Abstract
Vascular endothelial growth factors (VEGFs) and their tyrosine kinase receptors (VEGFR-1-3) are central mediators of angiogenesis and lymphangiogenesis. VEGFR-3 ligands VEGF-C and VEGF-D are produced as precursor proteins with long N- and C-terminal propeptides and show enhanced VEGFR-2 and VEGFR-3 binding on proteolytic removal of the propeptides. Two different proteolytic cleavage sites have been reported in the VEGF-D N-terminus. We report here the crystal structure of the human VEGF-D Cys117Ala mutant at 2.9 Å resolution. Comparison of the VEGF-D and VEGF-C structures shows similar extended N-terminal helices, conserved overall folds, and VEGFR-2 interacting residues. Consistent with this, the affinity and the thermodynamic parameters for VEGFR-2 binding are very similar. In comparison with VEGF-C structures, however, the VEGF-D N-terminal helix was extended by 2 more turns because of a better resolution. Both receptor binding and functional assays of N-terminally truncated VEGF-D polypeptides indicated that the residues between the reported proteolytic cleavage sites are important for VEGF-D binding and activation of VEGFR-3, but not of VEGFR-2. Thus, we define here a VEGFR-2–specific form of VEGF-D that is angiogenic but not lymphangiogenic. These results provide important new insights into VEGF-D structure and function.
Introduction
Vascular endothelial growth factor D (VEGF-D) is one of the 5 mammalian members of the VEGF family (VEGF-A, VEGF-B, VEGF-C, VEGF-D, and placenta growth factor). VEGF-D binds to and induces dimerization and tyrosine autophosphorylation of its endothelial cell-specific receptors VEGFR-2 and VEGFR-3.1 VEGFR-2 signals stimulate endothelial sprouting, proliferation, and survival, as well as vascular permeability, and VEGFR-3 signals stimulate similar processes in lymphatic endothelial cells.2,3 Whereas VEGF-A and VEGF-C are indispensable for embryonic vascular development, VEGF-D can be deleted without any obvious phenotype.4-9 However, recombinant VEGF-D is capable of inducing angiogenesis and lymphangiogenesis in several experimental conditions, suggesting that it is of potential therapeutic utility in regenerative medicine.10-12
The VEGF family ligands are antiparallel homodimers characterized by 8 conserved cysteine residues forming a cystine knot structure.13,14 The newly synthesized VEGF-D and VEGF-C have long N- and C-terminal propeptides flanking the VEGF homology domain (VHD).15 Proteolytic processing by furins cleaves between the VHD and the C-terminal propeptide, activating the VEGFR-3 binding activity and subsequent cleavage by extracellular serine proteases, including plasmin,16 produces the mature human VEGF-D (residues 89-205) that binds also VEGFR-2 (major form).17 Cleavage at a secondary N-terminal site results in an alternative, N-terminally shorter form comprising residues 100-205 (minor form).
VEGF-C and VEGF-D exist predominantly as noncovalently linked homodimers,15,17 although they both have the conserved cysteine residues that form the interchain disulfide bridges in the other VEGFs. Both have also an additional cysteine residue close to the interchain disulfide residues at the dimer interface as seen in the human VEGF-C crystal structure.18,19 This additional cysteine residue may interfere with the interchain bonding, explaining why its replacement with a small hydrophobic residue, including alanine, increased dimer stability and enhanced the activity of both VEGF-C and VEGF-D in cell culture, as well as the biologic activity of VEGF-C in vivo.11,19 However, the Cys137Ala mutation in VEGF-C did not affect VEGFR-3 or VEGFR-2 binding affinity,11,18 suggesting its effects were mediated by increased half-life of the active protein.
The extracellular parts of VEGFR-2 and VEGFR-3 share the same overall structure of 7 immunoglobulin-like domains. Structural and functional studies have yielded insights into how the distinct domains contribute to VEGFR activity. The VEGFR-2 ligand binding has been mapped to domains 2 and 3 (D23) by using deletion mutants20,21 and by determining the crystal structure of VEGF-C receptor complexes.18 VEGFR-2 D2 is the major ligand-binding domain, but D3 contributes important interactions for VEGF-C binding. In addition, in a recent electron microscopic study, VEGF-A binding to the VEGFR-2 D23 was shown to induce receptor dimerization with additional homotypic interactions between the membrane-proximal domains.22 In contrast to VEGFR-1 and VEGFR-2, VEGFR-3 ligand (VEGF-C) binding is D1 dependent, and the minimal construct needed for VEGF-C binding contains domains D1 and D2.21
We have solved the crystal structure of human VEGF-D, which showed a covalent homodimer with an extended N-terminal helix, unique to VEGF-C and VEGF-D. Interesting new data were obtained about the effect of the alternative N-terminal proteolytic cleavages on VEGFR-2 and VEGFR-3 binding and activation.
Methods
Protein expression and purification
Drosophila S2 expression constructs for the N- and C-terminal variants of VEGF-D were obtained by cloning polymerase chain reaction–amplified fragments of human VEGF-D cDNA (forward primers: 5′-CGGATCCATTTGCGGCAACTTTCTATGAC-3′ for variant D89-195, 5′-CGGATCCAACTTTCTATGACATTGAAACACT-3′ for variant D92-195, and 5′-CGGATCCAAAAGTTATAGATGAAGAATGGCAA-3′ for variants D100-195 and D100-205; reverse primers: 5′-TGAATTCAATGATGATGATGGTGATGGGCTGTTGGCAAGCACTTAC-3′ for variants D89-195, D92-195, and D100-195 and 5′-CATCTAGATCAATGATGATGATGGTGGTGTCTTCTGATAATTGAGTAAGGATGG-3′ for variant D100-205; template human VEGF-D cDNA containing the Cys117Ala mutation) as BamHI/blunt-end fragments into a BglII/EcoRV-opened modified pMTBiP-V5His-C vector (Invitrogen).23 The construct for the expression of wild-type VEGF-D was used by Achen et al1 : a polymerase chain reaction fragment (primers 5′-GTCAAGCTTAATGATGATGATGGTGATGGGGGGCTGTTGGC-3′ and 5′-GAGGATCCGTCAGCATCC-3′, template: wild-type human VEGF-D cDNA) was cloned into a pFASTBAC1 vector (Invitrogen) that had been modified to contain the mellitin signal peptide and multiple cloning site from pVT-Bac.1,24 Viral stocks were generated, and the protein was expressed in High 5 cells. Stably transfected S2 cell pools were prepared according to the instructions of the supplier (Invitrogen).
