLL-rearranged leukemia, of which 24 harbored a t(9;11) (Table 1). An ABI PRISM 7900HT sequence detector (Applied Biosystems) was used to validate the GEP results. Primers used for IGSF4 are described in supplemental Table 1. For expression analysis of IGSF4, SYBRgreen (Finnzymes) was used. The expression of the genes was compared with glyceraldehyde 3-phosphate dehydrogenase, with primers and probe as previously described (sequences are shown in supplemental Table 1).18 The average cycle threshold (Ct) value was used to calculate mRNA expression levels of IGSF4 relative to the expression level of the reference gene (glyceraldehyde 3-phosphate dehydrogenase) by use of the comparative cycle time (ΔCt) method.19
Western blot
For Western blot, 10 leukemia samples were selected on the basis of the availability of material, of which 3 harbored a t(9;11), 3 harbored another MLL-translocation, and 4 had a karyotype other than MLL (AML-other, containing a case with t(8;21), one with inv(16), one with t(15;17), and one with a normal karyotype). Cell pellets stored at −80°C were quickly thawed and resuspended in 100 μL of lysis buffer composed of 25mM Tris (tris[hydroxymethyl]aminomethane) buffer, 150mM NaCl, 5mM EDTA (ethylenediaminetetraacetic acid), 10% glycerol, 1% Triton X-100, 10mM sodium pyrophosphate, 1mM sodium orthovanadate, 10mM glycerolphosphate, 1mM dithiothreitol, 1mM phenylmethylsulfonyl fluoride, 1% aprotinin (Sigma-Aldrich), 10mM sodium fluoride, and 20 μL of freshly prepared sodium pervanadate. Subsequently, cell lysis was allowed for 30 minutes on ice. Cell lysates were cleared by centrifugation for 15 minutes at 10 000g (13 000 rpm) and 4°C. Protein concentration was determined by use of the bicinchoninic acid protein assay (Pierce Biotechnology) with different concentrations of bovine serum albumin as standards. Cell lysates containing 20 ng of protein were separated on 10% polyacrylamide gels and transferred onto nitrocellulose membranes (Schleicher & Schuell). Western Blots were probed with goat polyclonal IgG anti-TSLC1 (synonym of IGSF4, sc-25 077; Santa Cruz Biotechnology) and mouse antibeta-actin (ab6276; Abcam) antisera. Subsequently, the blots were labeled with peroxidase-conjugated antigoat antibody (sc-2020; Santa Cruz Biotechnology) or antimouse antibody (DAKO). Chemiluminescence (SuperSignal West Femto Maximum Sensitivity Substrate; Pierce Biotechnology) was used to detect luminescence using the Syngene chemigenius (Syngene).
Methylation-specific PCR
To investigate the methylation status of IGSF4 methylation-specific PCR (MS-PCR) was used. Fourteen leukemia samples were selected, 5 samples with a t(9;11) on the basis of their high IGSF4 expression with GEP and RT-qPCR. These samples with t(9;11) were compared with 5 MLL-rearranged samples with other translocation partners and 4 other AML samples. The primers as described by Overmeer et al20 were used; 3 different areas of the promoter (designated 1 M/U, 5 M/U, and 9 M/U) were used (sequences are shown in supplemental Table 1). Unmodified genomic DNA was used to test the specificity of the primers for bisulfite-converted DNA. One DNA sample was first treated with DNA methylase SSS1 and methyl donor SAM (M0226S; New England Biolabs) and then bisulfite converted, creating a hypermethylated sample (M) as a control for the methylation specific primers. As a control for the unmethylated specific primers, bisulfate-converted DNA of healthy, adult male donors was used (U).
The specificity of the methylation (M)- and unmethylation (U)-specific primers was tested on a dilution range with a mix of M and U DNA (supplemental Figure 1). The dilution ranges indicated that the combination of 9M and 9U is the most specific.
Demethylating agents
Cell lines ML-2 (MLL t(6;11)), HL-60 (AML-other), and MONO-MAC-1 (MLL t(9;11); DSMZ GmbH) were cultured with and without demethylating agent decitabine. ML-2 and HL-60 were selected for their low expression of IGSF4 on RT-qPCR, and MONO-MAC-1 was used as a control because it shows high IGSF4 expression. Decitabine concentration was chosen after an in vitro drug assay with decitabine concentrations ranging from 0.125μM to 4.0μM and was determined for each cell line to be the approximate 50% lethal concentration. ML-2 was cultured with a concentration of decitabine of 2μM, HL-60, and MONO-MAC-1 with a concentration of 4μM. Decitabine and culturing medium (RPMI 1640 with L-alanyl-L-glutamine; Invitrogen); 10% fetal calf serum (FCS; Integro); and penicillin 100 U/mL, streptomycin 100 μg/mL, and fungizone 0.125 μg/mL (PSF; Invitrogen) were refreshed daily. The experimental condition started with 100 × 106 cells. Cell counts were determined on a daily basis, and cells were maintained in culture at a concentration of 0.5 × 106 cells/mL. Cell samples of both test and control conditions were taken from the medium every other day for the first 6 days and daily thereafter. They were washed with phosphate-buffered saline, and samples for protein studies were frozen at −80°C as dry cell pellets, and for DNA and RNA extraction cells were lysed in Trizol reagent and stored at −80°C. The experiment ended as soon as all remaining experimental cells were apoptotic.
