Abstract
B-cell receptor (BCR) signaling has been inferred as an important mechanism for disease progression in chronic lymphocytic leukemia (CLL) and other B-cell malignancies. In response to BCR activation, CLL cells secrete the chemokine CCL3, which fosters interactions between CLL cells and the leukemia microenvironment. CCL3 secretion correlates with expression of the 70-kDa ζ-associated protein (ZAP-70) and responsiveness of the CLL clone to BCR stimulation. Here, we measured CCL3 plasma levels by enzyme-linked immunosorbent assay (ELISA) in 351 CLL patients and examined CCL3 levels for associations with established prognostic markers and time from diagnosis to initial therapy. We found that CCL3 plasma concentrations were strongly associated with established prognostic markers. In a Cox proportional hazards regression model, CCL3 as well as established prognostic markers (immunoglobulin heavy chain variable-region mutation status, CD38 or ZAP-70 cytogenetics, clinical stage) were significantly associated with time to treatment. Multivariable analysis revealed that CCL3 (hazard ratio [HR] = 2.33, P < .0001), advanced clinical stage (HR = 2.75, P = .0025), poor risk cytogenetics (del 17p, HR = 2.38; del11q, HR = 2.36, P = .001), and CD38 expression (HR = 1.43, P = .023) were independent prognostic markers. Collectively, CCL3 is a novel, robust, and independent prognostic marker in CLL that can easily and reliably be measured by ELISA. CCL3 therefore should become useful for risk assessment in patients with CLL.
Introduction
Chronic lymphocytic leukemia (CLL), the most common leukemia in adults, is a lymphoproliferative disorder with a highly variable clinical course. CLL is characterized by the clonal expansion of mature, antigen-stimulated CD5+/CD23+ B lymphocytes in blood, secondary lymphoid tissues, and the bone marrow.1 The clinical staging systems developed by Rai et al2 and Binet et al3 remain the standard methods for risk assessment in CLL, but they do not allow predictions about the risk of disease progression in early-stage disease patients, which is the majority of patients. A sizable number of studies investigated prognostic markers, which can be helpful for predicting the individual risk at an early stage of the disease. The most accepted and widely used prognostic markers in CLL are the mutation status of the immunoglobulin variable gene segments (IgVH),4,5 the expression of CD384 and the ζ-associated protein of 70-kD (ZAP-70),6,7 as well as cytogenetic risk groups.8
B-cell antigen receptor (BCR) signaling is increasingly recognized as a key factor promoting clonal expansion in various B-cell malignancies, such as diffuse large B-cell lymphoma (DLBCL)9 and CLL.1 Two CLL risk factors (IgVH mutation status and ZAP-70) have functional links to the BCR. The mutation status of the IgVH segments of the BCR distinguishes “mutated” from “unmutated” CLL, with a low or high risk for disease progression, respectively, each accounting for approximately 50% of the patients. This association between the amount of somatic mutations in the BCR and prognosis suggests that antigen stimulation plays a role in disease progression.1,10 ZAP-70 is predominantly expressed in “unmutated” CLL cases,11 and ZAP-70 expression is associated with enhanced BCR signaling.12 Further evidence that BCR-derived signals play a critical role in the pathogenesis and prognosis of CLL comes from the notion that CLL patients express restricted sets of BCRs, as determined by BCR sequencing. These BCRs have immunoglobulin (Ig) heavy chain variable (V) gene sequences that are identical or stereotyped in subsets of patients,13,14 suggesting that these BCRs bind similar antigens that are relevant to the pathogenesis of CLL. Moreover, cells from poor-prognosis CLL patients with unmutated IgVH genes display gene expression profiles that indicate activation downstream of the BCR.11
CLL cells proliferate in the tissues, rather than in the peripheral blood; and in these tissue compartments (marrow, lymphatic tissues), CLL cells interact with accessory cells, collectively referred to as the CLL “microenvironment.”15 CLL cell growth in these tissues accounts for a daily turnover of approximately 0.1% to 1% of the clone, as demonstrated by heavy water incorporation in CLL patients in vivo.16 Areas of CLL proliferation are called proliferation centers or pseudofollicles, a hallmark finding in CLL histopathology, also referred to as the proliferative compartment of this disease. In these areas, CLL cells engage with multiple accessory cells, such as T cells17 and CD68+ nurselike cells (NLCs),18,19 and display signs of BCR activation,20 suggesting that CLL proliferation is BCR- and T cell-driven.