For protein expression, the S2 cells were adapted to suspension culture at 27°C and induced at a density of 2-4 × 106 cells/mL for 5 days with 0.5mM CuSO4. The conditioned medium was harvested by centrifugation, and the VEGF-D variants were extracted by Ni2+-charged chelating sepharose (GE Healthcare) in batch. The resin was washed in phosphate-buffered saline containing 15mM imidazole, and the proteins were eluted with 400mM imidazole. Finally, the VEGF-D variants were purified by gel filtration on a Superdex 200 (GE Healthcare) column in HEPES balanced solution (HBS; 10mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid], 0.1M NaCl) at pH 7.5. VEGF-C was expressed and purified similarly.21 Soluble, Fc-tagged (human immunoglobulin G) VEGFR-2 domains 2 + 3 (R2D23; residues 120-326), VEGFR-3 domain 1 (R3D1, residues 1-133), VEGFR-3 domain 2 (R3D2, residues 132-229), VEGFR-3 domains 1 + 2 (R3D12, residues 1-229), VEGFR-3 domains 2 + 3 (R3D23, residues 132-329), VEGFR-3 domains 1-3 (R3D1-3, residues 1-329), and VEGFR-3 domains 1-7 (D17Fc, residues 1-776) were prepared as described.21 Fc-tagged VEGFR-1/VEGFR-3 chimera (R1/3D12) consists of VEGFR-1 D1 (residues1-129) and VEGFR-3 D2 (residues 134-228). All the receptor constructs were expressed in Sf21 insect cells with the use of baculovirus expression and were purified by Protein A-Sepharose (GE Healthcare) affinity step followed by gel filtration on a Superdex 200 column. A Factor Xa cleavage site allowed the proteolytic Fc-tag removal and the preparation of the monomeric VEGFR-3 D17 construct.
Reversed-phase chromatography and N-terminal sequencing
Protein separation by reversed-phase chromatography was performed on a 1 × 20mm TSKgel-250 trimethylsilane (10 μm, 250 Å) column with the use of the ETTAN liquid chromatography (GE Healthcare). Proteins were eluted with a linear gradient of acetonitrile (0%-100%) in 0.1% trifluoroacetic acid. N-terminal sequencing of the proteins collected from reversed-phase chromatography was performed with Edman degradation in a Procise 494A-HT sequencer (Applied Biosystems) on BioBrene Plus–treated glass fiber filters. N-terminal sequencing confirmed the correct VEGF-D sequences and indicated N-terminal Pro (D100-195) and Asp-Pro from the linkers.
Recombinant adenoassociated virus expression in vivo and immunohistochemistry of skeletal muscles
All mouse experiments were approved by the Provincial State Office of Southern Finland and carried out in accordance with institutional guidelines. Production of the recombinant adenoassociated viruses (rAAVs; serotype 8) and the transduction of mouse (NMRI, Balb/c) tibialis anterior muscles were done essentially as described.11 For the analysis of the Cys117Ala mutation in vivo (NMRI mice), the rAAVs tested encode the cDNAs of the major form of the mature human VEGF-D (residues 89-205; D89-205), the same VEGF-D with the Cys117Ala mutation and human serum albumin as a control. For the analysis of the N-terminal deletion in vivo (Balb/c mice), the rAAVs used encode the cDNAs of the wild-type VEGF-D D89-195 and D100-195. Two weeks after transfection, the muscles were isolated and frozen in OCT (TissueTek, Sakura Finetek). Cryosections (8 μm) were cut, acetone-fixed, and immunostained with the following antibodies: rat anti–platelet endothelial cell adhesion molecule 1 (PECAM-1; PharMingen), goat anti–Prox-1 (R&D Systems), hamster anti-podoplanin (Acris), mouse anti–smooth muscle actin (SMA)–cyanine 3 (Sigma), and rabbit anti–human lymphatic endothelial hyaluronan receptor-1. Secondary antibodies were Alexa Fluor–conjugated (Molecular Probes). The samples were analyzed with Axioplan microscope (Zeiss; objectives 10× NA = 0.3 WD 5.6 and 20× NA = 0.5 WD 2.0; camera Zeiss AxioCam HRm 14-bit grayscale CCD; acquisition software Zeiss AxioVision 4.6). Microvessel area density was quantified with ImageJ software (National Institutes of Health). Results are presented as mean values ± SD, calculated with analysis of variance for multiple comparisons.
Cell culture and the MTT assay
Porcine aortic endothelial (PAE) cells expressing VEGFR-2 or VEGFR-3 were a kind gift from Dr Lena Claesson-Welsh (University of Uppsala).25 Human dermal microvascular endothelial (HDME) cells were obtained from PromoCell. The BaF3-hVEGFR-3 and BaF3-mVEGFR-2 cell lines represent transfected derivatives of the murine pro-B cell line BaF3,1,6 which stably express a chimeric receptor containing the extracellular domain of human VEGFR-3 or VEGFR-2, respectively, fused to the transmembrane and cytoplasmic domains of the mouse erythropoietin receptor. These cells were maintained in Dulbecco minimal essential medium (DMEM) containing 10% fetal bovine serum. For maintenance, the cell cultures were supplemented with 2 ng/mL murine interleukin-3 (IL-3; Calbiochem) and 250 μg/mL Zeocin (Invitrogen). In the absence of IL-3, BaF3-VEGFR-3 cells grow only in the presence of VEGF-C or VEGF-D. The BaF3/VEGFR cell survival was quantified with the use of the mitochondrial MTT (3-(4,5-dimethyl thiazolyl-2)-2,5-diphenyltetrazolium bromide) substrate, resulting in a color development.11 The survival of VEGFR-2/BaF3 cells (VEGF-A, VEGF-C, or VEGF-D) was measured after incubation at 37°C for 48 hours in the presence of the human VEGF-D variants ≤ 500 ng/mL.
Antibodies
Immunoprecipitation assays
For the comparison of wild-type and Cys117Ala mutant forms of VEGF-D (residues 89-205) under reducing and nonreducing conditions, the constructs were transfected into 293T cells in DMEM supplemented with 10% fetal calf serum. Twenty-four hours later, the cell culture medium was replaced by DMEM 0.2% bovine serum albumin, and 48 hours later the cell culture medium was harvested, cleared by centrifugation, and immunoprecipitated with the VD1 antibody. The immunoprecipitates were boiled for 5 minutes with nonreducing sample buffer. After taking aliquots, β-mercaptoethanol was added to a final concentration of 2.5%, and samples were boiled again for 5 minutes. Nonreducing and reducing samples were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by Western blotting with the same antibody.
For the VEGFR-2 and VEGFR-3 phosphorylation assays, the cells were lysed in 1 mL of PLCLB lysis buffer (150mM NaCl, 5% glycerol, 1% Triton X-100, 1.5M MgCl2, 50mM HEPES, pH 7.5) supplemented with 1mM vanadate, 2mM phenylmethylsulphonyl fluoride, 2 μg/mL leupeptin, and 0.07 U/mL aprotinin. Cleared lysates were incubated with 2 μg of primary antibody for 2 hours. Subsequently, the immunocomplexes were captured with the use of protein G-sepharose and washed 3 times in the PLCLB buffer, and the proteins were separated by SDS-PAGE under reducing conditions and visualized by Western blotting.