siRNA transfection
The MONO-MAC-1, ie, t(9;11) and NOMO-1, ie, t(9;11) cell line (DSMZ) were cultured in RPMI-1640 medium supplemented with 10% FCS and PSF and grown as suspension cultures at 37°C in humidified air containing 5% CO2. Cells from both cell lines (10 × 106) were transfected by electroporation in 400 μL of RPMI 1640 with L-alanyl-L-glutamine (Invitrogen) containing 250nM of either a mix of equal amounts of IGSF4 siRNAs (Dharmacon ON-TARGETplus LQ-016565; Thermo Fisher Scientific) or Nontargeting siRNA (Dharmacon ON-TARGETplus D-001810-01-05; Thermo Fisher Scientific), in 4-mm electroporation cuvettes (Bio-Rad; sequences are described in supplemental Table 1).
Electroporation was performed by the use of an EPI 2500 gene pulser (Fischer) by applying a rectangle pulse of 300 V for 10 milliseconds. To compensate for the amount of cell death induced merely as a consequence of the electroporation procedure, control cells were electroporated in the absence of siRNA. After incubating for 15 minutes at room temperature, the cells were diluted in 10 mL pf RPMI 1640 supplemented with 10% FCS and PSF and incubated at 37°C and 5% CO2. They were maintained in culture for 72 hours. Cell counts were determined daily (t = 6 hours, t = 24 hours, t = 48 hours, and t = 72 hours). Cell samples of both test and control conditions were taken from the medium at every time point. They were washed with phosphate-buffered saline and lysed in Trizol reagent and stored at −80°C. DNA content and cell-cycle phase were assessed with the use of propidium iodide staining and measured by flow cytometry with a FACSCalibur (Becton Dickinson).
In vitro drug resistance
After transfection, in vitro drug resistance for daunorubicin (Cerubidine; Sanofi Aventis), cytosine arabinoside (Cytosar; Pharmacia), cladribine (2-chlorodeoxyadenosine; Leustatin), and etoposide (Pharmachemie) and as a control also vincristine (Pharmachemie), L-asparaginase (Paronal; Nycomed Christiaens), prednisolone (Bufa Pharmaceutical Products), and dexamethasone (Erasmus MC) was determined by use of the 2-, 3-, and 4-day 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assays as described previously.21 Six concentrations of each drug were tested in duplicate. The ranges of the final concentrations of these drugs were as follows: daunorubicin, 0.002-2.0 μg/mL; cytosine arabinoside, 0.01-10 μg/mL; cladribine, 0.0004-4 μg/mL; etoposide, 0.05-50 μg/mL; vincristine, 0.05-50 μg/mL; L-asparaginase, 0.003-10 IU/mL; prednisolone, 0.008-250 μg/mL; and dexamethasone, 0.0002-6 μg/mL.
Statistical analyses
Statistical workup of GEP data are described in “Gene expression profiling.” For comparison of the gene expression in different groups, the Mann-Whitney U test was used. For assessment of correlation of the results from gene expression profiling and RT-qPCR, Spearman Correlation coefficient was used. All MLL-rearranged de novo AML cases with available follow-up data were included for survival analysis. Probabilities of overall survival, EFS (events: nonremitter, relapse, secondary malignancy, death from any cause), and cumulative incidence of relapse (events: nonremitter, relapse) were estimated by the method of Kaplan and Meier and compared by use of the log-rank test. Median IGSF4 expression in the t(9;11) group was used to split all MLL cases in high and low IGSF4 expression. The Cox proportional hazards model analysis was applied to determine the association of IGSF4 overexpression with overall survival and EFS adjusted for prognostic factors as described for pediatric AML (white blood cell count, age, and karyotype). All analyses were performed with SPSS Statistics Version 16.0 (SPSS Inc). All used tests were 2-tailed, and a P value of less than .05 was considered significant.