The rationale for studying CCL3 and CCL4 plasma levels in CLL is based on our recent studies in which we analyzed the impact of the microenvironment on CLL gene expression in vitro.21 We found that CLL cells up-regulate and secrete CCL3 and CCL4 in response to BCR stimulation and in coculture with NLCs,21 a model system resembling the lymphatic tissue microenvironment.15,18 This BCR- and NLC-dependent induction of CCL3 and CCL4 was sensitive to inhibition of BCR signaling, using a spleen tyrosine kinase inhibitor.21,22
CCL3 and CCL4, previously called macrophage inflammatory protein-1α (MIP-1α) and MIP-1β are chemokines of the CC subfamily and inducible in a number of hematopoietic cells, particularly in those involved in adaptive immune responses (macrophages, dendritic cells, and B and T lymphocytes). CCL3 signals through the chemokine receptors CCR1 and CCR5, whereas CCL4 signals only through CCR5. CCL3 and CCL4 are chemoattractants for monocytes and lymphocytes.23 Previous in vitro and in vivo studies highlighted CCL3 as a key response gene up-regulated in normal and neoplastic B cells in response to BCR signaling,21,24,25 and repressed by Bcl-6.26
Previous studies from our group and other investigators demonstrated CCL3 and CCL4 overexpression by activated CLL cells21,27,28 and elevated CCL3 and CCL4 plasma levels in CLL patients.21,29 Therefore, we hypothesized that CCL3 and CCL4 plasma levels may function as surrogate markers for BCR-dependent CLL cell activation in vivo. To explore this hypothesis, we tested plasma samples from CLL patients for CCL3 and CCL4 concentrations and investigated the relationship of CCL3 and CCL4 levels with established prognostic factors and clinical outcome.
Methods
Patient selection and clinical characteristics
Informed consent was obtained under M. D. Anderson Cancer Center Institutional Review Board–approved protocols and in accordance with the Declaration of Helsinki, and then peripheral blood samples were collected from 351 consecutive patients fulfilling diagnostic and immunophenotypic criteria for CLL at the Leukemia Department, M. D. Anderson Cancer Center, Houston, TX. Samples used for the first dataset were obtained from patients during their first visit at our center. A validation set of plasma samples was obtained from patients before frontline chemoimmunotherapy with fludarabine, cyclophosphamide, and rituximab (FCR regimen, n = 53) at M. D. Anderson Cancer Center. Peripheral blood samples were drawn using BD Vacutainer K2-ethylenediaminetetraacetic acid tubes (BD Biosciences), and plasma was collected after centrifugation and stored at −80°C. Clinical data were collected using Clinic Station Version 3.4.4, an electronic medical record system at M. D. Anderson Cancer Center, and the CLL Research Consortium database. The following patient characteristics were collected and analyzed: clinical stage according to Rai et al,2 routine laboratory data (absolute lymphocyte count, β2-microglobulin), fluorescence in situ hybridization cytogenetics according to the hierarchical model,8 IgVH mutation status (unmutated: ≥ 98% homology; mutated < 98% homology to the germline sequence), CD38 expression (negative: < 30%; positive ≥ 30%), and ZAP-70 expression by flow cytometry (negative: < 20%; positive ≥ 20%7 ). The clinical characteristics are summarized in Tables 1 and 2.
Clinical characteristics of 351 patients with CLL: categorical variables
| Characteristic . | Category . | n (frequency, %) . |
|---|---|---|
| Sex | Female | 136 (38.7) |
| Male | 215 (61.3) | |
| RAI stage | 0 | 54 (16.3) |
| I | 138 (41.6) | |
| II | 73 (22) | |
| III | 26 (7.8) | |
| IV | 41 (12.3) | |
| Cytogenetics | del 13q | 135 (41.4) |
| Diploid | 72 (22.1) | |
| Trisomy 12q | 36 (11.0) | |
| del 11q | 45 (13.8) | |
| Dead | 11 (3.1) | |
| IgVH | Mutated | 142 (52.4) |
| Unmutated | 129 (47.6) | |
| CD38 | Negative | 252 (75) |
| Positive | 84 (25) | |
| ZAP-70 | Negative | 153 (53.5) |
| Positive | 133 (46.5) |
| Characteristic . | Category . | n (frequency, %) . |
|---|---|---|
| Sex | Female | 136 (38.7) |
| Male | 215 (61.3) | |
| RAI stage | 0 | 54 (16.3) |
| I | 138 (41.6) | |
| II | 73 (22) | |
| III | 26 (7.8) | |
| IV | 41 (12.3) | |
| Cytogenetics | del 13q | 135 (41.4) |
| Diploid | 72 (22.1) | |
| Trisomy 12q | 36 (11.0) | |
| del 11q | 45 (13.8) | |
| Dead | 11 (3.1) | |
| IgVH | Mutated | 142 (52.4) |
| Unmutated | 129 (47.6) | |
| CD38 | Negative | 252 (75) |
| Positive | 84 (25) | |
| ZAP-70 | Negative | 153 (53.5) |
| Positive | 133 (46.5) |
Clinical characteristics of 351 patients with CLL: continuous variables
| Characteristic . | Mean ± SD . | Median (range) . |
|---|---|---|
| Age, y | 60.7 ± 10.3 | 61 (30-86) |
| White blood cell count, × 109/L | 57.3 ± 57.7 | 35.3 (2.4-398.3) |
| Hemoglobin, g/dL | 13.1 ± 1.8 | 13.2 (4.6-16.7) |
| Platelets, × 109/L | 185 ± 87.9 | 172 (18-949) |
| β2-microglobulin, mg/L | 3.3 ± 1.8 | 2.9 (1.2-13) |
| LDH, IU/L | 574.7 ± 227.8 | 526 (276-2394) |
| CCL3, pg/mL | 32.4 ± 66 | 9.6 (0.1-685.7) |
| CCL4, pg/mL | 128.1 ± 243.2 | 58.3 (4.3-2229.3) |
| Characteristic . | Mean ± SD . | Median (range) . |
|---|---|---|
| Age, y | 60.7 ± 10.3 | 61 (30-86) |
| White blood cell count, × 109/L | 57.3 ± 57.7 | 35.3 (2.4-398.3) |
| Hemoglobin, g/dL | 13.1 ± 1.8 | 13.2 (4.6-16.7) |
| Platelets, × 109/L | 185 ± 87.9 | 172 (18-949) |
| β2-microglobulin, mg/L | 3.3 ± 1.8 | 2.9 (1.2-13) |
| LDH, IU/L | 574.7 ± 227.8 | 526 (276-2394) |
| CCL3, pg/mL | 32.4 ± 66 | 9.6 (0.1-685.7) |
| CCL4, pg/mL | 128.1 ± 243.2 | 58.3 (4.3-2229.3) |
LDH indicates lactate dehydrogenase.