Affinity measurements
Isothermal calorimetric titrations of the binding of human VEGF-D (Cys117Ala) variants to the soluble, Fc-tagged VEGFR-2 domains 2 + 3 (D23), VEGFR-3 deletion mutants (domains 1-3; R3D1–R3D1-3), VEGFR-1-D1/VEGFR-3-D2 chimera (R1/3D12) and to the untagged VEGFR-3 domains 1-7 (D1-7) were carried out at 25°C with the use of an ultrasensitive isothermal titration calorimeter (ITC; MicroCal). To control for heat dilution effects, all the protein buffers were adjusted to HBS at pH 7.5. The receptor constructs were used in the calorimeter cell at a concentration of 5-8μM, and the VEGF-D ligands in the syringe at a concentration of 0.10-0.20mM. After the ITC titrations, the samples were visually analyzed for aggregation. Data were processed with the MicroCal Origin 7.0 software.
Pull-down assay
Purified, Fc-tagged VEGFR-2 (D23), VEGFR-3 deletion mutants (R3D1-R3D1-3), and the VEGFR-1-D1/VEGFR-3-D2 chimera (R1/3D12) were incubated with VEGF-D (Cys117Ala) D92-195 in molar excess (50 μg + 20 μg, respectively) at room temperature for 1 hour. Protein complexes were precipitated with the use of Protein A-Sepharose (GE Healthcare), and the beads were collected with Ultrafree (Millipore) centrifugal filter units. After a wash with phosphate-buffered saline, the protein complexes were eluted with 0.2M glycine, pH 3.0, and analyzed by SDS-PAGE under reducing conditions. The gel was stained with Coomassie Blue.
Crystallization and structure determination
For crystallization, VEGF-D (Cys117Ala) D92-195 was concentrated to 2 mg/mL and the buffer (HBS) was supplemented with 0.1% (vol/vol) P8340 protease inhibitor cocktail (Sigma) and 0.01% (wt/vol) NaN3. Crystallization conditions were screened with the sitting-drop vapor-diffusion technique. A single VEGF-D crystal grew in 6 weeks at room temperature over a reservoir solution of 0.1M phosphate/citrate buffer at pH 4.2, 40% ethanol (vol/vol), and 5% polyethylene glycol 1000 (wt/vol). The hexagonal crystal belongs to spacegroup P6122 (a, b = 95.72 Å and c = 70.94 Å) with half of the covalent dimer per asymmetric unit and solvent content of 50%. For data collection, the crystal was frozen in liquid nitrogen in a 1 + 1 mixture of Paratone-N and mineral oils (Hampton Research).
A complete dataset to 2.9 Å resolution was collected from the single crystal at the beamline X06SA at the Swiss Light Source in Villigen, Switzerland (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Data were processed with XDS and the CCP4 suite of programs.28,29 The VEGF-D structure was solved by molecular replacement by Molrep, using a single VEGF-C chain (2X1W) as a search model.30 The phases were further improved by solvent flattening, and the VEGF-D model was completed by iterative refinement in Phenix and model-building in Coot.28,31,32 A subset of 10% of the diffraction data were omitted from refinement for calculating the free R factor (Rfree). The final VEGF-D model comprises residues 92-194, an N-terminal proline from cloning, 13 solvent molecules, and 2 glycan chains. The glycan chains, linked to Asn155 and Asn185, consist of 2 N-acetyl-glucosamines and 3 or one mannose moieties, respectively. Stereochemical properties were assessed by Molprobity.33 Figures were prepared using the program PyMol (http://pymol.sourceforge.net).
Results
Establishment of the angiogenic activity of human VEGF-D (Cys117Ala) in vivo
We have recently shown that the Cys117Ala mutation of human VEGF-D gives rise to improved VEGFR-2 and VEGFR-3 activation in cultured cells,19 presumably as a consequence of higher dimer stability of the protein. In vitro, the Cys117Ala mutant exists mainly as a covalent dimer, whereas the wild-type protein migrates as a monomer on a reducing gel (supplemental Figure 1). We confirmed here that this Cys117Ala mutant retains biologic activity in vivo. The wild-type and Cys117Ala mutants (major form consisting of residues 89-205) were fused with the sequence encoding the mouse IL-3 signal peptide and expressed by the rAAV vector in mouse tibialis anterior muscles. Analysis by immunohistochemical staining of blood vessel endothelial cells (PECAM-1) and perivascular smooth muscle cells (SMA) indicated that the angiogenic activity was retained in this mutant VEGF-D that we subjected to further structural analysis (supplemental Figure 2).
VEGF-D structure determination
The human VEGF-D (Cys117Ala) VHD, residues 92-195, fused to a C-terminal histidine tag was crystallized for structure determination. N-terminal sequence was confirmed for the recombinant VEGF-D expressed in Drosophila S2 cells. The structure was determined by x-ray crystallography at 2.9 Å resolution with the use of human VEGF-C18 as a search model in molecular replacement. The model was built into the electron density maps using Coot and was refined with Phenix to an R-factor of 25.4% and an Rfree of 33.3% (supplemental Table 1).31,32
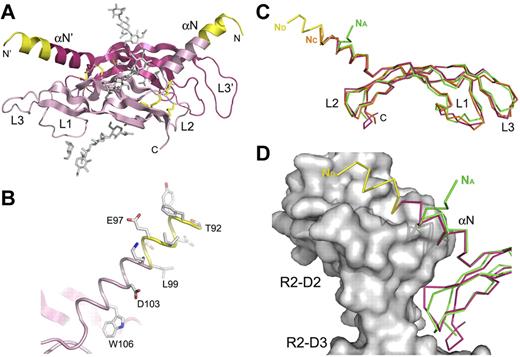
The asymmetric unit contains one VEGF-D monomer, and the VEGF-D covalent dimer (Figure 1A) is generated by a crystallographic 2-fold axis in the hexagonal spacegroup. The human VEGF-D residues are numbered according to the full-length protein. The final model contains residues 92-194, an N-terminal proline from the linker, 2 N-linked glycan chains (Figure 1A), and 13 solvent molecules. Overall, the electron density is of good quality, but several side chains, including loop 1 and 3 residues 124-129 and 169-173, respectively, had poor density, and part of the side chain atoms were omitted from the refinement.
Crystal structure of human VEGF-D and its comparison to the other VEGF family ligands. (A). A cartoon representation of the crystal structure of the covalent VEGF-D (Cys117Ala mutant) homodimer in magenta and pink. The N-terminal residues between the 2 reported proteolytic cleavage sites are colored in yellow.17 The sugar moieties and the disulfide bonds are shown in gray and yellow sticks, respectively. N- and C-termini, the N-terminal helix (αN), and the connecting loops 1-3 (L1-L3) are labeled where applicable. (B). Close-up of the N-terminal helix (αN) in the same orientation as in panel A. The helix is shown in a cartoon loop representation with the same coloring as in panel A. Asp103 and Trp106, the equivalents of the VEGFR-2 binding VEGF-C residues Asp123 and Trp126,18 and the first α-helical residues Thr92-Lys100 are shown as sticks. (C) Superposition of the VEGF-D (magenta and yellow) and VEGF-A (PDB code 1FLT; green) monomer structures with VEGF-C (PDB code 2X1W; orange) in the VEGFR-2 complex structure. The Cα-traces are shown as ribbon diagrams. Labeling is as in panel A, except that the N-termini are labeled according to the VEGF coloring in the figure. (D) The VEGF-D and VEGF-A monomer structures from panel C are superimposed with VEGF-C in the VEGF-C/VEGFR-2D23 complex structure. For clarity, VEGF-C is not shown. VEGFR-2 D2 (R2-D2) and D3 (R2-D3) are shown as a molecular surface model in gray. Coloring as indicated in panel C.