Results
Gene expression profiling
In a comparison of t(9;11)(MLL-AF9) AML with other MLL-rearranged AML cases, IGSF4 was among the highly differentially expressed genes. Recent literature described IGSF4 as a tumor suppressor gene in solid tumors, but so far no data are available about the function of IGSF4 in leukemia.22-24 Because this gene was never described to be expressed in pediatric AML, we decided to further study IGSF4 specifically.
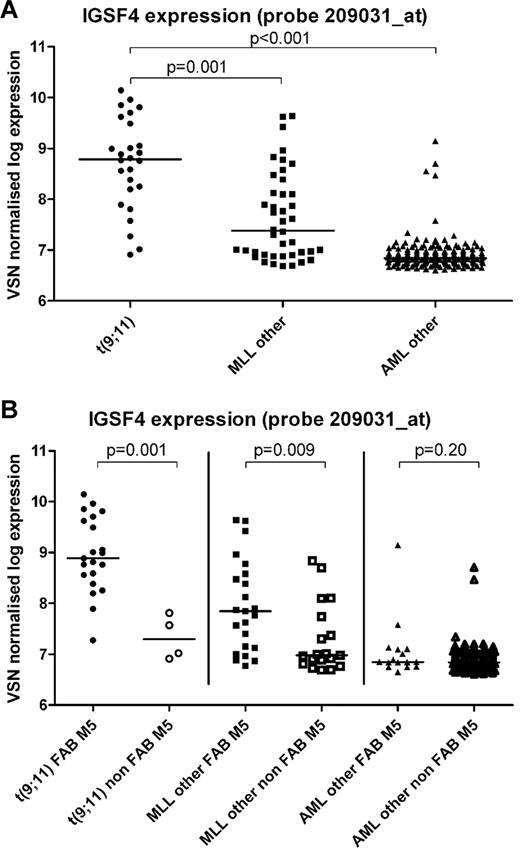
We chose probe set 209031_at, which revealed the most significant differences, to compare differential expression in AML subgroups. Patients with t(9;11), n = 26, had a 4.1-fold greater median IGSF4 mRNA expression compared with patients with other MLL-rearrangements (n = 42; 8.8 arbitrary units [AU] vs 7.4 AU, P < .001; Figure 1A). IGSF4 expression was also significantly greater in t(9;11) compared with non-MLL–rearranged AML cases with other karyotypes (n = 192; fold change 7.0, median expression of 8.8 vs 6.8 AU, P < .001; Figure 1A). Within the t(9;11) group, expression of IGSF4 was 4.9-fold greater in FAB-M5 (n = 21) versus other FAB-types (n = 4; median: 8.9 AU vs 7.3 AU, P = .001; Figure 1B). This difference, associated with FAB-classification, was also present in MLL-rearranged AML patients with other translocation partners (n = 23 vs n = 19; median 7.8 vs 7.0, fold change 2.4, P = .009), but not in the AML-other group (n = 16 vs n = 166; median 6.8 vs 6.8, P = .20; Figure 1B). All cases with unknown FAB-type were excluded from these analyses (t(9:11) n = 1, AML-other n = 10).
IGSF4 gene expression in pediatric AML as determined by gene expression arrays. Graphs showing the expression of probe set 209031_at, representing the IGSF4 gene, after log transformation. Bars represent the median expression in each group. (A) Significant differences are shown between patients with a t(9;11) (n = 26) and patients with another MLL rearrangement (MLL other, n = 42; 8.8 vs 7.4, P < .001) or AML patients without an MLL rearrangement (AML other, n = 192; 8.8 vs 6.8, P < .001). (B) Expression of probe set 209031_at with all groups divided on the basis of morphology, ie, FAB M5 versus other FAB-types (non-FAB M5). All cases with unknown FAB type were excluded from this analysis (t(9;11) n = 1, AML-other n = 10). We detected a significant difference for median expression within the patients with a t(9;11) (n = 21 vs n = 4; 8.9 vs 7.3, P = .001) and the patients with other MLL rearrangements (n = 23 vs n = 19; 7.8 vs 7.0, P = .009). This difference was not detected in the remaining patients without an MLL rearrangement (AML other, n = 16 vs n = 166; 6.8 vs 6.8, P = .20).