Analysis for CCL3 and CCL4 using an enzyme-linked immunosorbent assay
CCL3 and CCL4 plasma levels were measured using Quantikine Kits, according to the manufacturer's instructions (R&D Systems). High intra-assay and interassay precision has been established by the manufacturer. The absorbance was recorded by a microplate reader (ELx808, Bio-Tek Instruments), and data collection and analysis were performed using Gen5 software Version 1.08 (Bio-Tek Instruments).
Detection of CCL3 by immunostaining
Immunostaining on formalin-fixed, paraffin-embedded sections of lymph nodes infiltrated with CLL cells was performed using an antibody against CCL3 (polyclonal goat IgG, clone BAF270 from R&D Systems) and the Histostain plus kit (Zymed/Invitrogen), according to the manufacturer's protocol. In brief, after deparaffinization and antigen retrieval by pressure cooking, blocking was performed using rabbit serum (Zymed/Invitrogen) for 10 minutes, followed by incubation with the CCL3 antibody (1:50) for 1 hour, and then an incubation with the biotinylated secondary antibody (polyclonal rabbit antigoat, 1:400; Dako Cytomation) for 20 minutes, followed by incubation with an enzyme conjugate with streptavidin and horseradish peroxidase, and finally visualization with the diaminobenzidine system. After each incubation, the slides were washed for 5 minutes in phosphate-buffered saline 3 times. Immunofluorescence double staining was performed according to standard protocols. The images were evaluated using confocal laser scanning microscopy (Leica TSC).
Statistical analysis
Continuous variables were summarized with descriptive statistics, such as mean, SD, median, and range. Categorical variables were tabulated with frequency and percentage. The Fisher exact test was used to evaluate the association between categorical variables; the Wilcoxon rank-sum test was applied to compare each of the continuous variables between 2 different categories of dichotomized factors. Spearman correlation coefficients were computed to evaluate the correlation between 2 continuous variables. The time to first treatment (TTFT) was defined as the time from diagnosis to the date of first therapy if patients had received any or censored on the last visit date, and was estimated by the method of Kaplan and Meier and assessed by the log-rank test. For multivariable analysis, we used a Cox proportional hazards model with stepwise selection. Throughout this process, clinical and laboratory variables were retained only if their Wald P values were < .05. The statistical analyses were performed with the following software packages: SAS Version 9.0 (SAS Institute) and S-plus 8 (MathSoft).
Results
Correlation between CCL3 and established prognostic markers in CLL
For analysis of correlations between CCL3 and CCL4 and clinical and prognostic markers, patients first were dichotomized into groups of high or low CCL3 and CCL4, based on the approximate median CCL3 and CCL4 values (10 pg/mL and 60 pg/mL, respectively; Table 2). We found strong associations between high plasma levels of CCL3 and poor prognostic markers, such as advanced Rai stages, unmutated IgVH status, CD38 positivity, ZAP-70 positivity, levels of β2-microglobulin ≥ 4 mg/L, and high-risk cytogenetic abnormalities (del 11q and del 17p, Table 3). Eleven patients falling into the high CCL3 group were dead by the time of analysis, whereas no deaths occurred in the low CCL3 group.