Crystal structure of human VEGF-D and its comparison to the other VEGF family ligands. (A). A cartoon representation of the crystal structure of the covalent VEGF-D (Cys117Ala mutant) homodimer in magenta and pink. The N-terminal residues between the 2 reported proteolytic cleavage sites are colored in yellow.17 The sugar moieties and the disulfide bonds are shown in gray and yellow sticks, respectively. N- and C-termini, the N-terminal helix (αN), and the connecting loops 1-3 (L1-L3) are labeled where applicable. (B). Close-up of the N-terminal helix (αN) in the same orientation as in panel A. The helix is shown in a cartoon loop representation with the same coloring as in panel A. Asp103 and Trp106, the equivalents of the VEGFR-2 binding VEGF-C residues Asp123 and Trp126,18 and the first α-helical residues Thr92-Lys100 are shown as sticks. (C) Superposition of the VEGF-D (magenta and yellow) and VEGF-A (PDB code 1FLT; green) monomer structures with VEGF-C (PDB code 2X1W; orange) in the VEGFR-2 complex structure. The Cα-traces are shown as ribbon diagrams. Labeling is as in panel A, except that the N-termini are labeled according to the VEGF coloring in the figure. (D) The VEGF-D and VEGF-A monomer structures from panel C are superimposed with VEGF-C in the VEGF-C/VEGFR-2D23 complex structure. For clarity, VEGF-C is not shown. VEGFR-2 D2 (R2-D2) and D3 (R2-D3) are shown as a molecular surface model in gray. Coloring as indicated in panel C.
VEGF-D comparison with other VEGF family ligands and putative VEGFR-2 interactions
Similar to the other VEGF family ligands, human VEGF-D monomer structure consists of an antiparallel 4-stranded β-sheet, 3 connecting loops (L1-L3), and an N-terminal α-helix (αN) that folds on top of the second monomer (Figure 1A). The VEGF-D antiparallel homodimer is further stabilized with 2 intermolecular disulfide bridges between Cys136 and Cys145′. Like in VEGF-C, Ala117 (Cys117 in the native VEGF-D) is only a few angstroms from the intermolecular Cys136-Cys145 disulfide bridge, and its side chain points toward the dimer interface. The extended 18-residue N-terminal α-helix of VEGF-D starts from the very first VEGF-D residue (Thr92) in the construct and is well visible in the electron density (Figure 1B). The VEGF-D and VEGF-C monomer structures can be superimposed with a root mean square difference of 1.3 Å for 96 Cα-atoms (Figure 1C). VEGF-D and VEGF-A monomers also superimpose well with an root mean square difference of 1.1 Å for 91 Cα-atoms, but differences in the N-termini form the structural hallmarks of the 2 subfamilies (Figure 1C-D).
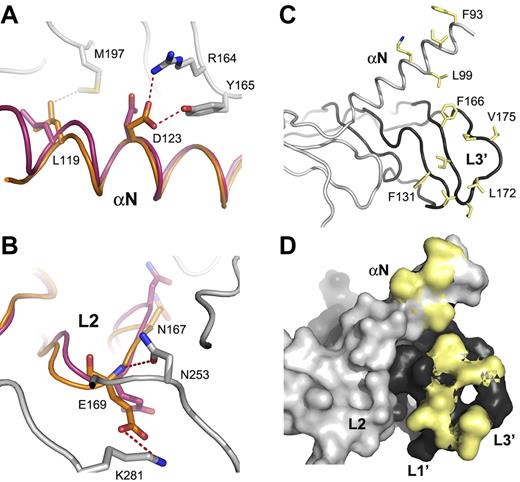
VEGF-D and VEGF-C VHD domains have 60% sequence identity, and VEGF-D retains essentially all of the VEGF-C residues involved in the VEGFR-2 interactions.18 Superposition of the VEGF-D monomer structure with a VEGF-C monomer in the VEGFR-2D23 complex structure (PDB code 2X1W) shows a highly similar loop 2 (L2) conformation (Figure 2A-B). VEGF-D Asn147 points up toward VEGFR-2 domain 2 and Glu149 down toward domain 3, like Asn167 and Glu169 in VEGF-C, and are thus capable of making the same VEGFR-2 interactions. VEGF-C Asp123 in the N-terminal α-helix, salt-bridged, and hydrogen-bonded to VEGFR-2 Arg164 and Tyr165, respectively, is also conserved in VEGF-D. VEGF-D Asp103 and Glu149 need just to adjust the side chain rotamers to accommodate the VEGFR-2 interactions visible in the VEGF-C complex. Of the VEGFR-2–interacting hydrophobic residues, VEGF-C Leu119 (Figure 2A) and Trp126 in the N-terminal helix; Phe151 in loop 1 (L1); and Phe186, Ile188, Val190, and Leu192 in loop 3 (L3) are conserved both in the VEGF-D sequence and in the structure (Figure 2C).18 These L1 and L3 residues, together with L3 Pro171, Val175, and Pro176 and the hydrophobic residues in the VEGF-D N-terminal helix comprise a large hydrophobic surface (Figure 2C-D).
VEGFR-2–interacting residues are conserved between VEGF-C and VEGF-D. (A). A close-up of the VEGF-D Leu99 and Asp103 in the VEGF-D superposition with VEGF-C in the complex with VEGFR2-D23. VEGF-D (magenta), VEGF-C (orange), and VEGFR-2 (gray) are shown as a cartoon loop representation. VEGF-C Leu119 and Asp123 interactions with VEGFR-2 are shown along with the VEGF-D counterparts Leu99 and Asp103. VEGF-C and VEGFR-2 numbering are used. Hydrophobic and hydrophilic interactions are shown in gray and red dashed line, respectively. (B) A close-up of the L2 residues as in panel A. VEGF-C Glu169 and Asn167 and its VEGF-D counterparts Glu149 and Asn147 are shown in sticks. VEGF-C Glu169 interactions with VEGFR-2 Asn253 and Lys281 are shown. (C) VEGF-D hydrophobic residues in L1 and L3 and in the N-terminal helix are shown as yellow sticks. The 2 VEGF-D monomers in the homodimer are shown in light and dark gray in a cartoon loop representation. (D) A molecular surface model of the same as in panel C. Only the side chain surface is shown for the hydrophobic (yellow) residues.