IGSF4 gene expression in pediatric AML as determined by gene expression arrays. Graphs showing the expression of probe set 209031_at, representing the IGSF4 gene, after log transformation. Bars represent the median expression in each group. (A) Significant differences are shown between patients with a t(9;11) (n = 26) and patients with another MLL rearrangement (MLL other, n = 42; 8.8 vs 7.4, P < .001) or AML patients without an MLL rearrangement (AML other, n = 192; 8.8 vs 6.8, P < .001). (B) Expression of probe set 209031_at with all groups divided on the basis of morphology, ie, FAB M5 versus other FAB-types (non-FAB M5). All cases with unknown FAB type were excluded from this analysis (t(9;11) n = 1, AML-other n = 10). We detected a significant difference for median expression within the patients with a t(9;11) (n = 21 vs n = 4; 8.9 vs 7.3, P = .001) and the patients with other MLL rearrangements (n = 23 vs n = 19; 7.8 vs 7.0, P = .009). This difference was not detected in the remaining patients without an MLL rearrangement (AML other, n = 16 vs n = 166; 6.8 vs 6.8, P = .20).
RT-qPCR
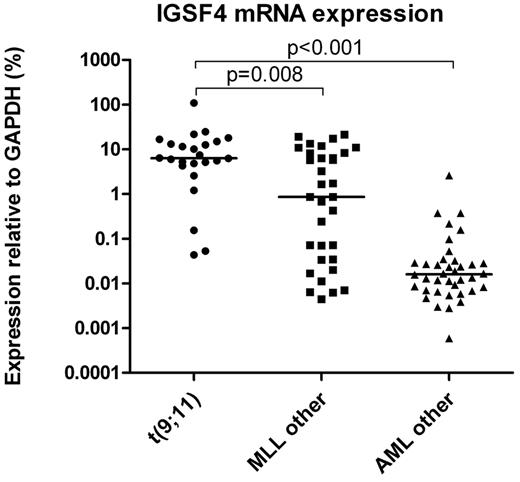
Gene expression results were confirmed with RT-qPCR in 78 cases. An additional 17 cases of which no GEP data were available were used to expand the number of cases. The median relative expression of IGSF4 in patients with t(9;11) was 7.4-fold greater compared with MLL-rearranged patients with another translocation (6.4% vs 0.9%, P = .008; Figure 2). Relative mRNA expression of IGSF4 in other AML patients was 396-fold lower than in t(9;11) patients (0.02% vs 6.4%, P < .001).
IGSF4 gene expression in pediatric AML as determined by RT-qPCR. Graph showing the expression of IGSF4 on mRNA level measured with RT-qPCR. Bars represent the median expression in each group. Significant differences are observed between patients with a t(9;11) (n = 24) and patients with another MLL-rearrangement (MLL other, n = 33; (6.4 vs 0.9, P = .008) or AML patients without a MLL rearrangement (AML other, n = 38; 6.4 vs 0.02, P < .001).
IGSF4 gene expression in pediatric AML as determined by RT-qPCR. Graph showing the expression of IGSF4 on mRNA level measured with RT-qPCR. Bars represent the median expression in each group. Significant differences are observed between patients with a t(9;11) (n = 24) and patients with another MLL-rearrangement (MLL other, n = 33; (6.4 vs 0.9, P = .008) or AML patients without a MLL rearrangement (AML other, n = 38; 6.4 vs 0.02, P < .001).
A correlation coefficient was calculated comparing the expression of IGSF4 by GEP and RT-qPCR. Because of the use of SYBRgreen in the RT-qPCR reaction, Ct values > 32 can be considered noise. The remaining 56 pairs resulted in a highly correlated Spearman R = 0.839 (P = .01; supplemental Figure 2).
Western blot
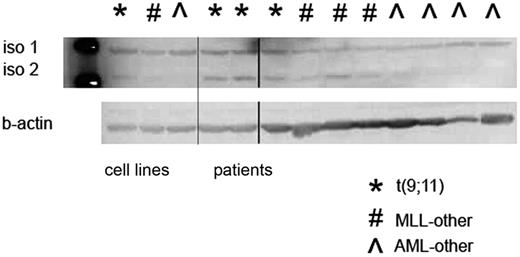
The IGSF4 antibody specifically identified 2 different isoforms that have previously been described.25 We did not find a difference in isoform 1 expression, but the expression of isoform 2 was greater in t(9;11) positive patients than in the other patients (Figure 3).
Protein expression analysis of IGSF4 with Western blot. Sections of Western blot showing data from 3 cell lines and 10 patient samples. Two described isoforms (iso 1 and iso 2) are shown at 48 kDa and 45 kDa, respectively. Bottom panel shows loading control with beta-actin. The protein expression of IGSF4 isoform 2 in patients with a MLL t(9;11) is greater than protein expression of IGSF4 isoform 2 in the other groups. The first lane shows the ladder, the other lanes contain cell lysates from cell lines and patients (separated by the thin line). At the thick line, one lane was spliced out.