Correlation between high CCL3 levels (> 10 pg/mL) and established prognostics markers in CLL
| Characteristic . | Category . | Frequency, % . | P . |
|---|---|---|---|
| Sex | Female | 44.9 | .1886 |
| Male | 52.6 | ||
| RAI stage | 0 | 31.5 | < .0001 |
| I | 39.9 | ||
| II | 63.0 | ||
| III | 84.6 | ||
| IV | 75.6 | ||
| IgVH | Mutated | 28.9 | < .0001 |
| Unmutated | 72.9 | ||
| CD38 | Negative | 41.7 | < .0001 |
| Positive | 75.0 | ||
| ZAP-70 | Negative | 35.3 | < .0001 |
| Positive | 69.9 | ||
| β2-microglobulin | < 4 mg/L | 37.8 | < .0001 |
| ≥ 4 mg/L | 86.2 | ||
| Cytogenetics | del 13q | 34.8 | < .0001 |
| Diploid | 52.8 | ||
| Trisomy 12q | 69.4 | ||
| del 11q | 71.1 | ||
| del 17p | 71.0 | ||
| CCL4, pg/mL | High (≥ 60) | 74.3 | < .0001 |
| Low (< 60) | 26.3 |
| Characteristic . | Category . | Frequency, % . | P . |
|---|---|---|---|
| Sex | Female | 44.9 | .1886 |
| Male | 52.6 | ||
| RAI stage | 0 | 31.5 | < .0001 |
| I | 39.9 | ||
| II | 63.0 | ||
| III | 84.6 | ||
| IV | 75.6 | ||
| IgVH | Mutated | 28.9 | < .0001 |
| Unmutated | 72.9 | ||
| CD38 | Negative | 41.7 | < .0001 |
| Positive | 75.0 | ||
| ZAP-70 | Negative | 35.3 | < .0001 |
| Positive | 69.9 | ||
| β2-microglobulin | < 4 mg/L | 37.8 | < .0001 |
| ≥ 4 mg/L | 86.2 | ||
| Cytogenetics | del 13q | 34.8 | < .0001 |
| Diploid | 52.8 | ||
| Trisomy 12q | 69.4 | ||
| del 11q | 71.1 | ||
| del 17p | 71.0 | ||
| CCL4, pg/mL | High (≥ 60) | 74.3 | < .0001 |
| Low (< 60) | 26.3 |
CCL3 was significantly associated (P < .0001) with death, advanced clinical stages, IgVH mutation status, CD38, ZAL-70, β2-microglobulin, high-risk cytogenetic subgroups, and CCL4.
In addition, we noticed that high levels of CCL3 were significantly associated with high levels of CCL4 (Table 3). Consequently, CCL4 levels also correlated with these established prognostic markers, although the correlations were less robust (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). We calculated the mean CCL3 levels in different prognostic subgroups and found significantly higher mean CCL3 levels in high-risk groups (Table 4). The distribution of CCL3 levels in the different prognostic subgroups is shown in supplemental Figure 3 (and the respective data for CCL4 in supplemental Figure 4). Furthermore, we calculated correlations between CCL3, CCL4, and laboratory values at the time of plasma collection. We found positive correlations between CCL3 and CCL4, and CCL3 or CCL4 and the white blood cell counts, β2-microglobulin, and lactate dehydrogenase, and negative correlations between CCL3 or CCL4 and hemoglobin and platelet counts (supplemental Table 2). Given that CCL3 turned out to be a more robust prognostic marker than CCL4, we provide our data related to CCL3 in the manuscript, whereas CCL4-related data are given in supplemental Table 1 and supplemental Figure 1.
CCL3 levels in different prognostic subgroups
| Characteristic . | CCL3, pg/mL . |
|---|---|
| RAI stage | |
| 0/I/II | 25.7 ± 45.8* |
| III/IV | 65.7 ± 114.9 |
| Mutational status | |
| IgVH mutated | 14.4 ± 28.5* |
| IgVH unmutated | 55.8 ± 95.7 |
| CD38 | |
| Negative | 22.5 ± 44.6* |
| Positive | 55.8 ± 77.6 |
| ZAP-70 | |
| Negative | 17 ± 36.1* |
| Positive | 53.1 ± 89 |
| β2-microglobulin | |
| < 4 mg/L | 17.4 ± 28.8* |
| ≥ 4 mg/L | 78.2 ± 111.2 |
| White blood cell count | |
| < 100 000 | 28.7 ± 63.9* |
| > 100 000 | 50.1 ± 73.2 |
| Cytogenetics | |
| del 13q | 16.1 ± 30.6† |
| Diploid | 38.0 ± 80.9 |
| Trisomy 12q | 44.5 ± 111.8 |
| del 11q | 51.5 ± 64.6 |
| del 17p | 55.1 ± 75.5 |
| Characteristic . | CCL3, pg/mL . |
|---|---|
| RAI stage | |
| 0/I/II | 25.7 ± 45.8* |
| III/IV | 65.7 ± 114.9 |
| Mutational status | |
| IgVH mutated | 14.4 ± 28.5* |
| IgVH unmutated | 55.8 ± 95.7 |
| CD38 | |
| Negative | 22.5 ± 44.6* |
| Positive | 55.8 ± 77.6 |
| ZAP-70 | |
| Negative | 17 ± 36.1* |
| Positive | 53.1 ± 89 |
| β2-microglobulin | |
| < 4 mg/L | 17.4 ± 28.8* |
| ≥ 4 mg/L | 78.2 ± 111.2 |
| White blood cell count | |
| < 100 000 | 28.7 ± 63.9* |
| > 100 000 | 50.1 ± 73.2 |
| Cytogenetics | |
| del 13q | 16.1 ± 30.6† |
| Diploid | 38.0 ± 80.9 |
| Trisomy 12q | 44.5 ± 111.8 |
| del 11q | 51.5 ± 64.6 |
| del 17p | 55.1 ± 75.5 |
Mean CCL3 levels were significantly higher in patients with poor prognostic features.
P < .01.
P < .05.