VEGFR-2–interacting residues are conserved between VEGF-C and VEGF-D. (A). A close-up of the VEGF-D Leu99 and Asp103 in the VEGF-D superposition with VEGF-C in the complex with VEGFR2-D23. VEGF-D (magenta), VEGF-C (orange), and VEGFR-2 (gray) are shown as a cartoon loop representation. VEGF-C Leu119 and Asp123 interactions with VEGFR-2 are shown along with the VEGF-D counterparts Leu99 and Asp103. VEGF-C and VEGFR-2 numbering are used. Hydrophobic and hydrophilic interactions are shown in gray and red dashed line, respectively. (B) A close-up of the L2 residues as in panel A. VEGF-C Glu169 and Asn167 and its VEGF-D counterparts Glu149 and Asn147 are shown in sticks. VEGF-C Glu169 interactions with VEGFR-2 Asn253 and Lys281 are shown. (C) VEGF-D hydrophobic residues in L1 and L3 and in the N-terminal helix are shown as yellow sticks. The 2 VEGF-D monomers in the homodimer are shown in light and dark gray in a cartoon loop representation. (D) A molecular surface model of the same as in panel C. Only the side chain surface is shown for the hydrophobic (yellow) residues.
VEGF-D N-terminal residues are important for VEGFR-3 activation
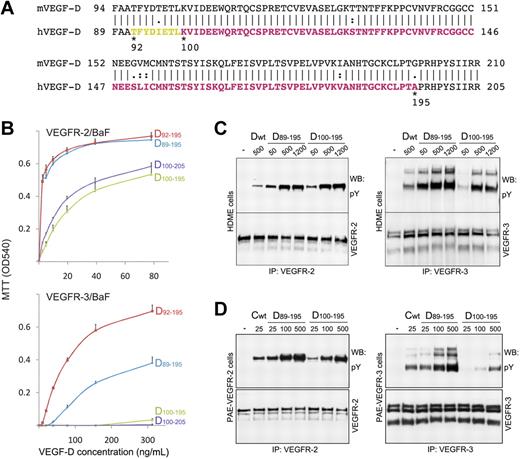
The bioactive VEGF-D short form (VHD domain) is generated on proteolytic processing at either 1 of the 2 different N-terminal proteolytic sites, corresponding to N-terminal residues 89 and 100.17 To better understand the effect of these sites on VEGF-D binding to and activation of VEGFR-2 and VEGFR-3, we generated a set of VEGF-D Cys117Ala variants with deletions at both ends (Figure 3A). The C-terminal site for deletion was selected according to the VHD domain boundary, and the additional N-terminal site, at residue 92, was designed to optimize the crystallization properties.
Characterization of the VEGF-D (Cys117Ala) N- and C-terminal variants. (A) Alignment of the human (h) and mouse (m) VEGF-D sequences. The amino acid residue differences are indicated. The N-terminal residues 89, 92, and 100 and the C-terminal residues 195 and 205 of the deletion variants of human VEGF-D are labeled. The residues visible in the crystal structure are colored in yellow and magenta. The residues colored in yellow are between the 2 proteolytic sites.17 (B) VEGFR-2/BaF and VEGFR-3/BaF cell survival induced with the VEGF-D variants. The variants are labeled according to the residue numbering. (C) Comparison of the wt VEGF-D short form (Dwt, residues 89-205 without the Cys117Ala mutation), VEGF-D D89-195 and D100-195 variant induced VEGFR-2 and VEGFR-3 phosphorylation in HDME cells. The VEGF-D concentrations (ng/mL) are indicated above the lanes. (D) Comparison of the wt VEGF-C short form (Cwt, residues 112-215),23 VEGF-D D89-195 and D100-195 variant induced VEGFR-2 and VEGFR-3 phosphorylation in PAE–VEGFR-2 and PAE–VEGFR-3 cells, respectively. The concentrations (ng/mL) of growth factors are indicated above the lanes.
Characterization of the VEGF-D (Cys117Ala) N- and C-terminal variants. (A) Alignment of the human (h) and mouse (m) VEGF-D sequences. The amino acid residue differences are indicated. The N-terminal residues 89, 92, and 100 and the C-terminal residues 195 and 205 of the deletion variants of human VEGF-D are labeled. The residues visible in the crystal structure are colored in yellow and magenta. The residues colored in yellow are between the 2 proteolytic sites.17 (B) VEGFR-2/BaF and VEGFR-3/BaF cell survival induced with the VEGF-D variants. The variants are labeled according to the residue numbering. (C) Comparison of the wt VEGF-D short form (Dwt, residues 89-205 without the Cys117Ala mutation), VEGF-D D89-195 and D100-195 variant induced VEGFR-2 and VEGFR-3 phosphorylation in HDME cells. The VEGF-D concentrations (ng/mL) are indicated above the lanes. (D) Comparison of the wt VEGF-C short form (Cwt, residues 112-215),23 VEGF-D D89-195 and D100-195 variant induced VEGFR-2 and VEGFR-3 phosphorylation in PAE–VEGFR-2 and PAE–VEGFR-3 cells, respectively. The concentrations (ng/mL) of growth factors are indicated above the lanes.
The binding behavior and activity of the VEGF-D variants toward VEGFR-2 and VEGFR-3 were assessed by ligand-dependent BaF3 cell proliferation (Figure 3B), VEGFR-2 and VEGFR-3 phosphorylation (Figure 3C), and receptor binding assays. All of the tested VEGF-D Cys117Ala variants induced strong BaF3/VEGFR-2 cell proliferation (Figure 3B). The variants D89-195 and D92-195 induced also strong BaF3/VEGFR-3 cell proliferation, whereas variants D100-195 and D100-205 did not. To confirm these results from the BaF3/VEGFR assays, we determined receptor activation in HDME cells and in VEGFR-2 or VEGFR-3 expressing PAE cells with the use of receptor immunoprecipitation followed by anti-phosphotyrosine immunoblotting. We tested 3 concentrations for the VEGF-D variants D89-195 and D100-195 in receptor phosphorylation assays in the HDME cells (Figure 3C). In line with the results of the BaF3/VEGFR assays, both of the variants induced strong VEGFR-2 phosphorylation, whereas the N-terminally truncated D100-195 failed to activate VEGFR-3 at the lowest concentration. Unlike the BaF3/VEGFR-3 cells, the HDME cells express both VEGFR-2 and VEGFR-3; thus, the ligand may, similarly to VEGF-C, induce also receptor heterodimerization, leading to cross-phosphorylation of VEGFR-2 and VEGFR-3 heterodimers.34,35 When phosphorylation was tested in PAE cells expressing either VEGFR-2 or VEGFR-3, both variants showed comparable VEGFR-2 autophosphorylation, but the D100-195 variant induced weak VEGFR-3 activation only at the highest concentration (Figure 3D).