Protein expression analysis of IGSF4 with Western blot. Sections of Western blot showing data from 3 cell lines and 10 patient samples. Two described isoforms (iso 1 and iso 2) are shown at 48 kDa and 45 kDa, respectively. Bottom panel shows loading control with beta-actin. The protein expression of IGSF4 isoform 2 in patients with a MLL t(9;11) is greater than protein expression of IGSF4 isoform 2 in the other groups. The first lane shows the ladder, the other lanes contain cell lysates from cell lines and patients (separated by the thin line). At the thick line, one lane was spliced out.
MS-PCR
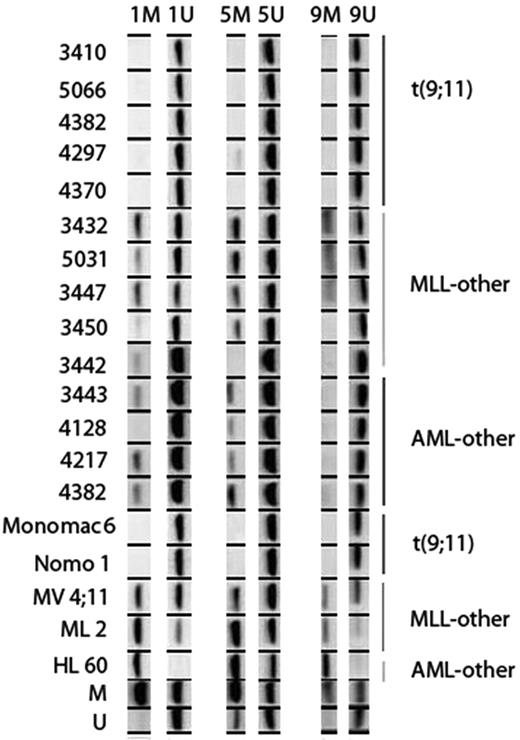
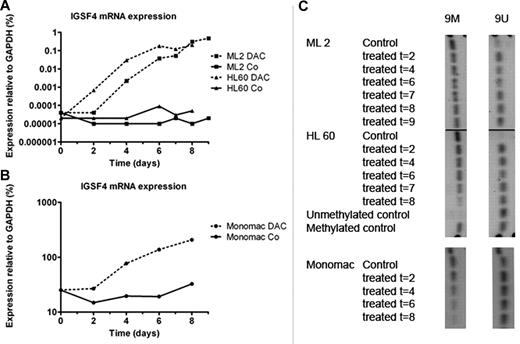
In the selected MLL-rearranged AML cases with t(9;11), the IGSF4 promoter was unmethylated. In contrast, in other MLL-rearranged cases and cases without an MLL-rearrangement the IGSF4 promoter was methylated (Figure 4). This difference between cytogenetic groups was also found in the cell lines (Figure 4).
Methylation status of IGSF4 tested by MS-PCR. Figure showing results of MS-PCR in AML patients and cell lines. Three separate regions of the promoter were investigated; region 1, 5, and 9. Each column shows results for a specific primer pair (M, methylated; U, unmethylated). The top of the figure shows the IGSF4 methylation status of several patients (indicated by number), and the bottom shows methylation status of cell lines and M and U control DNA. On the left the identity of each sample is indicated, and on the right the cytogenetic group each patient or cell line belongs to is shown. In patients with t(9;11) (n = 5), no bands are seen with the methylated-specific primers and heavy bands are seen with the unmethylated-specific primers. In contrast, the other MLL patients (n = 5) and other AML patients (n = 5) do show a band with the methylated primer. This difference is also seen in the cell lines.
Methylation status of IGSF4 tested by MS-PCR. Figure showing results of MS-PCR in AML patients and cell lines. Three separate regions of the promoter were investigated; region 1, 5, and 9. Each column shows results for a specific primer pair (M, methylated; U, unmethylated). The top of the figure shows the IGSF4 methylation status of several patients (indicated by number), and the bottom shows methylation status of cell lines and M and U control DNA. On the left the identity of each sample is indicated, and on the right the cytogenetic group each patient or cell line belongs to is shown. In patients with t(9;11) (n = 5), no bands are seen with the methylated-specific primers and heavy bands are seen with the unmethylated-specific primers. In contrast, the other MLL patients (n = 5) and other AML patients (n = 5) do show a band with the methylated primer. This difference is also seen in the cell lines.