Association between CCL3 and time from diagnosis to initial therapy
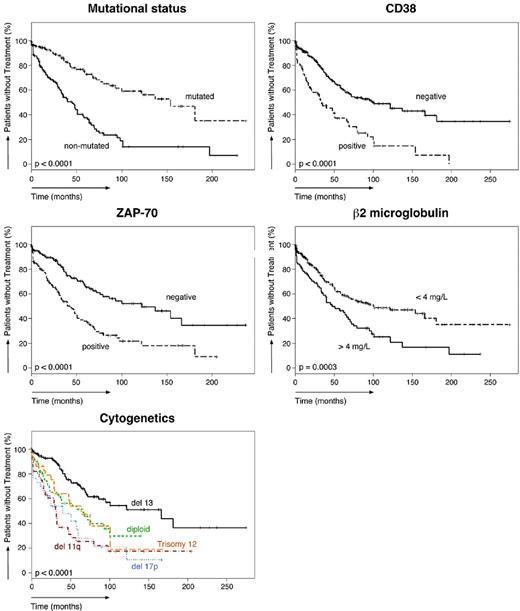
Of the 351 patients studied, 147 patients required treatment according to the National Cancer Institute Working Group criteria.30 The TTFT of the entire population was 72 months (95% confidence interval, 60-101). As shown in Table 5, the median TTFT in various subgroups differed significantly. The median TTFT in CLL patients with high levels of CCL3 was significantly shorter (40 months) than in patients with low CCL3 levels (> 250 months, P < .0001). The median TTFT was also significantly shorter in patients with established poor prognostic markers. The TTFT of patients with unmutated IgVH was 45 months, 34 months in CD38+ patients, 43 months in ZAP-70+ patients, and 47 months in patients with high β2-microglobulin levels. Figure 1A shows Kaplan-Meier curves that depict the TTFT in patients with high or low CCL3 (left panel) or CCL4 (right panel) levels. Figure 2 shows Kaplan-Meier curves for established prognostic markers studied.
Time to first treatment in the different prognostic subgroups
| Characteristic . | Category . | Total . | Patients treated . | Median TTFT, mo (95% CI) . | Treatment free at 5 y (95% CI) . | P . |
|---|---|---|---|---|---|---|
| All patients | 351 | 147 | 72 (60-101) | 0.56 (0.50-0.62) | ||
| IgVH | Mutated | 142 | 40 | 154 (100-> 250) | 0.73 (0.64-0.82) | < .0001 |
| Unmutated | 129 | 79 | 45 (33-58) | 0.37 (0.29-0.49) | ||
| CD38 | Negative | 252 | 89 | 101 (70-> 250) | 0.60 (0.53-0.68) | < .0001 |
| Positive | 84 | 55 | 34 (21-66) | 0.37 (0.27-0.51) | ||
| ZAP-70 | Negative | 153 | 51 | 122 (79-> 250) | 0.65 (0.57-0.75) | < .0001 |
| Positive | 133 | 78 | 43 (32-58) | 0.38 (0.29-0.49) | ||
| β2-microglobulin | < 4 mg/L | 262 | 92 | 101 (69-> 250) | 0.59 (0.52-0.67) | .0003 |
| ≥ 4 mg/L | 87 | 55 | 47 (36-72) | 0.46 (0.36-0.60) | ||
| CCL3 | Low | 177 | 35 | > 250 (181-> 250) | 0.75 (0.67-0.84) | < .0001 |
| High | 174 | 112 | 40 (32-59) | 0.40 (0.33-0.49) | ||
| CCL4 | Low | 180 | 49 | 181 (101-> 250) | 0.66 (0.58-0.76) | .0002 |
| High | 171 | 98 | 58 (40-72) | 0.47 (0.40-0.57) |
| Characteristic . | Category . | Total . | Patients treated . | Median TTFT, mo (95% CI) . | Treatment free at 5 y (95% CI) . | P . |
|---|---|---|---|---|---|---|
| All patients | 351 | 147 | 72 (60-101) | 0.56 (0.50-0.62) | ||
| IgVH | Mutated | 142 | 40 | 154 (100-> 250) | 0.73 (0.64-0.82) | < .0001 |
| Unmutated | 129 | 79 | 45 (33-58) | 0.37 (0.29-0.49) | ||
| CD38 | Negative | 252 | 89 | 101 (70-> 250) | 0.60 (0.53-0.68) | < .0001 |
| Positive | 84 | 55 | 34 (21-66) | 0.37 (0.27-0.51) | ||
| ZAP-70 | Negative | 153 | 51 | 122 (79-> 250) | 0.65 (0.57-0.75) | < .0001 |
| Positive | 133 | 78 | 43 (32-58) | 0.38 (0.29-0.49) | ||
| β2-microglobulin | < 4 mg/L | 262 | 92 | 101 (69-> 250) | 0.59 (0.52-0.67) | .0003 |
| ≥ 4 mg/L | 87 | 55 | 47 (36-72) | 0.46 (0.36-0.60) | ||
| CCL3 | Low | 177 | 35 | > 250 (181-> 250) | 0.75 (0.67-0.84) | < .0001 |
| High | 174 | 112 | 40 (32-59) | 0.40 (0.33-0.49) | ||
| CCL4 | Low | 180 | 49 | 181 (101-> 250) | 0.66 (0.58-0.76) | .0002 |
| High | 171 | 98 | 58 (40-72) | 0.47 (0.40-0.57) |
Relationship between CCL3, CCL4, and the time from diagnosis to initial therapy. (A) Kaplan-Meier curves show the probability of treatment-free survival according to the time since diagnosis. CLL patients are divided into groups of patients with low or high CCL3 (left-hand curve) or CCL4 (right-hand curve) plasma levels. These categories are based on CCL3 and CCL4 cutoff levels that correspond to the median CCL3 and CCL4 concentrations in our population (Table 2); the respective cutoff levels are displayed next to each of the curves. (B) Kaplan-Meier curves that display the proportion of untreated patients in 4 CLL subgroups, based on their CCL3 and CCL4 levels. Patients with high CCL3 (≥ 10 pg/mL) and low or high CCL4 show lower probabilities of treatment-free survival than patients with low levels of CCL3 (< 10 pg/mL) and high or low CCL4 levels.