VEGF-D with N-terminal deletion induces angiogenesis but not lymphangiogenesis in vivo
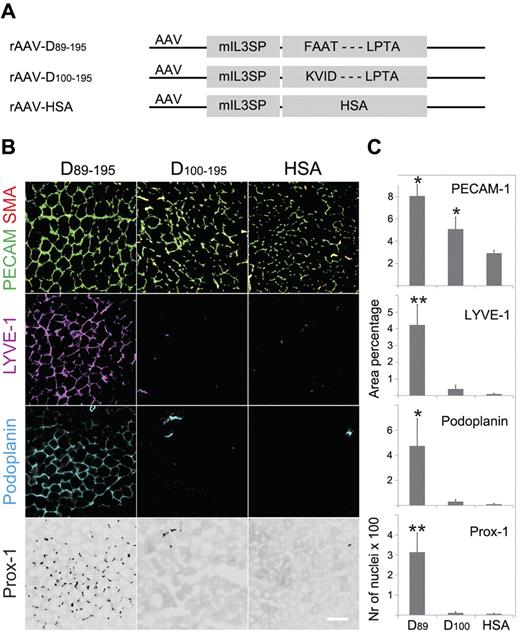
The biologic activity of the wild-type VEGF-D variants D89-195 and D100-195 was analyzed in mouse tibialis anterior muscles by rAAV delivery (Figure 4A). Two weeks after the injection of the rAAV vectors, the muscles were processed for immunohistochemical staining of PECAM-1 (endothelial cells) and SMA (smooth muscle cells and pericytes) and for the lymphatic endothelial cell markers lymphatic endothelial hyaluronan receptor-1, Prox-1, and podoplanin (Figure 4B-C). In comparison to rAAV-encoded human serum albumin, both of the VEGF-D variants induced angiogenesis, and VEGF-D D89-195 induced also lymphangiogenesis. Interestingly, VEGF-D D100-195 with the N-terminal deletion did not induce detectable lymphangiogenesis under these conditions, suggesting the lack of VEGFR-3 interactions in vivo. VD1 antibody staining was used to confirm the expression of both VEGF-D variants (supplemental Figure 4).27 Notably, part of the immunostaining of the major form D89-195 colocalized with podoplanin staining, suggesting VEGF-D binding to VEGFR-3 on lymphatic endothelium.
In vivo activity of the major and minor forms of wild-type mature VEGF-D. Tibialis anterior muscles of Balb/c mice were injected with rAAVs encoding the indicated cDNAs (D89-195, the N-terminal major form of the mature human VEGF-D, residues 89-195; D100-195, the N-terminal minor form of the mature human VEGF-D, residues 100-195; and HSA, human serum albumin, as a control) and analyzed 2 weeks later by immunohistochemistry of frozen sections. (A) Schematic representation of the rAAV vectors. (B) Representative images of the staining. (C) Quantification of stained area from ≥ 5 randomly chosen view fields (D89, D89-195;D100, D100-195). PECAM-1, SMA, and the lymphangiogenic antibodies lymphatic endothelial hyaluronan receptor-1 (LYVE-1), Prox-1, and podoplanin were used for immunostaining. *P < .05, **P < .01.
In vivo activity of the major and minor forms of wild-type mature VEGF-D. Tibialis anterior muscles of Balb/c mice were injected with rAAVs encoding the indicated cDNAs (D89-195, the N-terminal major form of the mature human VEGF-D, residues 89-195; D100-195, the N-terminal minor form of the mature human VEGF-D, residues 100-195; and HSA, human serum albumin, as a control) and analyzed 2 weeks later by immunohistochemistry of frozen sections. (A) Schematic representation of the rAAV vectors. (B) Representative images of the staining. (C) Quantification of stained area from ≥ 5 randomly chosen view fields (D89, D89-195;D100, D100-195). PECAM-1, SMA, and the lymphangiogenic antibodies lymphatic endothelial hyaluronan receptor-1 (LYVE-1), Prox-1, and podoplanin were used for immunostaining. *P < .05, **P < .01.
The N-terminal residues of VEGF-D account for high-affinity VEGFR-3 binding
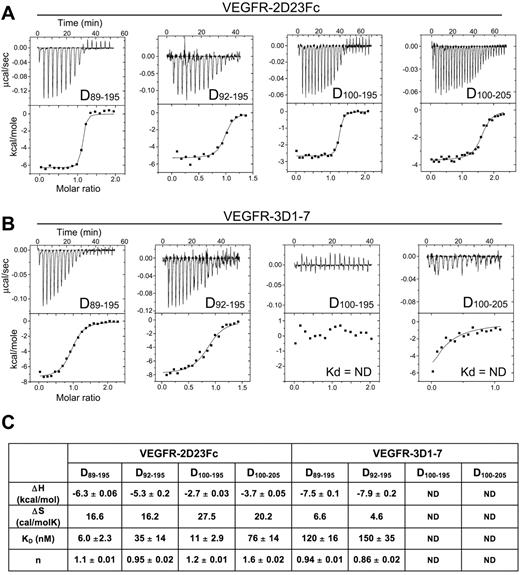
To further characterize the VEGF-D variants, we measured the binding affinity of the VEGF-D ligands for VEGFR-2D23 (VEGFR-2 domains 2 + 3; Figure 5A) and VEGFR-3D17 (VEGFR-3 domains 1-7; Figure 5B) using ITC. Consistent with the BaF cell assays (Figure 3), the binding data indicated that all of the tested VEGF-D variants are high-affinity ligands for VEGFR-2 (Figure 5C). The D89-195 and D92-195 variants with the long N-terminal helix were also VEGFR-3 ligands, whereas the D100-195 and D100-205 variants with N-terminal truncations showed no binding. The D89-195 and D92-195 variants showed almost identical thermodynamic parameters and affinities (Figure 5C), suggesting that the residues 92-99, visible in the D92-195 crystal structure, rather than the 3 residue difference (FAA, residues 89-91) are important for VEGFR-3 binding. The VEGFR-2 and VEGFR-3 binding isoterms indicated that binding of the dimer-stabilized VEGF-D is both enthalpically and entropically favorable and suggested a 2:2 ligand/receptor stoichiometry.
Thermodynamic analysis of VEGF-D interactions with VEGFR-2 and VEGFR-3. (A) Calorimetric titrations of the 4 VEGF-D (Cys117Ala) variants (D89-195, D92-195, D100-195, and D100-205) to the Fc-tagged VEGFR-2D23. (B) Titration of the 4 VEGF-D variants with VEGFR-3D17. (C) Summary of the enthalpy change (ΔH ± SD), entropy change (ΔS), binding affinities (Kd ± SD), and stoichiometry (n) of the ITC binding experiments. ND indicates not determinable.
Thermodynamic analysis of VEGF-D interactions with VEGFR-2 and VEGFR-3. (A) Calorimetric titrations of the 4 VEGF-D (Cys117Ala) variants (D89-195, D92-195, D100-195, and D100-205) to the Fc-tagged VEGFR-2D23. (B) Titration of the 4 VEGF-D variants with VEGFR-3D17. (C) Summary of the enthalpy change (ΔH ± SD), entropy change (ΔS), binding affinities (Kd ± SD), and stoichiometry (n) of the ITC binding experiments. ND indicates not determinable.