Treatment with demethylating agent
To study the role of promoter methylation in the regulation of IGSF4 expression, AML cell lines were cultured with and without decitabine. RT-qPCR showed an increase of IGSF4 RNA expression, up to at least 1000-fold on days 8-9 for the treated hypermethylated cell lines ML-2 (t(6;11)(q27;q23)) and HL-60 (AML-other) compared with their nontreated counterparts (Figure 5A). Bisulfite-treated DNA tested on MS-PCR showed methylation status changes, most significantly with selected primers 9M and 9U (Figure 5C). The control cell line MONO-MAC-1 (t(9;11)), which has a high IGSF4 mRNA expression and moderate methylation, showed a 10-fold increase of expression during treatment with decitabine (Figure 5B). In this cell line MS-PCR showed moderate methylation at the start of treatment that was lost during treatment (Figure 5C).
relative expression and promoter methylation of IGSF4 in cell lines after culture with decitabine. Graph showing IGSF4 mRNA expression in cell lines ML-2, HL-60, and MONO-MAC-1 at different time points during culture with demethylating agent decitabine (DAC). (A) Solid lines correspond to untreated conditions of cell lines ML-2 (t(6;11)) and HL-60 (AML-other), and dotted lines reflect values from treated conditions of the same cell lines. Shown is > 1000-fold up-regulation of IGSF4 expression during culture with decitabine, whereas in control conditions expression remained stable. (B) Treatment of cell line MONO-MAC-1 with decitabine. The solid line reflects values from untreated condition, and the dotted line from corresponding treated samples. A 10-fold up-regulation was found during a treatment period of 8 days. (C) Results for MS-PCR in cell lines cultured with demethylating agent decitabine for 2-9 days. Left column, results for 9M primers; right column, for 9U primers. Under control conditions ML-2 and HL-60 mainly show a methylated promoter region. Shortly after start of treatment unmethylated bands are visible, and methylated bands decrease in intensity. MONO-MAC-1 shows both bands at the start of the experiment, and the methylated band clearly weakens during treatment.
relative expression and promoter methylation of IGSF4 in cell lines after culture with decitabine. Graph showing IGSF4 mRNA expression in cell lines ML-2, HL-60, and MONO-MAC-1 at different time points during culture with demethylating agent decitabine (DAC). (A) Solid lines correspond to untreated conditions of cell lines ML-2 (t(6;11)) and HL-60 (AML-other), and dotted lines reflect values from treated conditions of the same cell lines. Shown is > 1000-fold up-regulation of IGSF4 expression during culture with decitabine, whereas in control conditions expression remained stable. (B) Treatment of cell line MONO-MAC-1 with decitabine. The solid line reflects values from untreated condition, and the dotted line from corresponding treated samples. A 10-fold up-regulation was found during a treatment period of 8 days. (C) Results for MS-PCR in cell lines cultured with demethylating agent decitabine for 2-9 days. Left column, results for 9M primers; right column, for 9U primers. Under control conditions ML-2 and HL-60 mainly show a methylated promoter region. Shortly after start of treatment unmethylated bands are visible, and methylated bands decrease in intensity. MONO-MAC-1 shows both bands at the start of the experiment, and the methylated band clearly weakens during treatment.
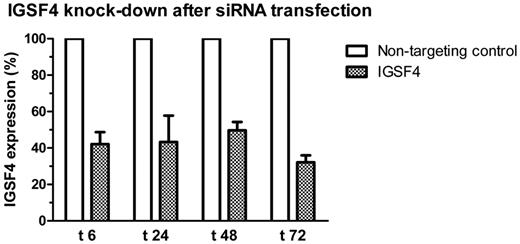
Transfection
Transfection of siRNAs targeting IGSF4 by electroporation in the cell line MONO-MAC-1 resulted in 50%-70% silencing of IGSF4 mRNA in repeated experiments (Figure 6). NOMO-1 was more difficult to transfect than MONO-MAC-1 and therefore was not used in further experiments. No significant differences were found between transfected and control conditions in MONO-MAC-1 cells, neither in apoptosis or cell-cycle arrest (supplemental Figure 3) nor in cell proliferation (data not shown).
IGSF4 knock-down in MONO-MAC-1 after transfection. The figure shows IGSF4 expression levels after transfection with IGSF4 siRNAs measured by RT-qPCR relative to levels measured in corresponding samples transfected with nontargeting siRNA. Time points are given in hours after transfection. Shown are the means of 4 experiments. Error bars represent standard error of the mean.
IGSF4 knock-down in MONO-MAC-1 after transfection. The figure shows IGSF4 expression levels after transfection with IGSF4 siRNAs measured by RT-qPCR relative to levels measured in corresponding samples transfected with nontargeting siRNA. Time points are given in hours after transfection. Shown are the means of 4 experiments. Error bars represent standard error of the mean.
In vitro drug resistance
No significant differences in drug toxicity were found after 2, 3, or 4 days of consecutive culturing of transfected MONO-MAC-1 cells with the most commonly used cytostatic drugs in AML and ALL (supplemental Figures 4-5).