Relationship between CCL3, CCL4, and the time from diagnosis to initial therapy. (A) Kaplan-Meier curves show the probability of treatment-free survival according to the time since diagnosis. CLL patients are divided into groups of patients with low or high CCL3 (left-hand curve) or CCL4 (right-hand curve) plasma levels. These categories are based on CCL3 and CCL4 cutoff levels that correspond to the median CCL3 and CCL4 concentrations in our population (Table 2); the respective cutoff levels are displayed next to each of the curves. (B) Kaplan-Meier curves that display the proportion of untreated patients in 4 CLL subgroups, based on their CCL3 and CCL4 levels. Patients with high CCL3 (≥ 10 pg/mL) and low or high CCL4 show lower probabilities of treatment-free survival than patients with low levels of CCL3 (< 10 pg/mL) and high or low CCL4 levels.
Relationship between established prognostic markers and the time from diagnosis to initial therapy. Five Kaplan-Meier plots indicate the probability of treatment-free survival in our cohort of CLL patients, based on the presence or absence of established prognostics markers (mutation status, CD38, ZAP-70, β2-microglobulin, and cytogenetics).
Relationship between established prognostic markers and the time from diagnosis to initial therapy. Five Kaplan-Meier plots indicate the probability of treatment-free survival in our cohort of CLL patients, based on the presence or absence of established prognostics markers (mutation status, CD38, ZAP-70, β2-microglobulin, and cytogenetics).
To better understand the differential impact of CCL3 versus CCL4 on TTFT, we categorized patients into 4 groups (Figure 1B): CCL3high/CCL4high, CCL3high/CCL4low, CCL3low/CCL4high, and CCL3low/CCL4low. The median TTFT among patients with high levels of CCL3 and high or low levels of CCL4 was significantly shorter (40 and 35 months, respectively) than among patients with low levels of CCL3 and high or low levels of CCL4 (> 250 and 181 months, respectively, Figure 1B). Collectively, these data suggest that CCL3, but not CCL4, has a robust impact on TTFT.
Multivariable analysis
Patient clinical characteristics, risk factors, and CCL3 and CCL4 levels were fitted into a Cox proportional hazards regression model with stepwise selection. When ZAP-70 and IgVH mutation status were fitted separately into a multivariable analysis, IgVH mutation status was selected by the process (P < .05) but not ZAP-70 (P = .07). We also included both ZAP-70 and IgVH mutation status into one multivariable model along with other clinical features (excluding CCL3 and CCL4). Again, IgVH mutation status was selected by the process. Finally, we included both ZAP-70 and IgVH mutation status along with other clinical features (including CCL3 and CCL4) into a multivariable model. Here, we found that the clinical stage, fluorescence in situ hybridization, cytogenetic abnormalities, CD38, and CCL3 were associated with time to first treatment (Table 6). Interestingly, CCL4, ZAP-70, and IgVH mutation status had no significant association with time to treatment. This model suggests that the instantaneous risk of requiring therapy in the group of patients with high levels of CCL3 is 2.33 times higher than in patients with low CCL3 levels.
Multivariable analysis of time from diagnosis to the first treatment
| Characteristic . | Category . | HR (95% CI) . | P . |
|---|---|---|---|
| RAI stage | 0 | Baseline | .0025 |
| I-IV | 2.75 (1.43-5.31) | ||
| Cytogenetics | del 13q | Baseline | .001 |
| Diploid | 1.69 (1.05-2.75) | ||
| Trisomy 12q | 1.41 (0.78-2.56) | ||
| del 11q | 2.36 (1.44-3.86) | ||
| del 17p | 2.38 (1.35-4.20) | ||
| CD38 | Negative | Baseline | .023 |
| Positive | 1.43 (1.0-2.05) | ||
| CCL3, pg/mL | Low (< 10) | Baseline | < .0001 |
| High (≥ 10) | 2.33 (1.56-3.46) |
| Characteristic . | Category . | HR (95% CI) . | P . |
|---|---|---|---|
| RAI stage | 0 | Baseline | .0025 |
| I-IV | 2.75 (1.43-5.31) | ||
| Cytogenetics | del 13q | Baseline | .001 |
| Diploid | 1.69 (1.05-2.75) | ||
| Trisomy 12q | 1.41 (0.78-2.56) | ||
| del 11q | 2.36 (1.44-3.86) | ||
| del 17p | 2.38 (1.35-4.20) | ||
| CD38 | Negative | Baseline | .023 |
| Positive | 1.43 (1.0-2.05) | ||
| CCL3, pg/mL | Low (< 10) | Baseline | < .0001 |
| High (≥ 10) | 2.33 (1.56-3.46) |
Distribution of CCL3 and CCL4 plasma levels and CCL3 serial samples
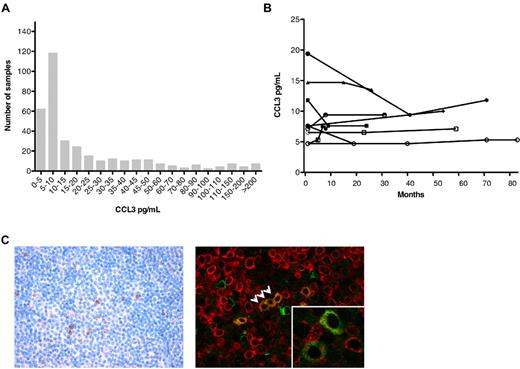
The distribution of CCL3 plasma concentrations in our cohort of 351 patients is shown in Figure 3A. A total of 177 samples (50.4%) had CCL3 levels that were below the median value of 10 pg/mL. Measurement of CLL3 levels in serial plasma samples obtained from 7 different patients at multiple time points revealed that one of 7 patients crossed that median 10-pg/mL threshold over time (Figure 3B). Similar results were found for CCL4 (supplemental Figure 2).