VEGFR-3 domain 1 is important for VEGF-D binding
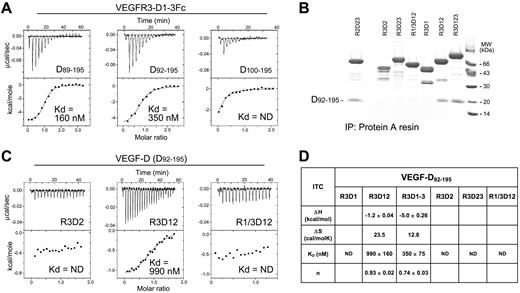
The VEGF-D binding site in VEGFR-3 was mapped by measuring the binding affinities of VEGF-D (Cys117Ala) for a set of VEGFR-3 domains and their combinations. VEGFR-3 domains 1-3 comprise the high-affinity binding site for the major form (Figure 6A). In a pull-down assay, only the VEGFR-3 domain combinations D1 and D2 (R3D12), and D1 to D3 (R3D1-3) were able to interact with the D92-195 variant (Figure 6B). Similarly, in the calorimetric titrations, only R3D12 of the single or the 2-domain VEGFR-3 constructs showed binding (Figure 6C-D). However, R3D12 affinity for D92-195 was decreased in comparison to R3D1-3. VEGFR-1/VEGFR-3 chimera (R1/3D12) showed no binding, confirming the importance of VEGFR-3 D1 for VEGF-D binding.
Characterization of the VEGF-D binding domains in VEGFR-3. (A) Thermodynamic analysis of the Fc-tagged VEGFR-3D1-3 (domains 1-3) interactions with the VEGF-D (Cys117Ala) variants D89-195, D92-195, and D100-195. The binding affinities (Kd) are indicated. ND indicates not determinable. (B) VEGF-D D92-195 complexation with soluble, Fc-tagged VEGFR-2 domains 2 and 3 (R2D23), VEGFR-3 deletion mutants (R3D1-R3D123), and VEGFR-1-D1/VEGFR-3-D2 (R1/3D12) chimera. SDS-PAGE analysis of protein A pull-down assays is shown with Coomassie Blue staining. (C) Representative thermodynamic titrations of the VEGFR-3 deletion mutants (R3D2 and R3D12) and the VEGFR-1/VEGFR-3 (R1/3D12) chimera with VEGF-D D92-195. (D) Summary of the VEGF-D D92-195 binding experiments with Fc-tagged VEGFR-3 deletion mutants and the VEGFR-1/VEGFR-3 (R1/3D12) chimera. The enthalpy change (ΔH ± SD), entropy change (ΔS), binding affinity (Kd ± SD), and stoichiometry (n) of the thermodynamic binding experiments are shown. ND indicates not determinable.
Characterization of the VEGF-D binding domains in VEGFR-3. (A) Thermodynamic analysis of the Fc-tagged VEGFR-3D1-3 (domains 1-3) interactions with the VEGF-D (Cys117Ala) variants D89-195, D92-195, and D100-195. The binding affinities (Kd) are indicated. ND indicates not determinable. (B) VEGF-D D92-195 complexation with soluble, Fc-tagged VEGFR-2 domains 2 and 3 (R2D23), VEGFR-3 deletion mutants (R3D1-R3D123), and VEGFR-1-D1/VEGFR-3-D2 (R1/3D12) chimera. SDS-PAGE analysis of protein A pull-down assays is shown with Coomassie Blue staining. (C) Representative thermodynamic titrations of the VEGFR-3 deletion mutants (R3D2 and R3D12) and the VEGFR-1/VEGFR-3 (R1/3D12) chimera with VEGF-D D92-195. (D) Summary of the VEGF-D D92-195 binding experiments with Fc-tagged VEGFR-3 deletion mutants and the VEGFR-1/VEGFR-3 (R1/3D12) chimera. The enthalpy change (ΔH ± SD), entropy change (ΔS), binding affinity (Kd ± SD), and stoichiometry (n) of the thermodynamic binding experiments are shown. ND indicates not determinable.
Discussion
The determination of the VEGF-D crystal structure completes the structural studies of the human VEGF family ligands and allows conclusions about their structural diversity. This analysis shows that VEGF-C and VEGF-D comprise a functional subfamily of VEGFR-3 ligands with long N- and C-terminal propeptides requiring proteolytic processing to produce mature forms that bind VEGFR-2 and VEGFR-3 with high affinity.15,17 All 5 VEGFs contain an antiparallel β-sheet with 3 connecting loops (L1-L3) and an N-terminal α-helix that form 2 equal receptor binding surfaces.14 The VEGF-D structure showed an extended N-terminal α-helix for the first 18 residues, whereas in the other members, such as in VEGF-A (Figure 1C-D; PDB code 1FLT), the N-terminal α-helix is short with preceding residues folding away from the receptor binding surface. In the VEGF-C structures the N-terminal residues are only partially visible, but the nature of an extended α-helix is clear (Figure 1C).18
In comparison to the other VEGF family members, VEGF-C and VEGF-D have an additional cysteine residue, and the crystal structures of both of these factors as covalent dimers were achieved only with the extra cysteine mutated to alanine. In comparison to wild-type VEGF-D, Toivanen et al19 reported the Cys117Ala mutant as a more potent VEGFR-2 and VEGFR-3 activator in vitro, and similar findings were published for VEGF-C by Anisimov et al.11 They also reported a rapid loss of VEGF-D activity in vitro connected with a conversion of covalent dimers to noncovalent forms. Consistent with our crystal structure, Toivanen et al19 suggested that the Cys117 is close to the inter-domain disulfide bridge and could regulate VEGF-D dimer stability and consequently the biologic activity in response to the redox environment. We show here that the wild-type VEGF-D and the Cys117Ala mutant induce similar amounts of angiogenesis in mouse skeletal muscle without a statistically significant difference (supplemental Figure 2). Compared with the in vitro results, this probably reflects different redox environments and the fact that the rAAV-infected myofibers provide continuous expression and supply of the virus-transduced protein. It is also of interest here that the functions of some secreted proteins and cell-surface receptors are controlled by cleavage of their disulfide bonds (reviewed by Hogg36 ). Although the mechanism and function of such cleavage in VEGF-C and VEGF-D are not yet known, both have a neighboring free thiol involved.11,19
Comparison of the human VEGF-D structure to human VEGF-C in the VEGFR-2D23 complex shows that the VEGFR-2 interacting residues, in particular the hydrophilic Asp123 and Glu169, in VEGF-C are structurally conserved in VEGF-D (Asp103 and Glu149) and seem to require only minor changes in the side chain conformations for VEGFR-2 binding. VEGF-D L1 and L3 bear multiple hydrophobic residues and comprise a large hydrophobic surface extending to the N-terminal helix. These residues include equivalents of the VEGF-C L1 and L3 hydrophobic residues shown to be important for VEGFR-2 binding and residues found at the VEGFR-2 interface in the VEGF-C complex structure.18,21 The deletion of the first N-terminal residues of VEGF-D (Phe89-Leu99; D100-195 and D100-205 variants) had essentially no effect in the VEGFR-2 activity or VEGFR-2D23 affinity assays, suggesting that these residues do not contribute to VEGFR-2 binding. This is consistent with the VEGF-C/VEGFR-2 complex structure (PDB code 2X1W) in which the VEGF-C N-terminal residues extending to the Glu97 residue of VEGF-D and the neighboring VEGFR-2 D2 residues 128-131 were disordered, indicating a lack of interactions. The structural comparison of VEGF-D and VEGF-C together with the very similar thermodynamic parameters for VEGFR-2 binding, including the affinity (Kd) and the changes in enthalpy (ΔH) and entropy (ΔS) (Figure 5)18 suggest that the 2 ligands share the structural determinants of VEGFR-2 specificity shown in the analysis of the VEGF-C/VEGFR-2 complex.