Outcome
Five-year overall survival of MLL-rearranged patients with high IGSF4 expression was 70%, which is significantly better than an overall survival of 37% in MLL-rearranged patients with low IGSF4 expression (n = 79, P = .03; Figure 7A-C). This group included 28 patients with t(9;11). When analyzed separately, this group proved to be too small to show significant survival differences (data not shown). With the use of the Cox proportional hazards model, we found that no correlation with outcome could be shown after adjustment for known prognostic factors (white blood cell count, age; data not shown).
Survival plots for patients with high and low IGSF4 expression. Plots showing overall survival (pOS) (A), EFS (pEFS) (B), and cumulative incidence of relapse (CIR; C) for all MLL-rearranged patients (n = 26 [IGSF4 high] vs n = 53 [IGSF4 low]). Median expression in the t(9;11) group was chosen as a cut-off for the division in high and low IGSF4 expression. The solid line represents patients with high IGSF4 expression, the dotted line represents patients with low IGSF4 expression. NR indicates nonremitter.
Survival plots for patients with high and low IGSF4 expression. Plots showing overall survival (pOS) (A), EFS (pEFS) (B), and cumulative incidence of relapse (CIR; C) for all MLL-rearranged patients (n = 26 [IGSF4 high] vs n = 53 [IGSF4 low]). Median expression in the t(9;11) group was chosen as a cut-off for the division in high and low IGSF4 expression. The solid line represents patients with high IGSF4 expression, the dotted line represents patients with low IGSF4 expression. NR indicates nonremitter.
Discussion
In pediatric MLL-rearranged AML, t(9;11) is the most common genetic aberration. Recently we showed that prognosis of this patient group largely depends on the morphologic FAB classification, that is, patients with t(9;11) with FAB M5 had a significantly better prognosis than patients with other FAB types.10 However, so far the biologic background for this survival difference is largely unknown. To study differentially expressed genes in this specific group, we performed GEP and identified IGSF4 as a discriminative gene.
In nonmalignant cells, IGSF4 is known to play a role in cell-cell adhesion, cell polarity, and as a signaling molecule for natural killer- and T-cell cytotoxicity.23,26 Recently, Kawano et al27 showed that IGSF4 participates in the ErbB2/ErbB3 pathway as a competitive antagonist of ErbB2 in complex formation with ErbB3. Loss of IGSF4 expression resulted in AKT pathway stimulation, which resulted in improved cell movement and survival.27 We could not confirm similar pathway activation in our pediatric AML dataset by using microarray analysis (data not shown). In nonsmall cell lung carcinoma, IGSF4 was found to be located in an area with a common loss of heterozygosity. Transferring this gene in A549 cells (nonsmall cell lung carcinoma cell line) inhibited tumor formation in nude mice.22 In neuroblastoma and cervical carcinoma, aberrant promoter methylation of IGSF4 influenced tumor growth.20,24
We found a high IGSF4 mRNA expression in MLL-rearranged pediatric patients with t(9;11) that was associated with increased protein expression, in combination with a hypomethylated promoter region of IGSF4. This finding indicates that indeed epigenetic regulation plays a major role in the expression of IGSF4 in pediatric AML, as was further illustrated by cell line studies with demethylating agents. The expected effect of IGSF4 on proliferation could not be shown in the siRNA experiments in a t(9;11) cell line. Our in vitro studies, however, do not represent the normal cell environment. Future studies that use a design including environmental factors (such as homing assays) are more potent to show proliferative advantage caused by differential expression of this cell surface protein. We are not the first group to report gene silencing by methylation in pediatric AML. CCAAT/enhancer binding protein is a well-known gene that is linked to mutations as well as methylation differences and whose expression predicts survival in AML.28,29 In this context, we might consider designing clinical studies to assess whether the outcome of patients with epigenetic silencing can be improved by adding demethylating agents.