CCL3 plasma levels in CLL: distribution and sequential samples, and CCL3 immunohistochemistry. (A) The distribution of the plasma levels of CCL3 among 351 patients with CLL. (B) Sequential plasma levels of CCL3 detected in serial samples of CLL patients. The lines connect the symbols of individual patients representing the CCL3 concentrations (y-axis) in any one patient over time (x-axis). (C) Conventional (left) and immunofluorescence (right) immunodetection of CCL3 in CLL lymph node sections in a representative case (original magnification ×400). Images were captured with the use of an Olympus BX50 microscope and a ColorView digital camera, and processed with cellSens imaging software (all Olympus). CCL3+ cells (brown stain on the left side) tended to accumulate in areas of proliferation centers (left). Immunofluorescence double staining with CD79a (red) and CCL3 (green) revealed colocalization of CD79a+ and CCL3+ CLL cells, as indicated by the arrows. (Inset) Coexpression of CCL3 and CD79a in another case. More immunohistochemistry data are available in supplemental Figure 2.
CCL3 plasma levels in CLL: distribution and sequential samples, and CCL3 immunohistochemistry. (A) The distribution of the plasma levels of CCL3 among 351 patients with CLL. (B) Sequential plasma levels of CCL3 detected in serial samples of CLL patients. The lines connect the symbols of individual patients representing the CCL3 concentrations (y-axis) in any one patient over time (x-axis). (C) Conventional (left) and immunofluorescence (right) immunodetection of CCL3 in CLL lymph node sections in a representative case (original magnification ×400). Images were captured with the use of an Olympus BX50 microscope and a ColorView digital camera, and processed with cellSens imaging software (all Olympus). CCL3+ cells (brown stain on the left side) tended to accumulate in areas of proliferation centers (left). Immunofluorescence double staining with CD79a (red) and CCL3 (green) revealed colocalization of CD79a+ and CCL3+ CLL cells, as indicated by the arrows. (Inset) Coexpression of CCL3 and CD79a in another case. More immunohistochemistry data are available in supplemental Figure 2.
CCL3 and CCL4 levels in plasma samples from CLL patients requiring therapy
As a validation set, we analyzed CCL3 and CCL4 levels in a series of serial plasma samples obtained from patients before start of FCR frontline therapy. The purpose of this analysis is to further study the hypotheses generated in the test set (our first 351 samples) to prove the validity of our initial hypothesis, a step vital in unbiased approaches.31 If our hypotheses are correct, patients requiring therapy would have CCL3 and CCL4 plasma levels that are greater than the median CCL3 and CCL4 plasma levels and hence above the cutoff levels that distinguish low- from high-risk patients (10 pg/mL and 60 pg/mL, respectively). We found that the mean CCL3 level in patients undergoing FCR therapy was 48.2 plus or minus 5.1 pg/mL (± SEM, n = 53) and the mean CCL4 level was 280 plus or minus 50.8 pg/mL (± SEM, n = 53). As such, these levels are above the median and the cutoff values for CCL3 and CCL4, defined in the first set of samples; therefore, these data support and validate our first set of data.
Detection of CCL3 in plasma samples by enzyme-linked immunosorbent assay and in lymph nodes by immunohistochemistry
Immunostaining for CCL3 was performed on a selected set of 38 sections of lymph nodes infiltrated with CLL cells. Immunohistochemistry revealed CLL3 expression in 13 of 38 cases (34%), in which few prolymphocytes and paraimmunoblasts in proliferation centers showed positive staining (Figure 3C). In an initial evaluation using immunofluorescence double staining, we detected single cells with colocalization of CD79a and CCL3 (Figure 3C; supplemental Figure 2), suggesting that indeed a subset of CLL cells express CCL3.
Discussion
BCR signaling is increasingly recognized as a central pathomechanism in B-cell malignancies, including CLL1,10 and DLBCL.9 New targeted agents that interfere with BCR signaling, such as spleen tyrosine kinase and Bruton tyrosine kinase inhibitors, are entering the clinical stage and show promising results in first clinical trials in patients with CLL and other B-cell malignancies.32,33 Because of these emerging new therapeutic options, it is important to identify patients who may benefit from these new approaches and/or become candidates for treatment at an early stage of their disease.