In VEGF-D, the N-terminal but not the C-terminal deletions affected VEGFR-3 binding and activity. D100-195 and D100-205 variants were both poor inducers of BaF/VEGFR-3 cell proliferation, and D100-195 induced weaker VEGFR-3 phosphorylation in HDME cells than D89-195. However, both were equally active in induction of VEGFR-2 phosphorylation in these cells. VEGF-C induced VEGFR-2/VEGFR-3 heterodimerization and cross-activation has been reported in endothelial cells.34,35 Possible VEGFR-2 and VEGFR-3 heterodimerization may explain the relatively high D100-195 activity toward VEGFR-3 in the HDME cells. In contrast, in VEGFR-3–expressing PAE cells, the D89-195 variant showed high VEGF-C–like activity already at the lowest concentration, whereas the D100-195 variant was active only at the highest concentration. D100-195 and D100-205 showed no binding in the ITC binding assays, suggesting that the Kd is higher than the receptor concentration (5-8μM) in the calorimeter cell, which represents the detection limit of the calorimetric titrations. Extension of the C-terminal residues (Pro196-Arg205) in the D100-205 protein did not rescue VEGFR-3 activity, suggesting that the N-terminal residues of VEGF-D forming the α-helix shown by the crystal structure are crucial for high-affinity binding and VEGFR-3 activity.
We show here that the rAAV-delivered major and minor forms of human VEGF-D induce angiogenesis in skeletal muscle. Consistent with our in vitro VEGFR-3 activation and binding assays, the major form also induced lymphangiogenesis at the 2-week time point, whereas the minor form (residues 100-195) with the N-terminal deletion did not (Figure 4). Thus, the minor form of VEGF-D may be considered as a VEGFR-2–specific angiogenic growth factor. The N-terminal truncation results from proteolytic cleavage of VEGF-D in the human HEK293 cells.17 This, and our data presented here, suggest that differential proteolytic processing of the VEGF-D N-terminus may provide additional control over VEGF-D activity toward VEGFR-2 and VEGFR-3.
In general, growth factors and their receptors display cross-species functionality, but mouse VEGF-D and mouse VEGFR-2 are an exception. Mouse VEGF-D does not bind to or activate mouse VEGFR-2 in vitro, although it binds to human VEGFR-2 and human VEGF-D binds mouse VEGFR-2. In our recent in vivo study, however, the mature mouse VEGF-D (rAAV-encoded residues 94-210) did induce angiogenesis in mouse skeletal muscle.11,37 With the VEGFR-2D2318 and our VEGF-D structures, it is now possible to analyze the sequence differences in detail. We superimposed the VEGF-D structure on the VEGF-C structure in the VEGFR-2 complex and mapped the differences in human and mouse sequences in these structures (supplemental Figure 3A). VEGFR-2 sequence differences were scattered with most amino acid changes residing in D3 and outside of the ligand binding site, suggesting that amino acid changes in the mouse VEGFR-2 residues are not responsible for the inability of mouse VEGF-D to bind the mouse VEGFR-2 (supplemental Figure 3B). Most of the differences in the VEGF-D sequences are located in the binding surface, including the Ile96 and the L2 triplet Ser150-Leu151-Ile152 counterparts (Figure 3A). Consistent with the mutagenesis data,37 we propose that these residues in mouse VEGF-D are responsible for its inability to bind mouse VEGFR-2. However, the reported angiogenic activity in mouse skeletal muscle suggests that the role of mouse VEGFR-2 in the in vivo activity of mouse VEGF-D should be further analyzed. In this regard, the poor stability of the covalent VEGF-D dimers should also be considered (supplemental Figure 1).11,19
VEGFR-3 ligand binding uses multiple domains and is domain D1 dependent. Unlike the ligand binding in VEGFR-1 and VEGFR-2, VEGFR-3 domain D2 and even domains D23 need the presence of domain D1 for VEGF-C binding.18,21,38-40 Similarly, we show here that also VEGF-D binding to VEGFR-3 is D1 dependent (Figure 6). The combination of VEGFR-3 domains D1 and D2 is the minimal VEGF-D binding construct, and the presence of D3 increases the affinity for VEGF-D. All the 3 first VEGFR-3 domains are thus involved, and VEGFR-3 ligand binding is probably centered on D2 after the general scheme of type III and V receptor tyrosine kinases.18,41 The extended VEGF-D N-terminal α-helix, pointing upward, presumably toward D1, could interact with this domain or with the D1/D2 junction. Importantly, the VEGF-D structure reported here represents a truncated form of the originally described mature form lacking 3 N-terminal residues (FAA). Thus, our data suggest an important role for the N-terminal residues for VEGFR-3 binding, and the proposed proximity with D1 in the ligand-receptor complex suggests that the VEGF-D N-terminal residues could also interact with D1 or with the D1/D2 junction. In general, the extended N-terminal α-helix of VEGF-D comprises part of the VEGFR binding surface, and our data clearly show that the N-terminal residues are crucial for VEGFR-3 binding and activation. Additional structural studies, in particular on the VEGFR-3 complexes, are needed to fully understand the mechanism of VEGFR-3 ligand binding and activation.
The coordinates and structure factors have been deposited in the Protein Data Bank (PDB ID code 2XV7).
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Andrea Prota (Paul Scherrer Institut, Villigen, Switzerland) and Dr Anuschka Pauluhn (Swiss Light Source, Villigen, Switzerland) for help with the data collection.
This work was supported by the Sigrid Juselius Foundation, the Finnish Cancer Research Organizations, the Louis-Jeantet Foundation, and the Swiss National Science Foundation (grant 31003A-112455). The x-ray data collection was performed on the X06SA (PXI) beamline at the Swiss Light Source, Paul Scherrer Institut, Villigen, Switzerland.
Authorship
Contribution: V.-M.L., M.J. and K. Alitalo designed research and wrote the manuscript; V.-M.L., M.J., A.A., D.T., K. Aho, and N.K. performed research and analyzed data; P.T. and S.Y.-H. analyzed data and edited the manuscript; and K.B.-H. contributed analytical tools and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for K. Aho is Department of Biosciences, University of Helsinki, Helsinki, Finland.
Correspondence: Kari Alitalo, Molecular/Cancer Biology Laboratory, Biomedicum Helsinki, POB 63, University of Helsinki, FI-00014, Helsinki, Finland; e-mail: kari.alitalo@helsinki.fi
References
Author notes
V.-M.L. and M.J. contributed equally to this study.