So far only the authors of one study in AML cell lines reported on IGSF4, showing hypermethylation of its promoter region in MLL-rearranged AML cell lines versus cell lines without an MLL rearrangement.30 In adult T-cell leukemia, IGSF4 overexpression resulted in a proliferation advantage.31 However, the precise role of IGSF4 in hematopoiesis and leukemogenesis is currently unknown. It remains to be determined whether cell-cell adhesion plays a role in IGSF4+ leukemia like it does in solid tumors. The finding by Boles et al26 that expression of IGSF4 protein on the cell surface is a trigger for natural killer cell– and CD8+ T cell–mediated cytotoxicity, might support our finding of overexpression of IGSF4 in a group with a more favorable outcome. Normally, circulating leukocytes do not express high levels of IGSF4. If Boles' hypothesis proves to be true in pediatric AML, we would expect blasts with high IGSF4 expression to be more easily recognized by the immune system. Blasts with low IGSF4 expression are able to evade this mechanism. Because low expression is often derived from promoter hypermethylation, these patients might benefit from demethylating agents. In conclusion, we hypothesize that silencing of IGSF4 could be considered as a secondary event, causing the leukemic blasts to be immunologically silent and thereby allowing longer survival. The ErbB-RAC-AKT pathway, influenced by IGSF4 interaction with ErbB3, could also be of interest for leukemias, because this pathway is linked to proliferation and apoptosis.27
We found that IGSF4 was mainly and most apparently expressed in monoblastic t(9;11)-rearranged patients. Because this subgroup of MLL-rearranged pediatric AML has recently been identified as important prognostic group,10 the role of IGSF4 deserves further attention. Interestingly, the IGSF4 expression also seems to be determined by the cell type (M5) in which the maturation arrest occurs. This reflects a unique novel collaboration of a specific (epi-)genetic aberration and type II mutations (ie, MLL) together with maturation status in pediatric AML.
In this retrospective study, which included AML samples from differently treated pediatric patients, we show there was no significant difference for EFS between cases with high and low IGSF4 expression. However, there was a significant difference in overall survival favoring patients with high IGSF4 expression as the result of a better salvage rate after relapse in these patients. Further studies in larger prospective cohorts will be necessary to determine the full role of IGSF4 in pediatric AML.
In conclusion, we found IGSF4 mRNA and protein to be differentially expressed in MLL-rearranged pediatric monoblastic AML patients with the greatest expression in t(9;11) M5 AML. This expression seems to be largely regulated by promoter hypermethylation. Further studies are needed to be able to determine the biologic role and prognostic relevance of IGSF4 expression in pediatric AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Pediatric Oncology Foundation Rotterdam (KOCR) has funded part of the work of E.A.C. and B.V.B. The work of B.V.B. was also funded by the NWO (Netherlands Organization for Scientific Research). The work of J.E.K. and A.A.D.-v.O. was supported by a grant from the Foundation KiKa, Amstelveen, The Netherlands. R.M. has been partially funded by grant 107819 from Deutsche Krebshilfe.
Authorship
Contribution: J.E.K., E.A.C., B.V.V., and A.A.D.-v.O. designed the study and performed the research; J.S., A.B., V.d.H., E.S.J.M.d.B., D.R., G.J.L.K., and J.C. contributed materials and clinical data; J.E.K., E.A.C., B.V.B., M.L.d.B., R.P., C.M.Z., and M.M.v.d.H.-E. analyzed data and wrote the paper; R.M. and C.M. performed LDI-PCR to identify the MLL-rearrangements; and R.P., C.M.Z., and M.M.v.d.H.-E. supervised the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marry M. van den Heuvel-Eibrink, MD, PhD, Associate Professor in Pediatric Oncology/Hematology, Erasmus MC/Sophia Children's Hospital, Department of Pediatric Oncology/Hematology, Room Sp2568, Dr Molewaterplein 60, PO Box 2060, 3000 CB Rotterdam, The Netherlands; e-mail: m.vandenheuvel@erasmusmc.nl.
References
Author notes
J.E.K. and E.A.C. contributed equally to this work.

![Figure 7. Survival plots for patients with high and low IGSF4 expression. Plots showing overall survival (pOS) (A), EFS (pEFS) (B), and cumulative incidence of relapse (CIR; C) for all MLL-rearranged patients (n = 26 [IGSF4 high] vs n = 53 [IGSF4 low]). Median expression in the t(9;11) group was chosen as a cut-off for the division in high and low IGSF4 expression. The solid line represents patients with high IGSF4 expression, the dotted line represents patients with low IGSF4 expression. NR indicates nonremitter.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/117/3/10.1182_blood-2010-05-286138/4/m_zh89991064670007.jpeg?Expires=1767741324&Signature=iOEwE-f7fdK23C9mjuvAk1De~DeCKzr4Ov4BKg8xThknKtUf8AX81EGimXZsNqwynV3qH27nx4-vqDmZ0UASpez4BbjIygxHWWSp-~KL~6VzG9jKOAmVVPkPe0OwIZm4jC5s1NM-sUDxK1izs8Rg51eSpE461wfsi~pya50HzDOuSVK1hdrrORW1ZJfwxreReQepXPaCi-dYPhSijcfKjRH6Hj0XRHz6-9~NZXl9koAtR1SHJ8UgzK6Ss4mi-GMmq1QlV8MvIE4Z33NprDg510kDxk4J5IqaK~ao1n9Di7EeakXt23JUfeINa-EfbkgZrTvq816XLtIKvw21QXmvkA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)