Here, we describe CCL3 as a novel, robust prognostic marker in CLL that can easily and reliably be measured in plasma samples by enzyme-linked immunosorbent assay. Compared with other prognostic markers in this disease, these technical aspects are in favor for CCL3. Measurement of some of the established prognostic markers in CLL is labor- and cost-intensive (mutation status, cytogenetics), whereas others (ZAP-70) are difficult to standardize for routine assessment. Based on the median CCL3 levels in our cohort, patients with low and high CCL3 levels can be distinguished at a cut-off point of 10 pg/mL, accounting for low- versus higher-risk patients. Compared with CCL3, CCL4 turned out to be comparably less predictive and did not add additional prognostic information in discordant cases. CCL3 is secreted by CLL cells (and normal B cells) in response to BCR signaling, which mainly occurs in the lymphatic tissues. Therefore, CCL3 plasma levels presumably reflect the status of BCR-derived activation of the CLL clone. This is supported by comparative gene expression profiling of CLL cells isolated from the different tissue compartments (blood, marrow, lymphatic tissues), which revealed CCL3 and CCL4 as lymphatic tissue “signature” genes up-regulated in lymphoid tissue CLL cells, as determined by gene expression profiling.20 Stable levels of CCL3 and CCL4 in most patients who initially show low levels of CCL3 and CCL4 (Figure 3B; supplemental Figure 1) suggest that low-risk patients have relatively stable CCL3 and CCL4 levels over time. However, a detailed analysis of a larger number of sequential samples in correlation to clinical and other prognostic factors needs to be done to better understand the relationship between CCL3 and CCL4 levels and disease activity.
Previous in vitro and in vivo findings highlighted CCL3 as a key response gene up-regulated in normal and neoplastic B cells in response to BCR signaling.21,24,25 SCYA3, the gene encoding for CCL3, is part of the activated B-cell signature34 in DLBCL and functions as a predictor for poor survival in patients with DLBCL.35 Lossos et al35 proposed measurement of SCYA3, along with LMO2, BCL6, FN1, CCND, and BCL2 gene expression for risk stratification in DLBCL. From a functional standpoint, it is intriguing that the combination of low BCL6 and high SCYA3 expression were indicators for poor prognosis in this model,35 given that BCL6 binds to cis-elements in the CCL3 promoter, where it functions as a critical repressor of CCL3 expression.26
The exact function of CCL3 in lymphomagenesis remains unclear. Based on the postulated function of B cell-derived CCL3 in normal immune responses, we hypothesized that increased CCL3 secretion by CLL cells may induce trafficking and homing of accessory cells to the malignant B cells in the tissue microenvironments.21 It is well recognized that CLL cells in the proliferative compartment are interspersed with T cells17,36 and cells of monocyte/macrophage lineage, termed NLCs.15 Conceivably, CLL cell-derived CCL3 may attract these accessory cells, thereby creating a favorable microenvironment that allows CLL cells to interact with T cells and NLC to receive survival and proliferation signals. This is supported by in vitro24 and in vivo37,38 studies, which indicated that CCL3 and CCL4 show a specific function in the lymphatic tissues. These studies demonstrated that B-cell activation within lymphoid tissues results in CCL3 and CCL4 secretion, leading to the recruitment of CCR5+ regulatory T cells for cognate interactions with B cells and antigen-presenting cells.37,38
Collectively, our findings along with the gene expression data in DLBCL indicate that CCL3 functions as a robust prognostic marker in selected B-cell neoplasias, where BCR signaling and increased CCL3 expression are a common theme. Our study demonstrates the feasibility and validity of measurement of CCL3 plasma levels for prognostication in CLL. The simplicity of the assay, the significance of the prognostic information, the relationship to BCR signaling, and the emerging therapies related to this pathway indicate that CCL3 will become a highly useful prognostic marker in CLL and potentially other B-cell malignancies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Theodora Nedeva and Sabine Roth for expert technical assistance.
This work was supported by CLL Global Research Foundation grants (W.W., A.R., and J.B.), an ASCO Career Development Award (J.B.), and National Institutes of Health CLL Research Consortium (grant PO1-CA81534).
National Institutes of Health
Authorship
Contribution: M.S. performed CCL3/4 serum analysis and data analysis and designed the figures and tables; E.H. performed immunohistochemistry for CCL3 and reviewed data and the manuscript; T.J.K. and L.R. provided samples, analyzed data, and reviewed the manuscript; D.K., S.L., and R.L. collected and organized data; L.X., X.H., and L.W. analyzed and interpreted the data; D.N., H.K., S.O., W.G.W., and M.J.K. provided samples, helped with data interpretation, and reviewed the manuscript; A.R. performed immunohistochemistry and reviewed data and the manuscript; and J.A.B. designed the research, supervised the study, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jan A. Burger, Department of Leukemia, Unit 428, University of Texas M. D. Anderson Cancer Center, PO Box 301402, Houston, TX 77230-1402; e-mail: jaburger@mdanderson.org.